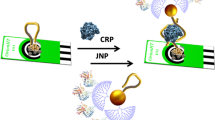
Abstract
We report on a sensitive aptamer-antibody interaction-based assay for cytochrome c (Cyt c) using electrochemical impedance. 4-Amino benzoic acid is used for the oriented immobilization of aminated aptamers onto multi-walled carbon nanotubes on the surface of a screen-printed electrode via electrochemical grafting. Impedance was measured in a solution containing the redox system ferro/ferricyanide. The change in interfacial charge transfer resistance (Rct) experienced by the redox marker was recorded to confirm the formation of a complex between aptamer and the target (Cyt c). A biotinylated antibody against cytochrome c was then used in a sandwich type of assay. The addition of streptavidin conjugated to gold nanoparticles and signal enhancement by treatment with silver led to a further increase in Rct. Under optimized conditions, a detection limit as low as 12 pM was obtained. Cross-reactivity against other serum proteins including fibrinogen, BSA and immunoglobulin G demonstrated improved selectivity.

Sensitive and selective assay for cytochrome c protein using aptamer linked to multi-walled carbon nanotube screen printed electrode via diazonium electrochemical grafting and specific biotinylated antibody to improve selectivity. Detection can be based on electrochemical impedance spectroscopy, or using a streptavidin-gold nanoparticle conjugate.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cytochrome c (Cyt c) is a heme-containing metalloprotein located in the intermembrane space of mitochondria. It plays a central role in electron transport chain and it is also an intermediate in apoptosis. When mitochondria are injured under pathological conditions, Cyt c is released into the cytosol of the cell. This translocation of Cyt c from mitochondria to cytosol is a decisive event in the activation of intracellular signaling; it results in a cascade of caspase activation and leads to programmed cell death (apoptosis). For this reason, the quantification of Cyt c may be of great importance in clinical diagnostics and therapeutic research [1].
Aptamers are oligonucleotides (DNA or RNA) that possess properties comparable to those of protein monoclonal antibodies, and thus are clear alternatives to well established antibody-based diagnostic or other biotechnological tasks for research [2, 3], therapy [4, 5] and diagnostics [6, 7]. This kind of functional nucleic acids can fold into complex three-dimensional shapes, thus forming binding pockets and cavities able for specific recognition. Therefore, aptamers are able to obtain a high affinity binding of any given molecular target, from metal ions and small chemicals structures to large proteins and higher order proteins complexes, even whole cells, viruses or parasites [8]. Aptamers are generated by an in vitro selection process called SELEX (Systematic Evolution of Ligands by Exponential Enrichment), which was first reported in 1990 [9, 10]. This method has permitted the identification of unique DNA/RNA fragments, from large sets of random sequence oligomers (DNA or RNA libraries), which may bind to a specific target molecule with very high specificity and affinity [11]. Due to their numerous advantages versus antibodies, aptamers have been increasingly used in biosensing in the recent years [12–17].
Among the different electrochemical techniques available, electrochemical impedance spectroscopy (EIS) [18] has been used in numerous studies [19–21]. This technique is very sensitive to changes in the interfacial properties of the modified electrodes caused by biorecognition events at the electrode surface [22, 23]. For this reason, EIS is becoming an attractive electrochemical technique for numerous applications such as immunosensing [24], enzyme activity determination [25], genosensing [26, 27], studies of corrosion [28] and other surface phenomena [29].
Signal amplification based on biofunctional nanomaterials is attracting significant attention due to the need for ultrasensitive bioassays. Among nanomaterials, gold nanoparticles have been widely used thanks to their excellent properties, such as high biocompatibility, distinctive size-related electronic and optical behavior, high electrical conductivity and high catalytic activity [30]. For example, Deng et al. [31] used AuNPs stabilized with sodium dodecylsulfate to amplify the impedimetric signal for the detection of thrombin, Zheng et al. [32] used network-like thiocyanuric acid/gold nanoparticles to amplify the signal for the detection of thrombin, etc.
