Abstract
This article reports on a new square-wave anodic-stripping voltammetric method for sensitive determination of nucleic acids. It is based on a target-induced strand displacement reaction with blocker DNA (labeled with a CdS quantum dot) from a biotinylated hairpin DNA. First, a hairpin-blocker DNA duplex was immobilized on the surface of the well in a microtiter plate via biotin-streptavidin interaction. On addition of target DNA to the well, the CdS-labeled blocker DNA is displaced by target DNA from the hairpin-blocker duplex to form a new target-blocker DNA duplex. This is accompanied by the release of CdS-labeled blocker DNA. Next, cadmium ions are released from the Q-dots (by adding 1 M nitric acid) and then quantified by anodic stripping voltammetry using an in-situ prepared mercury film electrode. The voltammetric signal increases with the concentration of target DNA in the 5.0 pM to 1.0 nM concentration range, and the detection limit is as low as 1.2 pM. The assay has a good repeatability and displays an intermediate precision of down to 10 %.

The voltammetric nucleic acid assay is based on a target-induced strand displacement reaction between hairpin DNA and CdS quantum dot-labeled blocker DNA, and subsequent release and quantitation of Cd(II) ions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Methods for detecting and quantifying sequence-specific nucleic acids are important in biological studies, clinical diagnostics, and biodefense applications [1, 2]. Typically, DNA sequences of interest are present in very small amount. So, exploring some new amplification techniques that enable the detection of trace levels of a specific sequence is very crucial [3]. Ongoing efforts have been made by exploiting bioactive enzymes, indicators and nanomaterials for the amplification of detectable signal [4–6]. Existing DNA-based amplification techniques mainly consist of polymerase-ligase chain reaction-based thermal cycling and strand-displacement polymerase-based isothermal cycling [7, 8]. Unfavorably, the increasing amount of the product during the thermal cycling is acquired via the repeated thermal cycling in an exponential way. Therefore, the thermal cycling protocols are time-consuming, sometimes nonspecific, and limited to a thermostable enzyme and a laboratory setting [9]. However, the bioactive enzymes are susceptible to interference and changes in assay conditions during the signal generation stage. An alternative approach that can substitute the classical amplification method and does not require the participation of bioactive enzyme or enzyme labeling would be advantageous.
The rapid emergence of strand-displacement reaction-based isothermal amplification and bionanotechnology opens a new horizon for advanced development of new analytical tools and instrumentation [10]. Recent research has looked to develop innovative and powerful novel biofunctionalized nanostructures, controlling and tailoring their properties in a very predictable manner to meet the requirement of specific applications [11]. Zhu et al. developed an isothermal enzyme-free amplification and label-free graphene oxide-based SYBR Green I fluorescence platform for detection of miRNA [12]. Wong et al. reported ultrasensitive and closed-tube colorimetric loop-mediated isothermal amplification assay using carboxyl-modified gold nanoparticles [13]. Despite some advances in this field, there is still the quest for new schemes and strategies for improvement of the sensitivity and simplicity of enzyme-free isothermal amplification methods.
Quantum dots (QDs, a kind of semiconductor nanocrystals) have shown great potential for applications in biology and medicine, as well as in solar cells and photovoltaics, security inks, photonics and telecommunications because of their unique optical, electronic, and catalytic properties [14]. Inspiringly, the new electrochemical coding bioassay depends on the utilization of different inorganic-colloid nanocrystal tracers, whose metal components yield well-resolved highly sensitive stripping voltammetric signals for the corresponding targets [15]. Kokkinos et al. designed a microfabricated tin-film electrode for protein and DNA sensing based on stripping voltammetric detection of Cd(II) released from quantum dots labels [16]. Zhang et al. reported an anodic-stripping voltammetric immunoassay for ultrasensitive detection of low-abundance proteins using quantum dot aggregated hollow microspheres [17]. In this regard, our motivation is to combine the unique properties of quantum dots with strand-displacement reaction-based enzyme-free isothermal amplification for the construction of DNA sensing strategy.
Herein we report the proof-of-concept of a new and powerful electrochemical sensing strategy for sensitive detection of target DNA by coupling with target-induced displacement reaction with CdS QD-labeled blocker DNA. Two oligonucleotides including biotinylated hairpin DNA and QD-labeled blocker DNA are used in this work. Initially, the biotinylated hairpin DNA hybridizes with the designed blocker DNA to form the DNA duplex for the preparation of DNA sensing platform on the surface of the wells of microtiter plate via the biotin-avidin reaction. In the presence of target analyte, target DNA displace the hairpin probe from the hairpin-block duplex, resulting in the formation of hairpin DNA on the well surface and target-blocker duplex in solution. The formed target-blocker duplex with QDs is subsequently determined by using square-wave anodic stripping voltammetry at an in situ prepared mercury-film electrode under acidic condition. The voltammetric signal obtained increases with the increasing concentration of target DNA in the sample.
