Abstract
The authors describe an electrochemical strategy for highly sensitive determination of ATP that involves (a) aptamer-based target recognition, (b) enzyme-free dendritic DNA nanoassembly amplification with multiplex binding of the biotin-strepavidin system, and (c) enzyme-amplified differential pulse voltammetric readout. In the presence of ATP, binding of ATP to the aptamer releases trigger DNA from the double-stranded complex between ATP aptamer and trigger DNA. The single-stranded thiolated capture probe, chemisorbed on the gold electrode surface, captures the released trigger DNA via hybridization. The toehold of the trigger DNA is recombined with one end of the first substrate DNA (1) which is on its other end biotinylated and blocked, with loops, by a counterstrand. The latter is removed by a complementary single-stranded helper (1) exposing two toeholds and two identical complimentary sequences for a second biotinylated substrate DNA (2). The latter, which is double-stranded except for the toehold, binds to one of these two sites. It is then stripped from its counter strand by another single-stranded helper DNA 2, exposing a toehold to bind another substrate DNA 1. On this substrate, another cycle with dentrimeric bransching can start.
Substrate 1 with its two binding sites for substrate 2 initiates the assembly of dendritic DNA on the surface of the gold electrode, which finally possesses numerous biotins at the terminal ends of both of the associated substrate DNAs. Subsequent multiplex binding of streptavidinylated alkaline phosphatase and enzyme-amplified electrochemical readout leads to a highly sensitive electrochemical ATP aptasensor. If operated in the DPV mode, the current as measured at a typical working potential of 0.25 V (vs. Ag/AgCl) increases linearly over the 10 nM to 10 μM logarithmic ATP concentration range, and the detection limit is 5.8 nM (at an S/N ratio of 3). The assay is highly specific and reproducible. It was successfully applied to the detection of ATP in spiked human serum samples.

Schematic of the electrochemical strategy for adenosine triphosphate detection using aptamer-based target recognition and dendritic DNA nanoassembly amplification
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Adenosine triphosphate (ATP) is a multifunctional nucleoside that acts as the ‘molecular currency’ of intracellular energy transfer in all living cells [1]. It plays a critical role in the regulation of cellular metabolism and biochemical pathways in cell physiology [2, 3]. It has also been used as an indicator for cell viability and cell injury [4, 5]. In addition, the level of ATP is connected with many diseases such as hypoxia, hypoglycemia, ischemia, Parkinson’s disease, and some malignant tumors [6, 7]. Therefore, sensitive and reliable detection of ATP is of great importance in biochemical study as well as clinic diagnosis. Up to now, various techniques have been used for the detection of ATP, such as ultra performance liquid chromatography [8], fluorescence [9, 10], electrochemistry [11] and electrochemiluminescence [12]. Among these techniques, electrochemical biosensors have attracted great interest, due to their combined advantages of miniaturization capability, low cost, sensitivity and high stability.
Nowadays, new designs of ATP detection have emerged based on the advancement of analytical technologies employing aptamers [13, 14]. Aptamers are single-stranded DNA or RNA oligonucleotides, which are synthesized by systematic evolution of ligands by exponential enrichment (SELEX) technology [15, 16]. Aptamers can fold into special structures and possess high recognition ability to specific targets ranging from metal ions, small molecule, proteins and even whole viruses or cells [17]. An ATP aptamer can recognize and bind with ATP to form G-quadruplex structure with high affinity and selectivity [18], and is facile to integrate with electrochemical biosensing strategies for ATP detection [11, 19]. Aiming at obtaining higher sensitivity of aptamer based biosensors for ATP detection, many nucleic acid amplification strategies are employed to amplify the ATP recognition event and enhance signal readout, such as exonuclease III-assisted signal amplification [4, 19, 20], nicking endonuclease-induced target recycling [17] and rolling circle amplification [21]. These enzyme-assisted nucleic acid amplification strategies have enhanced the sensitivity of ATP aptasensors. But the disadvantages of enzymatic nucleic acid amplification, such as complex operation, specific reaction conditions and the reaction time dependent enzyme activity, compromise the simplicity and practicability of the designed aptasensors.