In this work, we report a sensitive impedimetric aptamer-antibody sandwich assay for Cyt c detection using a highly specific amplification strategy with the use of streptavidin gold nanoparticles and silver enhancement treatment. The employed transducer consisted of a multi-walled carbon nanotube (MWCNT) screen-printed electrode which surface allowed the immobilization of aptamer binding cytochrome c (AptCyt c) by covalent bond via prior electrochemical grafting. As a transducer material, MWCNTs are used for promoting electron-transfer between the electroactive species and electrode and provide a novel method for fabricating biosensors. The change of interfacial charge transfer resistance (Rct) experimented by the redox marker, was recorded to confirm the aptamer complex formation with target protein, cytochrome c (Cyt c). After that, a biotinylated anti-cytochrome c antibody (Cyt c Ab) is used to form the sandwich. The addition of strep-AuNPs and silver enhancement treatment led to a further increment of Rct and the subsequent achievement of significant signal amplification, high sensitivity and improvement of selectivity.
Experimental
Reagents and solutions
Potassium dihydrogen phosphate, potassium ferricyanide K3[Fe(CN)6], potassium ferrocyanide K4[Fe(CN)6], sodium monophosphate, 4-aminobenzoic acid (ABA), sodium nitrite, N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), gold (III) chloride solution (HAuCl4), N-hydroxysuccinimide (NHS), streptavidin gold nanoparticles, fibrinogen, immunoglobulin G and the target protein cytochrome c (Cyt c), were all purchased from Sigma (St. Louis, MO, USA, www.sigmaaldrich.com). Poly(ethylene glycol) 1000 (PEG), sodium chloride, hydroxylamine hydrochloride (NH2OH∙HCl) and potassium chloride were purchased from Fluka (Buchs, Switzerland). Polyclonal biotinylated anti-cytochrome c antibody (Cyt c Ab) was purchased from BioLegend (San Diego, California, www.biolegend.com). LI silver enhancement kit was obtained from Nanoprobes (Yaphank, New York, www.nanoprobes.com). All reagents were analytical reagent grade. The aptamer used in this study was synthesized by TIB-MOLBIOL (Berlin, Germany, www.tib-molbiol.de). Stock solutions of aptamers were diluted with sterilized and deionised water, separated into fractions and stored at −20 °C until required. Aptamer solutions were prepared in phosphate buffer pH 7 from stock solutions. A well-known aptamer for thrombin (AptThr) was used for negative control purposes. Base sequences of both aptamers were the following:
-
AptCyt c: 5′–NH2–AGTGT GAAAT ATCTA AACTA AATGT GGAGG GTGGG ACGGG AAGAA GTTTA TTTTC ACACT–3′
-
AptThr: 5′–AGTCC GTGGT AGGGC AGGTT GGGGT GACT–Biotin–3′
All solutions were prepared using MilliQ water from MilliQ System (Millipore, Billerica, MA, USA, www.emdmillipore.com). The buffers employed were: phosphate buffer (187 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4·2H2O, 1.76 mM KH2PO4, pH 7.0) and triethylammonium bicarbonate (0.6 M).
Biosensing protocol
The steps of the experimental protocol for Cyt c analysis, described in detail below, are represented in Fig. 1.
Aptamer immobilization
MWCNT screen-printed electrodes were modified with aminobenzoic acid by means of a one step procedure. Firstly, 30 mg of ABA were dissolved in 3 mL of 1 M HCl and ice-cooled. Then, the diazonium salt was prepared by adding 570 μL of 2 mM NaNO2 aqueous solution dropwise to the 4-aminobenzoic acid solution, with constant stirring. The electrode was immersed in this solution, and 10 successive voltammetric cycles ranging between 0.0 and −1.0 V (v = 200 mV∙s−1) were performed [24], generating a carbon-carbon bond and eliminating the azonium group. The modified electrodes (benzoic acid modified) were washed thoroughly with water and methanol and dried at room temperature. Finally, 60 μL of aptamer solution (together with 1 mg of EDC and 0.5 mg of NHS) was placed on the modified electrode and left to react for 12 h, with the goal of covalent immobilization of the aminated aptamer through the amide formation. This step was followed by two 10 min washing steps with phosphate buffer.