Experimental
Material and reagent
Oligonucleotides designed in this work were synthesized and purified through HPLC by Sangon Biotechn. Co., Ltd. (Shanghai, China, www.sangon.com), and their sequences were as follows:
-
Biotinylated hairpin DNA: 5′-bio-TTTGATACCT ACGGGAGACGAAGTAATGTCAGAAAGGTATC-3′
-
Blocker DNA: 5′-NH2-TAATGCGTTTGTAATAACTAAGTC CATTACTTC GTCTCCCGT-3′
-
Ureagene target DNA: 5′-GAAGTAATGGACTTAGTTATTACAAACGCATTA-3′
The underlined sequence of hairpin DNA represents the stem portion; the bold letters of blocker DNA are complementary to the bold letters of hairpin DNA; the italic letters of blocker DNA are the sequence complementary to target DNA). All high-binding polystyrene 96-well microplates used in this study were purchased from Greiner Bio-One GmbH (Frickenhausen, Germany, www.greinerbioone.com). Streptavidin was purchased from Amyjet Scientific Inc. (Wuhan, China, amyjet.bioon.com.cn). N-Hydroxysulfosuccinimid sodium salt (NHS), N-(3-dimethylaminopropyl)-N′-ethyl-carbodiimide hydrochloride (EDC) and 3-mercaptopropionic acid (MPA, >99 %) were purchased from Alfa Aesar® (China, www.alfachina.cn). Bis(2-ethylhexyl) sodium sulfosuccinate (AOT), phosphate-buffered saline (PBS), Tris–HCl buffer (0.3 M, pH 7.4), 10 × NEB buffer 2 (1 × NEB buffer 2: 50 mM NaCl, 10 mM Tris–HCl, 10 mM MgCl2, 1.0 mM dithiothreitol, pH 7.9), and HEPES buffer (25 mM, pH 7.4) were the product of Sinopharm Chem. Re. Inc. (Shanghai, China, www.reagent.com.cn). All other chemicals were of analytical grade. A Millipore purification system was used in all runs (18.2 MΩ cm; Milli-Q, from Millipore; www.merckmillipore.com). The washing buffer was prepared with 0.02 M Tris–HCl buffer containing 0.1 % Tween-20, 0.5 % BSA, and 0.15 M NaCl.
Synthesis of water-soluble, carboxylated CdS QD
Water-soluble, carboxylated CdS QDs were synthesized according to the literature with minor modification [17]. Prior to synthesis, a homogeneous mixture was first prepared by addition of 14-g AOT into 4-mL distilled water containing 196-mL n-hexane under vigorous stirring. Following that, Cd(NO3)2 (0.48 mL, 1.0 M) and Na2S (0.32 mL, 1.0 M) were added into 120 and 80 mL of the above-prepared reverse micelle solution, respectively. Afterward, two solutions were mixed together under vigorous stirring, and reacted for 60 min under the protection of N2 to yield CdS QDs. Subsequently, cysteamine (0.34 mL, 0.32 M) and 3-mercaptopropionic acid (70 μL, 3.0 M) were injected to the mixture and reacted for another 24 h under the same conditions. Finally, the suspension was evaporated in vacuo and washed with pyridine, hexane, acetone and methanol in sequence, and further centrifuged at 15,000 g for 30 min at 4 °C to obtain the carboxylated QDs.
Conjugation of carboxylated QDs with aminated blocker DNA
Conjugation of aminated blocker DNA with carboxylated QD were prepared through a typical carbodiimide coupling [18]. Initially, 1.86 mg of carboxylated CdS QDs was dissolved into 0.5 mL of distilled water, and the pH was then adjusted to 7.3 with 3 mM HCl. After that, NHS (11.0 g) and EDC (15.0 g) were dissolved in the suspension followed with continuous stirring for 45 at room temperature. Following that, 50 μL of 100-μM aminated blocker DNA was added dropwise to the mixture and incubated for 6 h at 4 °C. Subsequently, the suspension was centrifuged at 13,000g for 20 min at 4 °C. Finally, QD-labeled blocker DNA obtained was dispersed into 100-μL HEPES buffer for further use.