DNA nanoassembly is an enzyme-free process that when initiated with a DNA chain hybridization event, leads to kinetically controlled assembly of DNA nanostructures. Hybridization chain reaction (HCR) is a typical and well-known strategy of DNA nanoassembly, which is triggered by a target or initiator sequence and leads to the polymerization of oligonucleotides into a long linear nicked dsDNA molecule [22]. As such, HCR has been incorporated into biosensing strategies as an efficient enzyme-free signal amplification approach [23, 24]. However, the traditional HCR only provides linear detection signal amplification. Nonlinear HCR has demonstrated a dendritic DNA nanoassembly strategy [25–28]. This approach leads to chain-branched DNA by successive assembly of double-stranded DNA substrates. Moreover, this amplification strategy has exponential growth kinetics and well-controlled system leakage, which shows excellent amplification ability. Compared with traditional hairpin probes that utilize DNA nanoassembly strategies, this hairpin-free system has few secondary structures, making the whole reaction faster, which is rather suitable for biosensors.
Herein, an electrochemical aptasensor was established for the detection of ATP by combining aptamer-based target recognition, enzyme-free dendritic DNA nanoassembly on sensor surface with multiplex binding of the biotin-strepavidin system and enzyme-amplified electrochemical readout. The target induced dendritic DNA nanoassembly significantly amplified the ATP recognition event and facilitated subsequent multiplex binding of streptavidin-alkaline phosphatase and enzyme-amplified electrochemical readout. This aptasensor demonstrates excellent analytical performance towards ATP, and can be applied to the determination of ATP in real samples. Therefore, this biosensing strategy has great potential in ATP detection for medical research and early clinical diagnosis.
Materials and methods
Reagents
Adenosine triphosphate (ATP), cytosine triphosphate (CTP), guanosine triphosphate (GTP) and uridine triphosphate (UTP) were purchased from Sangon Inc. (Shanghai, China, http://www.sangon.com). 6-Mercapto-1-hexanol (MCH), streptavidin-alkaline phosphatase (ST-AP), α-naphthyl phosphate (α-NP), bovine serum albumin (BSA) and salmon sperm DNA were purchased from Sigma-Aldrich (St. Louis, MO, USA, http://www.sigmaaldrich.com). DL500 DNA Marker and agarose were purchased from Takara (Dalian, China, http://www.takara.com). GoldView was purchased from SBS Genetech (Beijing, China, http://www.sbsbio.com). All other reagents were of analytical grade, and Millipore-Q water (≥18 MΩ.cm) was used in all experiments. Hybridization buffer (pH 7.4) contained 30 mM Sodium Citrate and 300 mM NaCl. Tris–HCl buffer as washing buffer (pH 7.4) contained 20 mM Tris, 0.10 M NaCl, 5.0 mM MgCl2 and 0.05% Tween-20. Diethanolamine (DEA) buffer (pH 9.6) contained 0.1 M DEA, 1 M MgCl2 and 100 mM KCl. Piranha solution contained 98% H2SO4 and 30% H2O2 (3:1 by volume). Caution: Piranha solutions are extremely aggressive. The use of safety goggles and a hood is strongly recommended.
DNA oligonucleotides were designed and optimized according to the literatures [24, 25]. All oligonucleotides were synthesized and purified by Sangon Inc. (Shanghai, China, http://www.sangon.com). The base sequences are listed in Table S1 (Supporting information). All oligonucleotides were dissolved in tris-ethylenediaminetetraacetic acid (TE) buffer (pH 8.0, 10 mM Tris-HCl, 1 mM ethylene diamine tetraacetic acid) and stored at −20 °C, which were diluted in hybridization buffer prior to use.
Apparatus
All electrochemical measurements were performed on a CHI 660D electrochemical workstation (Shanghai Chenhua Instruments Co., Ltd., China, http://www.chinstruments.com) with a conventional three electrode system composed of platinum wire as auxiliary, Ag/AgCl/saturated KCl electrode as reference, and a 3 mm-diameter gold electrode as working electrode. The gel electrophoresis was imaged on a Bio-Rad ChemDoc XRS (Bio-Rad Laboratories, USA, http://www.bio-rad.com).
Preparation of probes
The complex of ATP aptamer (10 μM) and trigger DNA (10 μM) were obtained by heating mixtures of ATP aptamer and trigger DNA to 95 °C for 5 min and cooling to room temperature slowly. Substrate 1 and Substrate 2 were prepared separately by heating mixtures of corresponding A strand (1 μM) and B strand (1.5 μM) to 95 °C for 5 min and cooling to room temperature slowly, respectively. Then the probes were stored at 4 °C for further use.