Blocking step
To minimize any possible nonspecific adsorption of secondary species, 60 μL of PEG were dropped onto the electrodes and left to incubate during 15 min. This was followed by two washing steps using phosphate buffer for 10 min.
Cytochrome c detection
Sixty microliter of a solution with the desired concentration of Cyt c were dropped onto the electrodes. The incubation took place for 15 min. Then, the biosensors were washed twice with phosphate buffer for 10 min.
Sandwich formation
In order to achieve the aptamer–antibody sandwich formation, electrodes were dropped 60 μL of Cyt c Ab, from a 1/500 dilution of the stock solution in phosphate buffer. The incubation took place for 15 min. This was followed by two washing steps using phosphate buffer for 10 min.
Addition of strep-AuNPs
Sixty microliter of strep-AuNPs, from a 1/100 dilution of the stock solution in phosphate buffer were dropped onto the electrodes [33]. This step was followed by two gentle washing steps in phosphate buffer for 10 min at 25 °C. In order to obtain a negative control, for the strep-AuNPs addition step, an aptamer without affinity (AptThr) was used instead of Apt Cyt c
Silver enhancement of strep-AuNPs
Twenty microliter of a solution obtained by the combination of 10 μL of enhancer and 10 μL of initiator (commercial solutions) were deposited onto the electrode surface and left for 7 min to facilitate the reaction [33]. After the catalytic silver reduction, the electrodes were thoroughly washed with deionized water to stop the reaction. The silver enhancing solution was prepared immediately before each use. For silver enhancement treatment, the negative control used was a biotinylated AptThr as aptamer without affinity.
Gold enhancement of strep-AuNPs
The MWCNT screen-printed electrodes modified with sandwich and strep-AuNPs were immersed in a solution containing a mixture of 0.01 % HAuCl4 and 0.4 mM NH2OH∙HCl (pH 6.0) for 2 min at 25 °C, rinsed, and then treated for 2 additional min. In order to prevent the non-specific background of fine gold particles, the electrodes were rinsed with a solution of 0.6 M triethylammonium bicarbonate buffer after each amplification. Solutions for amplification were freshly prepared in a lightproof container before each use.
Different selectivity experiments were carried out to verify selectivity characteristics of the assay with potentially interfering proteins instead of Cyt c.
Spiked samples preparation
Cytochrome c serum samples were prepared by adding three different concentrations of Cyt c to undiluted serum samples. All experimental conditions were the same as for the target detection.
Equipment
AC impedance measurements were performed using an Autolab PGStat 20 (Metrohm Autolab B.V, Utrecht, The Netherlands, www.metrohm-autolab.com). FRA software (Metrohm Autolab) was used for data acquisition and control of the experiments. A three electrode configuration was used to perform the impedance measurements: a platinum-ring auxiliary electrode (Crison 52–67 1, Barcelona, Spain), a Ag/AgCl reference electrode and the MWCNT-screen printed electrode as the working electrode (Dropsens, Oviedo, Spain, www.dropsens.es). A scanning electron microscope (SEM) (Merlin, Zeiss, Germany, www.zeiss.com) was used to visualize gold enhanced strep-AuNPs on the electrode surface.