Hybridization of biotinylated hairpin DNA with QD-labeled blocker DNA
5-μL of 100-μM hairpin DNA and 5-μL of the above-prepared QD-labeled block DNA were initially mixed in 90-μL HEPES buffer containing 20 mM KCl, 200 mM NaCl, 0.05 wt% Triton X-100 and 1.0 wt% dimethyl sulfoxide. And then, the mixture was denatured for 4 min at 90 °C, annealed for 5 min at 50 °C, and finally cooled to room temperature. During this process, the QD-labeled blocker DNA opened the hairpin DNA to form a hairpin-blocker DNA duplex. The un-hybridized hairpin DNA was removed by centrifugation at 13,000g for 20 min at 4 °C and dispersed into 100-μL HEPES buffer (Note: At this step, the un-hybridized blocker DNA might be also collected, but it would be removed during the following immobilization and washing process on the well of microplate).
Strand-displacement reaction process and stripping voltammetric measurement
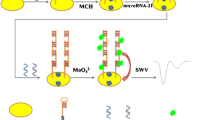
Scheme 1 displays target-induced strand-displacement reaction process and measurement mechanism. A high-binding polystyrene microplate was coated overnight at 4 °C with 50 μL per well of streptavidin (10 mg mL−1 in pH 7.4 Tris–HCl buffer) by covering with adhesive plastics plate sealing film. After being washed three times with PBS, the microplate was incubated with 300 μL per well of blocking buffer for 1 h at 37 °C with shaking. The plates were then washed as before. Afterward, 50 μL (per well) of biotinylated hairpin-blocker DNA with QDs prepared above was added to the microplate, and reacted for 4 h (optimized, please see our recent report [19]) at 37 °C. During this process, the hairpin-blocker DNA with CdS QD was immobilized onto the microplate through the labeled biotin on hairpin DNA based on biotin-avidin chemistry (Note: The microplate should be washed in order to remove the un-hybridized QD-labeled blocker DNA). Following that, 50 μL of Ureagene target DNA standards with various concentrations was added into the microplate, and incubated for 40 min at 37 °C under shaking (Note: At this step, target DNA-induced strand-displacement reaction was carried out, and CdS QD-labeled blocker DNA was dissociated to the solution accompanying with target DNA). The resulting suspension (~50 μL) was transferred in a 5-mL beaker, and a 50-μL aliquot of 1.0 M HNO3 was added to release cadmium ions from QD-labeled blocker DNA (~4 min). Subsequently, 2 mL of acetate buffer (0.2 M, pH 5.6) containing 10 μg mL−1 of mercury ions (from HgII acetate) was injected to the beaker. The square wave anodic stripping voltammetric (SWASV) measurement was conducted on a CHI 760 Electrochemical Workstation (Chenhua, Shanghai, China) with a conventional three-electrode system comprising a Pt-wire counter electrode, an Ag/AgCl reference electrode, and an in situ formed mercury drop electrode (on a glassy carbon electrode surface, effective working area: 3.14 mm2). The stripping process contained a 1.0-min pretreatment at + 0.6 V and 2.0-min accumulation at - 1.4 V. The SWASV measurement was carried out after a 15-s rest period (without stirring) at an applied potential range of −1.0 to −0.3 V with a potential step of 4 mV, a frequency of 25 Hz, and amplitude of 25 mV. A baseline correction of the resulting voltammogram was performed with CHI 760 software.
Schematic illustration of target-induced strand-displacement reaction with QD-labeled blocker DNA for sensitive voltammetric detection of nucleic acid: (a) formation of hairpin-blocker DNA duplex between QD-labeled blocker DNA and biotinylated hairpin DNA, (b) target-induced strand- displacement reaction in streptavidin-modified microplate, and (c) measurement process by using square- wave anodic-stripping voltammetry
Results and discussion
Choice of materials
Typically, one important concern on the successful development of a new electrochemical DNA sensor is to adopt a simple and sensitive signal-transduction method. Anodic-stripping voltammetry is a powerful analytical tool for quantitative detection of specific ionic species. Various nanomaterials including gold, silver, polymers and QDs have been used in the electrochemical sensors [14–17]. Among them, QD-based electrochemical sensors have shown great potential owing to their broad excitation spectra, robust, narrow-band emission and versatility in surface modification. More importantly, these metal components can yield well-resolved stripping voltammetric peak. In this work, the detectable stripping voltammetric signal mainly derives from the QDs on the blocker DNA. Initially, the hairpin-blocker DNA duplex is formed through the principle of complementary base pairing between biotinylated hairpin DNA and aminated blocker DNA during the denaturation and renaturation process, which can be readily immobilized onto the surface of the wells of microtiter plate through the biotin-avidin interface chemistry. In the presence of target DNA, the strand-displacement reaction between hairpin DNA and blocker DNA can be carried out owing to the difference of the folding free energy. In this regard, the QD-labeled blocker DNA is dissociated from the hairpin DNA and released to the solution accompanying target DNA (i.e., reforming the target-blocker DNA duplex). Under acidic conditions, the carried QDs accompanying target-blocker DNA duplex can be converted to cadmium ions, which can be determined by square-wave anodic-stripping voltammetry. In the absence of target DNA, the CdS QD-labeled blocker DNA can not be displaced from the microplate, and the QDs can not be released in the detection solution, thereby resulting in a weak voltammetric signal. The signal increased with the increasing target DNA in the sample. Logically, an important concern arises as to whether the strand-displacement can be readily implemented between hairpin-blocker duplex DNA and target DNA. Prior to experiments, we utilized RNA folding prediction software to simulate these two different duplex DNA structures (see http://rna.urmc.rochester.edu/software.html). Computational results revealed that the folding free energy of hairpin-blocker DNA duplex was ~ −23.4 kcal mol−1, whilst that of target-blocker DNA duplex was ~38.8 kcal mol−1. Hence, the strand-displacement reaction can be executed because target-blocker DNA duplex was more stable that that of hairpin-blocker DNA.