Preparation of electrochemical biosensor
The bare gold electrode was polished with 0.05 mm alumina slurries and ultrasonically treated in ultrapure water for a few minutes, followed by soaking inpiranha solution for 10 min to eliminate other substances. Then, the pretreated electrode was rinsed with ultrapure water and allowed to dry at room temperature. 10 μL of 200 nM thiolated capture probe was dropped on to the pretreated gold electrode surface and incubated overnight at 4 °C. After washing with washing buffer, the electrode was treated with 1 mM MCH for 1 h to obtain well-aligned DNA monolayer and occupy the left bare sites, and further immersed in salmon sperm DNA and 1% BSA for 30 min respectively to avoid nonspecific adsorption of DNA and enzyme on the electrode surface. The electrochemical biosensor was rinsed with the washing buffer and used for following operation.
ATP detection protocol
The annealed Substrate 1 and Substrate 2 were first mixed with the corresponding helpers (2 μM) separately and incubated for 30 min. Excess single stranded B strands in the two substrate solutions were removed by forming byproducts with the helpers, obtaining solution 1 and solution 2, respectively. 10 μL various concentrations of ATP were mixed with 10 μL ATP aptamer-trigger DNA complex at room temperature for 1 h to release the trigger DNA from the complex. Then 1 μL upper mixture, 3 μL solution 1, and 6 μL solution 2 were mixed, giving final concentrations of about 0.1 μM Substrate 1, 0.2 μM Helper 1, 0.2 μM Substrate 2, and 0.4 μM Helper 2 in the reaction solution. Then, 10 μL freshly resulted reaction mixture was dropped on the above prepared electrodes and incubated at 37 °C for 1 h. Following rinsed by DEA buffer solution, the aptasensor reacted with 10 μL of 1.25 μg. mL−1 ST-AP at 37 °C for 30 min, and washed thoroughly with DEA buffer. The differential pulse voltammetry (DPV) measurement was performed in DEA buffer containing 0.75 mg.mL−1 of α-NP substrate with modulation time of 0.05 s, interval time of 0.017 s, step potential of 5 mV, modulation amplitude of 70 mV and potential scan from 0.0 to +0.6 V.
Native polyacrylamide gel electrophoresis (PAGE)
A 8% PAGE analysis of the dendritic DNA nanoassembly process was carried out in 1 × TBE buffer (90 mM Tris–HCl, 90 mM boric acid, 2 mM EDTA, pH 7.9) at 120 V for about 40 min. The gel was photographed by Bio-Rad digital imaging system.
Results and discussion
Principle of the electrochemical assay
The basic principle of the ATP electrochemical aptasensor is shown in Scheme 1. The probes and their functions were listed in Table S2. In the presence of ATP, trigger DNA is released from the complex of ATP aptamer-trigger DNA through the target recognition of ATP. Then one terminal of trigger DNA hybridized with specifically designed capture DNA, and the other terminal will trigger the dendritic DNA nanoassembly process.
Schematic representation of ATP electrochemical detection based on aptamer-based target recognition and dendritic DNA nanoassembly amplification. (i) Toehold domain on A strand of Substrate 1; (i,) Toehold domain on A strand of Substrate 2; (ii) Bulge loop domain on A strand of Substrate 1; (ii,) Bulge loop domain on A strand of Substrate 2; (iii) Newly exposed toehold domain on B strand of Substrate 1; (iii,) Newly exposed toehold domain on B strand of Substrate 2
The dendritic DNA nanoassembly system employs two double-stranded substrates (Substrate 1 and Substrate 2) and two single-stranded helpers (Helper 1 and Helper 2). Two substrates both possess protruding toehold domain (i and i,) and bulge loop domain (ii and ii,) by annealing two partially complementary single-stranded DNA (A and B) together, and the longer strands were biotin labeled, named Substrate 1-A and Substrate 2-A, respectively. The short strands were named Substrate 1-B and Substrate 2-B. Complementarity relationships between different domains of Substrate 1, Substrate 2, Helper 1 and Helper 2 were carefully exploited. Substrate 1 possesses two identical sequences, which are complementary to the toehold domain on substrate 2, and vice versa. The helper strands are designed to be complementary to specific regions on the B strands of corresponding substrates. In the absence of ATP, all the possible binding sites of these sequences are hidden.