EIS detection
Impedance experiments were performed at an applied potential of 0.17 V (vs. Ag/AgCl reference electrode), with a range of frequency of 50 kHz–0.05 Hz, an AC amplitude of 10 mV and a sampling rate of 10 points per decade above 66 Hz and 5 points per decade at the lower range. All measurements were performed in phosphate buffer containing 0.01 M K3[Fe(CN)6]/K4[Fe(CN)6] (1:1) mixture, used as a redox marker. The impedance spectra were plotted in the form of complex plane diagrams (Nyquist plots, −Zim vs. Zre) and fitted to a theoretical curve corresponding to the equivalent circuit with Zview software (Scribner Associates Inc., USA). The equivalent circuit was formed by one resistor/ capacitor element in series with a resistance; the Warburg term was circumvented as the diffusion processes were not relevant in this study. For all performed fittings, the chi-square goodness-of-fit test was thoroughly checked to verify the calculations. In all cases, calculated values for each circuit remained in the range of 0.0003–0.15, which was much lower than the tabulated value for 50 degrees of freedom (67.505 at 95 % confidence level). The most important parameter in this work is the electron transfer resistance (Rct), which reflects the resistance to charge transfer between the redox probe and the electrode surface. In order to compare the results obtained from the different electrodes used, and to obtain independent and reproducible results, a relative transformation of signals was needed [34]. Thus, the Δratio value was defined according to the following equations:
Where Rct(AptCyt c/Cyt c/Cyt c Ab/strep-AuNPs/silver enhancement) was the electron transfer resistance value measured after sandwich formation and silver treatment; Rct(AptCyt c) was the electron transfer resistance value measured after aptamer immobilization on the electrode, and Rct(electrode-buffer) was the electron transfer resistance of the blank electrode in buffer.
Results and discussion
The fundamentals of the developed assay are illustrated in Fig. 1. Firstly, MWCNT screen-printed electrodes were modified with amino benzoic acid. Briefly, diazotation of ABA was performed with sodium nitrite in hydrochloride acid, the resulting 4-carboxybenzenediazonium ion solution was dropped onto the MWCNT electrode surface and the potential was cycled as described in “Experimental” Section. Figure 2a shows the Nyquist plots obtained by electrochemical impedance spectroscopy. As can be seen, modification of the MWCNT electrode with ABA gave rise to a large increase in the electron transfer resistance as a consequence of the electrostatic repulsion between the redox maker and the negatively charged carboxylate groups. Thereafter, surface-confined carboxyl groups were activated with EDC/NHS to form amide bonds with amino terminated aptamer. After aptamer incubation, the Rct decreased in relation to the modification surface with ABA due to reduction of the negative charge density of the electrode surface, as can be observed in Fig. 2b. Afterwards, the Rct value diameter of the semicircle increased after each performed step. The addition of a target protein, Cyt c, and Cyt c Ab to form a complex AptCyt c–Cyt c and a sandwich respectively, resulted in a less marked increment of the resistance value compared to the value after the modification with ABA. These increments were due to the augmented quantity of negative charges and to the hidrance caused by the formation of a double layer. After the addition of strept-AuNPs we can observe a further increment of charge transfer resistance because of the increased hindrance due to the formed conjugates. In the second amplification step, the silver enhancement treatment [35, 36], a significant increment of Rct value was also observed and attributable to the silver deposition on gold.
a Nyquist diagrams of: (black circle) Bare electrode, (white circle) Electrochemical grafting treatment and (black down-pointing triangle) Aptamer immobilization. b Nyquist diagrams of: (black circle) Bare electrode, (white circle) aptamer immobilization, (black down-pointing triangle) Aptamer with Cyt c, (white up-pointing triangle) sandwich complex with antibody, (black square) sandwich complex modified with gold-nanoparticles and (white square) sandwich complex modified with gold-nanoparticles and silver enhancement treatment. All experiments were performed in phosphate buffer and all EIS measurements were performed in PBS solution containing 0.01 M K3[Fe(CN)6]/ K4[Fe(CN)6]
Optimization of the experimental concentrations involved in the aptamer-antibody sandwich assay response to Cyt c
All concentrations involved in the analytical performance of the aptasensor for detection of Cyt c were optimized by constructing its relative response curve. For this, increasing concentrations of AptCyt c and PEG were used to determine the immobilization and surface blocking, respectively, evaluating the changes in the Δp. Figure 3a shows the curve of AptCyt c immobilization onto the electrode surface. It can be observed that the difference in resistance (Δp) increased up to a value. This is due to the physical adsorption of the aptamer onto the electrode surface, which followed a Langmuir isotherm. Embodied by the variation of Rct which increases to reach a saturation value, the optimal concentration was chosen as initial value to reach it. This value corresponded to a concentration of aptamer of 1.5 μM. Concerning blocking agent, and as shown in Fig. 3b, a different behavior was obtained. The optimal concentration of blocking agent was chosen as 30 mM because it was the concentration point where a small plateau was observed.