Figure 1a shows transmission electron microscope (TEM, Tecnai G2 F20, USA, www.fei.com) of CdS QDs. The average diameter was about 4.6 ± 0.5 nm. Moreover, the CdS QDs can be homogeneously dispersed in the distilled water, which provided the facilitation for the labeling of aminated blocker DNA. Furthermore, we also used Zetasizer (Zetasizer APS, Malvern Instruments Ltd, www.malvern.com.cn) to investigate CdS QDs before and after modification with aminated blocker DNA. Experimental results indicated that the zeta potentials were −1.8 mV for carboxylated CdS QDs and −3.1 mV for QD-labeled blocker DNA, respectively. The more negative zeta potential on the modified quantum dots might be most likely as a consequence of the conjugation of negatively charged blocker DNA molecules on the QDs.
a TEM image of CdS quantum dots, and b gel electrophoresis (lane 1: DNA marker; lane 2: 0.1 μM biotinylated hairpin DNA; lane 3: 0.1 μM aminated blocker DNA; lane 4: 0.1 μM target DNA; lane 5: 0.1 μM hairpin DNA + 0.1 μM blocker DNA; lane 6: 50 pM target DNA + 0.1 μM hairpin DNA + 0.1 μM blocker DNA)
Another two questions to be answered herein were whether (i) the hairpin-blocker DNA duplex can be successfully formed and (ii) the strand-displacement reaction could be readily carried out between hairpin-blocker DNA and target DNA. To this end, we used gel electrophoresis to characterize the reaction and displacement processes (Fig. 1b). Lanes 2–4 represent the gel electrophoresis images of biotinylated hairpin DNA, aminated blocker DNA and target DNA, respectively. Although the base numbers of our design for hairpin DNA (41 nt) and blocker DNA (42 nt) were almost the same, the slow migration for hairpin DNA might be ascribed to the labeled biotin at the 5′ end. When 0.1 μM hairpin DNA and 0.1 μM blocker DNA were adequately reacted, a strong spot could be observed (lane 5), indicating that the blocker DNA opens the hairpin DNA to form hairpin-blocker DNA duplex. Significantly, the reaction of 0.5 nM target DNA with lane 5′s sample causes the formation of two spots (lane 6). These two spots were almost in accordance with those of biotinylated hairpin DNA and hairpin-blocker DNA duplex, respectively. Relative to lane 5, however, the tailing spot at the upper of lane 6 mainly derived from the unreacted hairpin-blocker DNA. The result of lane 6 revealed that target-induced strand-displacement reaction could be carried out based on our design.
Control tests and voltammetric characteristics
During the immobilization of hairpin-blocker DNA duplex on the microplate, we considered that the un-hybridized QD-labeled blocker DNA might be non-specifically adsorbed onto the well even if the washing process was thoroughly implemented. To monitor its possible influence during the measurement on the voltammetric signal, QD-labeled blocker DNA alone was direct employed for conjugation with streptavidin-modified microplate (including washing and blocking subsequently). The as-prepared microplate was used for the detection of zero analyte and 50 pM target DNA based on the same assay mode, respectively (Fig. 2a). As seen from curve ‘a’, a weak peak current was observed in the absence of target DNA. After the modified microplate was incubated with 50 pM target DNA, significantly, the obtained voltammetric signal (curve ‘b’) was almost the same as that of curve ‘a’. The results indicated that the immobilized QD-labeled blocker DNA on the surface of the wells of the microplate were not released regardless of the presence of target DNA. So, the non-specifically adsorbed QDs in the microplate did not affect the detectable signal of the bioassay.