When ATP is introduced, the trigger DNA is released and it binds with the exposed toehold of Substrate 1, displaces part of the 1-A strand from the 1-B strand and opens the first loop (ii). The opened loop exposes the displaced part of the 1-B strand as a new toehold. Helper 1 then invades the newly exposed toehold (iii), displaces the 1-B strand from the 1-A strand, and releases Byproduct 1, subsequently opening the second loop. Now the 1-A strand exposes two identical sequences, which simultaneously hybridizes with toeholds of two Substrates 2. Soon after Helper 2 helps to dissociate Byproduct 2, two single-stranded regions consisting of the same sequences as the trigger DNA are exposed and can enter a new round of reactions. These two target sequences are capable of initiating more rounds of similar reactions, resulting in dendritic growth of the DNA nanostructure on the electrode surface. The finally assembled DNA dendrimer carries numerous terminal biotin molecules. The ST-AP links to the biotin and AP catalyzes the irreversible conversion of substrate α-NP to an electroactive product for generating amplified electrochemical signal readout for quantitative detection of ATP.
Characterization of electrode surface modification
In order to validate the designed dendritic DNA nanoassembly strategy, several reaction products were characterized by agarose gel electrophoresis. As shown in Fig. 1a, In the presence of ATP, new bands with much lower mobility appeared and the band representing Substrate 1 disappeared (lane 1), indicating the assembly of Substrate 1 into products with higher molecular weights and the formation of dendritic DNA nanostructure. On the contrary, in the absence of ATP, assembly products with high molecular weight were almost invisible (lane 2), showing a very good nonreacting stability between substrates and helpers. These results indicated the feasibility of the designed target-induced dendritic DNA nanoassembly strategy directly.
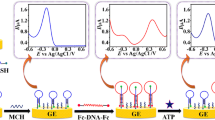
a The PAGE results of dendritic DNA nanoassembly. Lanes 1: dendritic DNA nanoassembly system with target ATP; Lane 2: dendritic DNA nanoassembly system without target ATP; Lane 3: Byproduct 2; Lane 4: Byproduct 1; Lane 5: Substrate 2; Lane 6: Substrate 1; Lane 7: 500 bp DNA ladder marker. b EIS in 5 mM [Fe(CN)6]3−/4- containing 1 M KCl at bare electrode (a), capture DNA modified electrode (b), MCH and BSA immobilized on the electrode surface (c), dendritic DNA nanoassembly system without target ATP (d), and with target ATP (e). c Typical DPV curves of the sensor responding to blank control (a), traditional sandwich assay, with ATP and Substrate 1 introduced (b), dendritic DNA nanoassembly system with target ATP (c)
Electrochemical impedance spectroscopy (EIS) measurements were adopted to characterize the established electrochemical biosensor. The EIS curves were obtained in 0.4 M KCl containing 0.5 mM Fe(CN)6 3−/4- and the semicircle diameter was equal to electron-transfer resistance (Ret) (Fig. 1b). In 0.4 M KCl containing 0.5 mM Fe(CN)6 3−/4-, bare electrode exhibited an almost straight line (curve a), reflecting the outstanding electrochemical conductivity. When the thiolated capture DNA was selfassembled onto the bare electrode via Au-thiol binding, the Ret increased (curve b). This is attributed to the negatively charged phosphate backbone of the oligonucleotides immobilized on the gold electrode, which prevented the negative charged redox probe [Fe(CN)6]3−/4− from reaching the gold electrode and inhibited interfacial charge transfer. Furthermore, after MCH and BSA were immobilized on the electrode surface, the Ret substantially increased (curve c), which was attributed to biomacromolecules obstructing electron transfer. Then, in the absence of ATP, with the amplification system introduced, the Ret slightly increased (curve d). Finally, with the participation of ATP and amplification system, the Ret increased significantly (curve e). The increase of Ret was ascribed to the fact that the dendritic DNA nanoassembly was successfully initiated and a large amount of DNA nanostructures were formed on the electrode surface.