In addition, in order to obtain the optimal concentration of Cyt c Ab to be used in the biosensing protocol, response was evaluated with increasing concentration of antibody. The optimal concentration was chosen as maximum ∆ratio value, 1/500 stock dilution, Figure S1 (Supplementary Information).
Scanning electron microscope examination
Screen-printed MWCNT electrode surfaces were investigated by SEM after the gold enhancement treatment. HAuCl4 was employed in order to achieve an adequate amplification of strept-AuNPs present on sensor surface to allow their direct observation by SEM. SEM images taken at an acceleration voltage of 3 kV are shown in Fig. 4, illustrating a positive experiment with sandwich protocol and strep-AuNPs conjugation. As can be observed in Fig. 4a, the distribution of gold enhanced-gold nanoparticles is quite homogeneous. This also implies a regular distribution of MWCNT and well-organized formation of sandwich complex onto the electrode surface. This high density distribution also demonstrates the proper functionality of the MWCNT platform and the immobilization of the biomolecule. Comparing this experiment with the negative control that did not use the biotinylated Cyt c Ab, Fig. 4b, a surface with almost absent nanoparticles can be observed.
Analytical performance of the aptamer-antibody sandwich assay for detection of Cyt c
After experimental concentration optimizations, the aptasensor was then used following the sandwich protocol, plus amplification employing the strep-AuNPs and silver enhancement treatment. Figure 5 shows calibration curves with increasing concentrations of Cyt c and their respective regression lines in the logarithmic scale, at the different steps of the protocol: (1) AptCyt c–Cyt c, (2) sandwich formation between AptCyt c, Cyt c and Cyt c Ab, (3) aptamer sandwich modified with strep-AuNPs, and (4) aptamer sandwich modified with strep-AuNPs and silver enhancement treatment. Although reproducibility was not ideal in all cases, the calibration curves obtained showed a good RSD. As can be seen in Figure S2 (Supplementary Information), all calibration curves increased until the value of 100 pM of Cyt c, this could be due to the fact that concentrations larger than 100 pM caused a saturation on the sensor surface. As can be observed in Table 1, the use of silver enhancement treatment led to the highest sensitivity and signal amplification, resulting in 108 % increase compared to the simple biosensing scheme. This demonstrates that the silver deposition on gold nanoparticles, basically increases the sterical hindrance, producing an increment of observed impedance, given this conductive silver is not wired to the electrode surface. Together with the higher sensitivity, the detection limit obtained in this case was 12 pM (calculated as the intersection with the horizontal reference line) and it is slightly improving the one obtained with only strep-AuNPs (15 pM). Without sandwich amplification, signal is of the same magnitude than associated errors, making the assay of impractical use. This results confirmed that the best method, showing a low detection limit and an ultrahigh sensitivity for the detection of Cyt c involves the sandwich and amplification protocol.
Calibration and regression curves of: (1) (black circle) AptCyt c and Cyt c, (2) (white circle) sandwich complex, (3) (black triangle) sandwich complex modified with strep-AuNPs, (4) (white triangle) sandwich complex modified with strep-AuNPs and silver enhancement treatment. All experiments were performed in PBS solution and all EIS measurements were performed in phosphate buffer containing 0.01 M K3[Fe(CN)6]/K4[Fe(CN)6]. Uncertainty values corresponding to replicated experiments (n = 5)
Selectivity of the aptamer-antibody sandwich assay
Control experiments were conducted to investigate the specificity of aptamer-antibody assay. In this work, majority serum proteins, such as human IgG, fibrinogen and albumin at serum physiological levels were tested to operate the aptasensor instead of Cyt c under the same experimental conditions. As can be seen in Fig. 6, the presence of interfering proteins such as albumin, fibrinogen and immunoglobulin G, at serum concentration level exhibits negligible response compared with 100 pM Cyt c in the amplified sandwich protocol, even at concentrations four or five orders of magnitude higher than typical Cyt c concentrations. Figure 6 also demonstrates that the sandwich protocol displays as clear advantage, more than the signal amplification, the marked decrease of interfering effects that are still remarkable in the simple biosensing protocol. This is especially remarkable for IgG protein (human), which shows appreciable interference in the assay without amplification, although it becomes practically negligible with the sandwich variant.