To further characterize the feasibility of the combination of dendritic DNA nanoassembly amplification with electrochemical biosensor for ATP detection, DPV measurements were performed. As shown in Fig. 1c, in the presence of ATP with the dendritic DNA nanoassembly system, a dramatically high DPV signal was obtained, which corresponded to the formed dendritic DNA possessing terminal-end biotin. The ST-AP linked to the biotin and produced enzymatic electrochemical DPV signal readout (curve c). On the contrary, in the absence of ATP, only a negligible DPV signal was observed (curve a), confirming the good stability and negligible cross-reactivity between substrates and helpers. And in the traditional sandwich assay, with ATP and Substrate 1 introduced, a higher DPV signal than that of blank was produced, but much smaller than that gained by the dendritic DNA nanoassembly amplification (curve c). The results shown here clearly demonstrate the remarkable signal amplification performance of the designed target-induced dendritic DNA nanoassembly and the feasibility of the electrochemical biosensing platform for sensitive detection of ATP.
Optimization of method
The following parameters were optimized: (a) toehold number of the substrates; (b) concentration of Substrate 1; (c) reaction time. Respective data and figures are given in the supporting information. From Fig. S1, we found the following experimental conditions to give best results: (a) a toehold number for substrates of 6; (b) a Substrate 1 concentration of 100 nM; (c) a reaction time of 60 min.
Analytical performance of ATP detection
To evaluate the analytical performance of the established electrochemical aptasensor, DPV was exploited to characterize aptasensor. The DPV responses of the aptasensor to different concentrations of ATP were obtained under optimal conditions. As shown in Fig. 2a, The DPV peak current increased with the increase of ATP concentration. The calibration plots showed a good linear relationship between the peak currents and the logarithm of ATP concentrations in the range from 10 nM to 10 μM (Fig. 2b), the resulting linear equation was ip (A) = 7.04 E−7 log CATP (μM) + 1.81 E−6 with a correlation coefficient of 0.9968. Based on the 3σ rule, the limits of detection (LODs) for target ATP was estimated to be 5.8 nM. To further highlight the merits of the designed aptasensor, the sensitivity of the current assay is compared with previously reported assays for ATP detection (Table 1). Such a detection limit is comparable to the other electrochemical ATP assays [4, 17]. This strategy also has a comparable sensitivity with the nanomaterial-based or the enzyme-assisted signal amplification approaches [5, 21, 29]. Nanomaterials and enzyme-assisted approaches are widely used as effective amplification techniques for the detection of ATP for their strong catalytic abilities, but they are time-consuming, and have the issues of high nonspecific binding. The current enzyme-free strategy not only has wide linear range and low limit of detection, but also has controllable and stable reaction with low background. Therefore, this enzyme-free dendritic DNA nanoassembly strategy can be applied for detection of ATP with high sensitivity.
Specificity and reproducibility of this sensor
The specificity of the aptasensor was investigated by detecting three ATP analogues: GTP, CTP and UTP. The concentrations of ATP, GTP, CTP and UTP were 1 μM, respectively, and they were analyzed under the same experimental conditions. As shown in Fig. 3, the DPV currents of three ATP analogues all similar to that of the blank. However, ATP caused an obvious increase of DPV signal. It indicated the biosensor had good specificity to ATP. Such high selectivity can be ascribed to the high specificity of the ATP binding aptamer to ATP and sequence-specific discrimination of the target induced dendritic DNA nanoassembly. It is known that factors of ionic strength, temperature and pH play important roles in the dendritic DNA nanoassembly, especially the ionic strength (salt concentration) that high ionic strength may enhance probe-target binding but also compromise specificity in the meanwhile. Therefore, according to the literatures [30, 31], a suitable hybridization buffer was chosen and a satisfied signal-to-noise ratio was obtained (Fig. S1). In addition, for further evaluating the reproducibility of the aptasensor, five electrodes were prepared for the detection of 1 μM ATP under the same condition. The relative standard deviation (RSD) of five replicates measurements was 5.6%, which showed the biosensor had good reproducibility for ATP detection.
Detection of ATP in complex samples
The sensor was used for the detection of ATP in human serum samples. With a standard addition method, the ATP levels in three human serum samples were detected to be 0.93 μM, 0.99 μM and 0.97 μM, respectively, which was similar to the result obtained with the reported method [32]. After these serum samples were spiked with 1 μM ATP (1 μL of 200 μM ATP added into 200 μL human serum sample), as shown in Table 2, the recoveries for three detections were 96%, 105% and 98%, respectively, indicating acceptable accuracy. These good results owned to the diluted serum samples and several washing and separation steps to remove the interfering substances. It demonstrated that this method can be used for quantification of ATP in complex biological fluids.