Comparison graph of responses towards different proteins present in serum, with simple biosensing scheme (white bar) and with the sandwich/amplification protocol (grey bar). The concentrations of the proteins were: 100 pM Cyt c, 7.35 μM Fbr, 100 μM IgG and 0.72 mM BSA. Uncertainty values corresponding to replicated experiments (n = 5)
Detection of Cyt c in spiked serum samples
The applicability of the biosensor was tested by the analysis of spiked samples of human serum samples. For that purpose, human serum samples were spiked with three different concentrations of Cyt c. As shown in Table 2, the recoveries of spiked samples were between 94.6 % and 107.8 · % when using this method, which showed a satisfactory result. In addition, a good reproducibility of the blanks was obtained, 5.5 % RSD. These findings imply that the developed methodology has a promising feature for the analytical application in complex biological samples.
Once we had confirmed that our biosensor was able to detect Cyt c with excellent analytical performance also in real samples, its analytical features were compared to other detection methodologies for Cyt c detection, as described in the literature. Details are shown in Table 3, where sensitivity info is presented as the LOD of each biosensor compared. The lowest detection limit value among all the analytical techniques corresponded to ref [39], but this Cyt c biosensor used ICP-MS as the transduction technique. This methodology displays high sensitivity but is not simple, portable or easy to use. However, our biosensor showed the second lowest LOD value and it was suitable for real samples, considering that the lowest level of Cyt c in serum samples is about 10 pM [43].
Conclusions
An ultrasensitive aptamer-antibody sandwich assay for cytochrome c detection using electrochemical impedance technique was reported. Due to the signal amplification with strep-AuNPs and silver deposition, it was possible to increase the sensitivity of the assay. Additionally, for a comparable amount of AptCyt c–Cyt c, the signal resulted in 108 % amplification, compared to results recorded with simple biosensing AptCyt c–Cyt c. Furthermore, the limit of detection obtained was 12 pM. An acceptable linear range, between 25 and 100 pM, and high selectivity with respect to different serum proteins at serum concentration level were also achieved with this protocol thanks to the double recognition scheme utilized. Finally, the suitability of the biosensor for measurement in real samples was checked by determining Cyt c in human serum samples. Obtained recovery values in the range 94.6–107.8 %, demonstrated a promising feature for the analytical application in complex biological samples.
References
Alleyne T, Joseph J, Sampson V (2001) Cytochrome-c detection: a diagnostic marker for myocardial infarction. Appl Biochem Biotechnol 90(2):97–105
Clark SL, Remcho VT (2002) Aptamers as analytical reagents. Electrophoresis 23(9):1335–1340
Radi A, Acero Sánchez JL, Baldrich E, O’Sullivan CK (2005) Reusable impedimetric aptasensor. Anal Chem 77(19):6320–6323
Biesecker G, Dihel L, Enney K, Bendele RA (1999) Derivation of RNA aptamer inhibitors of human complement C5. Immunopharmacology 42(1–3):219–230
Hicke BJ, Marion C, Chang YF, Gould T, Lynott CK, Parma D, Schmidt PG, Warren S (2001) Tenascin-C aptamers are generated using tumor cells and purified protein. J Biol Chem 276(52):48644–48654
Cox JC, Ellington AD (2001) Automated selection of anti-protein aptamers. Bioorg Med Chem 9(10):2525–2531