Conclusion
This work has demonstrated a sensitive and specific electrochemical biosensor for ATP detection based on specific target recognition of aptamer, enzyme-free dendritic DNA nanoassembly in situ and multiplex binding of the biotin-strepavidin system with enzymatic electrochemical readout. However, this dendritic DNA nanoassembly amplification system needs several kinds of DNAs with well defined base sequences, which requires skills. On the other hand, now most commercial ATP assay kits can realize multiple sample test in 1 h, so the measurement time of this work should be further shortened. Although the biosensor has a certain defect, it shows high sensitivity for the detection of ATP with the dynamic linear range from 10 nM to 10 μM and the detection limit down to 5.8 nM. In addition, the designed aptasensor possesses the advantages of excellent specificity, acceptable reproducibility, and is applicable for handy in analysis of ATP in human serum samples. Moreover, this electrochemical ATP biosensor can be conveniently applied for the detection of many other molecules by changing the corresponding aptamer sequences. Therefore, the method provides an innovative platform for the detection of ATP and many other molecules.
References
Wang J, Jiang YX, Zhou CS, Fang XH (2005) Aptamer-based ATP assay using a luminescent light switching complex. Anal Chem 77(11):3542–3546
Tian JN, Wang Y, Chen S, Jiang YX, Zhao YC, Zhao SL (2013) Mass-amplifying quantum dots in a fluorescence polarization-based aptasensor for ATP. Microchim Acta 180:203–209
Gourine AV, Llaudet E, Dale N, Spyer KM (2005) ATP is a mediator of chemosensory transduction in the central nervous system. Nature 436(7047):108–111
Liu SF, Wang Y, Zhang CX, Lin Y, Li F (2013) Homogeneous electrochemical aptamer-based ATP assay with signal amplification by exonuclease III assisted target recycling. Chem Commun 49(23):2335–2337
Liu YT, Lei JP, Huang Y, Ju HX (2014) “Off-on” electrochemiluminescence system for sensitive detection of ATP via target-induced structure switching. Anal Chem 86(17):8735–8741
Imamura H, Huynh Nhat KP, Togawa H, Saito K, Iino R, Kato-Yamada Y, Nagai T, Noji H (2009) Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. PNAS 106(37):15651–15656
Zhou ZX, Du Y, Dong SJ (2011) Double-strand DNA-templated formation of copper nanoparticles as fluorescent probe for label-free aptamer sensor. Anal Chem 83(13):5122–5127
Zhou L, Xue XF, Zhou JH, Li Y, Zhao J, Wu LM (2012) Fast determination of adenosine 5′-triphosphate (ATP) and its catabolites in royal jelly using ultraperformance liquid chromatography. J Agric Food Chem 60(36):8994–8999
Li X, Peng Y, Chai YQ, Yuan R, Xiang Y (2016) A target responsive aptamer machine for label-free and sensitive non-enzymatic recycling amplification detection of ATP. Chem Commun 52(18):3673–3676
Qiu HZ, Liu ZE, Huang ZJ, Chen M, Cai XH, Weng SH, Lin XH (2015) Aptamer based turn-off fluorescent ATP assay using DNA concatamers. Microchim Acta 182:2387–2393
Chen XJ, Ge LN, Guo BH, Yan M, Hao N, Xu L (2014) Homogeneously ultrasensitive electrochemical detection of adenosine triphosphate based on multiple signal amplification strategy. Biosens Bioelectron 58:48–56
Liu ZY, Zhang W, Qi WJ, Gao WY, Hanif S, Saqib M, Xu GB (2015) Label-free signal-on ATP aptasensor based on the remarkable quenching of tris(2,2′-bipyridine)-ruthenium(II) electrochemiluminescence by single-walled carbon nanohorn. Chem Commun 51(20):4256–4258
Huo Y, Qi L, Lv XJ, Lai T, Zhang J, Zhang ZQ (2016) A sensitive aptasensor for colorimetric detection of adenosine triphosphate based on the protective effect of ATP-aptamer complexes on unmodified gold nanoparticles. Biosens Bioelectron 78:315–320
He YF, Liao LF, Xu CH, Wu RR, Li SJ, Yang YY (2015) Determination of ATP by resonance light scattering using a binuclear uranyl complex and aptamer modified gold nanoparticles as optical probes. Microchim Acta 182:419–426
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249(4968):505–510
Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346(6287):818–822
Hu TX, Wen W, Zhang XH, Wang SF (2016) Nicking endonuclease-assisted recycling of target–aptamer complex for sensitive electrochemical detection of adenosine triphosphate. Analyst 141(4):1506–1511
Huizenga DE, Szostak JW (1995) A DNA aptamer that binds adenosine and ATP. Biochemistry 34(2):656–665
Bao T, Shu HW, Wen W, Zhang XH, Wang SF (2015) A sensitive electrochemical aptasensor for ATP detection based on exonuclease III-assisted signal amplification strategy. Anal Chim Acta 862:64–69
Liu XQ, Freeman R, Willne I (2012) Amplified fluorescence aptamer-based sensors using exonuclease III for the regeneration of the analyte. Chem Eur J 18(8):2207–2211
Cho EJ, Yang LT, Levy M, Ellington AD (2005) Using a deoxyribozyme ligase and rolling circle amplification to detect a non-nucleic acid analyte, ATP. J Am Chem Soc 127(7):2022–2023
Dirks RM, Pierce NA (2004) Triggered amplification by hybridization chain reaction. Proc Natl Acad Sci 101(43):15275–15278
Liu HY, Bei XQ, Xia QT, Fu Y, Zhang S, Liu MC, Fan K, Zhang MZ, Yang Y (2016) Enzyme-free electrochemical detection of microRNA-21 using immobilized hairpin probes and a target-triggered hybridization chain reaction amplification strategy. Microchim Acta 183:297–304
Li XM, Wang Y, Wang LL, Wei QL (2014) A surface plasmon resonance assay coupled with a hybridization chain reaction for amplified detection of DNA and small molecules. Chem Commun 50(39):5049–5052
Xuan F, Hsing IM (2014) Triggering hairpin-free chain-branching growth of fluorescent DNA dendrimers for nonlinear hybridization chain reaction. J Am Chem Soc 136(28):9810–9813
Xuan F, Fan TW, Hsing IM (2015) Electrochemical interrogation of kinetically-controlled dendritic DNA/PNA assembly for immobilization-free and enzyme-free nucleic acids sensing. ACS Nano 9(5):5027–5033
Chang CC, Chen CY, Chuang TL, Wu TH, Wei SC, Liao H, Lin CW (2016) Aptamer-based colorimetric detection of proteins using a branched DNA cascade amplification strategy and unmodified gold nanoparticles. Biosens Bioelectron 78:200–205
Jia LP, Shi SS, Ma RN, Jia WL, Wang HS (2016) Highly sensitive electrochemical biosensor based on nonlinear hybridization chain reaction for DNA detection. Biosens Bioelectron 80:392–397
Li J, Fu HE, Wu LJ, Zheng AX, Chen GN, Yang HH (2012) General colorimetric detection of proteins and small molecules based on cyclic enzymatic signal amplification and hairpin aptamer probe. Anal Chem 84(12):5309–5315
Luo CH, Tang H, Cheng W, Li Y, Zhang DC, Ju HX, Ding SJ (2013) A sensitive electrochemical DNA biosensor for specific detection of Enterobacteriaceae bacteria by Exonuclease III-assisted signal amplification. Biosens Bioelectron 48:132–137
Zhai Q, He Y, Li XL, Guo J, Li SQ, Yi G (2015) A simple and ultrasensitive electrochemical biosensor for detection of microRNA based on hybridization chain reaction amplification. J Electroanal Chem 758:20–25
Gorman MW, Feigl EO, Buffington CW (2007) Human plasma ATP concentration. Clin Chem 53(2):318–325
Acknowledgments
This work was supported by the National Natural Science Foundation of China (21475068 and 81572080), the Achievement Transfer Project of Institutions of Higher Education in Chongqing (KJZH14205), Application Development Plan Project of Chongqing (cstc2014yykfB10003), and the Natural Science Foundation Project of CQ (cstc2014kjrc-qnrc10001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The author(s) declare that they have no competing interests.
Additional information
Xiaojuan Ding and Yihua Wang contributed equally to this work.
Electronic supplementary material
ESM 1
(DOC 458 kb)
Rights and permissions
About this article
Cite this article
Ding, X., Wang, Y., Cheng, W. et al. Aptamer based electrochemical adenosine triphosphate assay based on a target-induced dendritic DNA nanoassembly. Microchim Acta 184, 431–438 (2017). https://doi.org/10.1007/s00604-016-2026-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-016-2026-x