Cai H, Lee TM-H, Hsing IM (2006) Label-free protein recognition using an aptamer-based impedance measurement assay. Sensors Actuators B Chem 114(1):433–437
Jayasena SD (1999) Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem 45(9):1628–1650
Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346(6287):818–822
Tuerk C, Gold L (1990) Systemic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249(4968):505–510
Tombelli S, Minunni M, Mascini M (2005) Analytical applications of aptamers. Biosens Bioelectron 20(12):2424–2434
Zhang L, Cui P, Zhang B, Gao F (2013) Aptamer-based turn-on detection of thrombin in biological fluids based on efficient phosphorescence energy transfer from Mn-doped ZnS quantum dots to carbon nanodots. Chem Eur J 19(28):9242–9250
Chang M, Kwon M, Kim S, Yunn NO, Kim D, Ryu SH, Lee JB (2014) Aptamer-based single-molecule imaging of insulin receptors in living cells. J Biomed Opt 19(5):051204
Loo AH, Bonanni A, Pumera M (2012) Impedimetric thrombin aptasensor based on chemically modified graphenes. Nanoscale 4(1):143–147
Ho MY, D’Souza N, Migliorato P (2012) Electrochemical aptamer-based sandwich assays for the detection of explosives. Anal Chem 84(10):4245–4247
Lei P, Tang H, Ding S, Ding X, Zhu D, Shen B, Chang Q, Yan Y (2015) Determination of the invA gene of Salmonella using surface plasmon resonance along with streptavidin aptamer amplification. Microchim Acta 182:289–296
Shen B, Li J, Cheng W, Yan Y, Tang R, Li Y, Ju H, Ding S (2015) Electrochemical aptasensor for highly sensitive determination of cocaine using a supramolecular aptamer and rolling circle amplification. Microchim Acta 182:361–367
McDonald JR (1987) Impedance spectroscopy. Wiley, New York
Bonanni A, Ambrosi A, Pumera M (2012) On oxygen-containing groups in chemically modified graphenes. Chem-Eur J 18(15):4541–4548
Bonanni A, Ambrosi A, Pumera M (2012) Nucleic acid functionalized graphene for biosensing. Chem-Eur J 18(6):1668–1673
Ocaña C, del Valle M (2014) A comparison of four protocols for the immobilization of an aptamer on graphite composite electrode. Microchim Acta 181:355–363
Bardea A, Patolsky F, Dagan A, Willner I (1999) Sensing and amplification of oligonucleotide-DNA interactions by means of impedance spectroscopy: a route to a Tay-Sachs sensor. Chem Comm 1:21–22
Loo AH, Bonanni A, Ambrosi A, Poh HL, Pumera M (2012) Impedimetric immunoglobulin G immunosensor based on chemically modified graphenes. Nanoscale 4(3):921–925
Moreno-Guzmán M, Ojeda I, Villalonga R, González-Cortés A, Yáñez-Sedeño P, Pingarrón JM (2012) Ultrasensitive detection of adrenocorticotropin hormone (ACTH) using disposable phenylboronic-modified electrochemical immunosensors. Biosens Bioelectron 35(1):82–86
Zhang Y, Li Y, Wu W, Jiang Y, Hu B (2014) Chitosan coated on the layers’ glucose oxidase immobilized on cysteamine/Au electrode for use as glucose biosensor. Biosens Bioelectron 60:271–276
Bonanni A, Esplandiu MJ, del Valle M (2010) Impedimetric genosensing of DNA polymorphism correlated to cystic fibrosis: a comparison among different protocols and electrode surfaces. Biosens Bioelectron 26(4):1245–1251
Bonanni A, Pumera M, Miyahara Y (2010) Rapid, sensitive, and label-free impedimetric detection of a single-nucleotide polymorphism correlated to kidney disease. Anal Chem 82(9):3772–3779
Blin F, Koutsoukos P, Klepetsianis P, Forsyth M (2007) The corrosion inhibition mechanism of new rare earth cinnamate compounds—electrochemical studies. Electrochim Acta 52(21):6212–6220
Liao Y-M, Feng Z-D, Chen Z-L (2007) In situ tracing the process of human enamel demineralization by electrochemical impedance spectroscopy (EIS). J Dent 35(5):425–430
Bonanni A, del Valle M (2010) Use of nanomaterials for impedimetric DNA sensors: a review. Anal Chim Acta 678(1):7–17
Deng C, Chen J, Nie Z, Wang M, Chu X, Chen X, Xiao X, Lei C, Yao S (2008) Impedimetric aptasensor with femtomolar sensitivity based on the enlargement of surface-charged gold nanoparticles. Anal Chem 81(2):739–745
Zheng J, Feng W, Lin L, Zhang F, Cheng G, He P, Fang Y (2007) A new amplification strategy for ultrasensitive electrochemical aptasensor with network-like thiocyanuric acid/gold nanoparticles. Biosens Bioelectron 23(3):341–347
Bonanni A, Esplandiu MJ, del Valle M (2008) Signal amplification for impedimetric genosensing using gold-streptavidin nanoparticles. Electrochim Acta 53(11):4022–4029
Bonanni A, Esplandiu MJ, Pividori MI, Alegret S, del Valle M (2006) Impedimetric genosensors for the detection of DNA hybridization. Anal Bioanal Chem 385(7):1195–1201
Cai H, Wang YQ, He PG, Fang YH (2002) Electrochemical detection of DNA hybridization based on silver-enhanced gold nanoparticle label. Anal Chim Acta 469(2):165–172
Hanaee H, Ghourchian H, Ziaee AA (2007) Nanoparticle-based electrochemical detection of hepatitis B virus using stripping chronopotentiometry. Anal Biochem 370(2):195–200
Loo FC, Ng SP, Wu C-ML, Kong SK (2014) An aptasensor using DNA aptamer and white light common-path SPR spectral interferometry to detect cytochrome-c for anti-cancer drug screening. Sensors Actuators B: Chem 198:416–423
Lau IPM, Ngan EKS, Loo JFC, Suen YK, Ho HP, Kong SK (2010) Aptamer-based bio-barcode assay for the detection of cytochrome-c released from apoptotic cells. Biochem Biophys Res Commun 395:560–564
Liu JM, Yan XP (2011) Ultrasensitive, selective and simultaneous detection of cytochrome c and insulin based on immunoassay and aptamer-based bioassay in combination with Au/Ag nanoparticle tagging and ICP-MS detection. J Anal At Spectrom 26:1191–1197
Ocaña C, Arcay E, Del Valle M (2014) Label-free impedimetric aptasensor based on epoxy-graphite electrode for the recognition of cytochrome c. Sensors Actuators B Chem 191:860–865
Pandiaraj M, Benjamin AR, Madasamy T, Vairamani K, Arya A, Sethy NK (2014) A cost-effective volume miniaturized and microcontroller based cytochrome c assay. Sensors Actuators A Phys 220:290–297
Pandiaraj M, Madasamy T, Gollavilli PN, Balamurugan M, Kotamraju S, Rao (2013) Nanomaterial-based electrochemical biosensors for cytochrome c using cytochrome c reductase. Bioelectrochemistry 91:1–7
Sakaida I, Kimura T, Yamasaki T, Fukumoto Y, Watanabe K, Aoyama M, Okita K (2005) Cytochrome c is a possible new marker for fulminant hepatitis in humans. J Gastroenterol 40(2):179–185
Acknowledgments
This research was partly supported by the Research Executive Agency (REA) of the European Union under Grant Agreement number PITN-GA-2010-264772 (ITN CHEBANA), by the Ministry of Science and Innovation (MCINN, Madrid, Spain) through the project CTQ2013-41577-P and by the Catalonia program ICREA Academia. Cristina Ocaña thanks the support of Ministry of Science and Innovation (MICINN, Madrid, Spain) for the predoctoral grant.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 78 kb)
Rights and permissions
About this article
Cite this article
Ocaña, C., Lukic, S. & del Valle, M. Aptamer-antibody sandwich assay for cytochrome c employing an MWCNT platform and electrochemical impedance. Microchim Acta 182, 2045–2053 (2015). https://doi.org/10.1007/s00604-015-1540-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-015-1540-6