We also considered whether target DNA may induce the strand-displacement reaction in the hairpin-blocker DNA duplex-modified microplate and cause the release of CdS QD-labeled blocker DNA from the microplate. To demonstrate this point, the hairpin-blocker DNA-modified microplate was also used for zero analyte and 50 pM target DNA, respectively (Fig. 2b). As seen from curve ‘a’ in Fig. 2b, the peak current was almost the same as that of curve ‘a’ in Fig. 2a, indicated that the hairpin-blocker DNA was firmly immobilized onto the microplate. In contrast, a strong voltammetric peak was appeared when the modified microplate reacted with 50 pM target DNA (curve ‘b’), suggesting that target DNA triggers the strand-displacement reaction. Based on the results in Fig. 2a and b, we might conclude that our design should be feasible for the detection of target DNA based on the strand-displacement reaction with quantum dot-labeled DNA.
Optimization of experimental conditions
To acquire a strong voltammetric signal, the conjugated amount of aminated blocker DNA on the QDs was optimized. Too small an amount of blocker DNA on the QDs was not favorable for the formation and immobilization of hairpin-blocker DNA duplex, whilst too much amount elevates the detection limit. As seen from Fig. 3a, the peak currents initially increased with the increasing blocker DNA, and then decreased when the volume was more than 50 μL (50 pM target DNA used in this case). The optimal current was obtained at 50 μL. Thus, 50 μL of 100-μM aminated blocker DNA was used for the labeling of 1.86 mg QDs in 0.5 mL distilled water.
Another important factor influencing the analytical performance of the developed bioassay was the displacement reaction between hairpin-blocker DNA duplex and target DNA. Usually, it takes some time for target DNA to displace QD-labeled blocker DNA from DNA duplex. As shown from Fig. 3b, the current increased with the increasing hybridization time, and tended to level off after 40 min. A long incubation time did not cause significant increase in the detectable signal. To save the assay time, 40 min was selected as the hybridization time for the strand-displacement reaction.
Analytical performance
Under optimal conditions, we investigated the sensitivity and dynamic working range of the electrochemical DNA sensor toward target DNA standards with different concentrations by using hairpin-blocker DNA duplex-modified microplate with square-wave anodic stripping voltammetry based on the developed protocol. Experimental results indicated that the peak currents increased with the increasing target DNA concentration in the sample solution (data not shown). A good linear dependence between the peak currents and target DNA concentration was obtained in the range from 5 pM to 1.0 nM. The linear regression equation was y (μA) = 0.0434 × C [target DNA] + 2.29624 (pM) (R 2 = 0.9983, n = 21) with a detection limit (LOD) of 1.2 pM, as calculated in terms of the rule of 3 × standard deviation over the blank signal. To clarify the merits of the displacement assays for DNA, the analytical properties were compared with other methods and strategies (Table 1). Apparently, the LOD of CdS QD-based voltammetric assay was comparable with those of other DNA detection methods.
To test the specificity of electrochemical DNA sensors, we used various oligonucleotides as matched, mismatched, deleted and inserted targets (Fig. 4). Only the matched DNA causes the strand-displacement reaction, resulting in a strong SWASV peak current. Hence, the designed scheme was highly selective for completely complementary DNA. The precision and reproducibility of detections were evaluated by repeatedly determining two target DNA levels including 10 pM and 0.5 nM with identical batches of QD-labeled blocker DNA. Experimental results revealed that the standard relative deviations (RSDs) of the bioassay using the same-batch QD-labeled blocker DNA were 8.3 and 7.4 % for 10 pM and 0.5 nM, respectively. The batch-to-batch reproducibility was also monitored, and the RSDs were 9.8 and 8.4 %, respectively, toward the aforementioned concentrations. Thus, the precision and reproducibility of the electrochemical DNA sensor was acceptable.
SWASV responses of electrochemical DNA sensors toward a matched target DNA, mismatched target DNA, a target DNA with a deleted nucleotide, and a target DNA with an inserted nucleotide (all 50 pM). Target sequence: 5′-GAAGTAATGGACTTAXTTATTACAAACGCATTA-3′; T (matched): X = G; T (mismatched): X = T; T (deleted): no X; T (inserted): X = GG
Preliminary application for spiking serum samples
To investigate the possible application of QD-based electrochemical DNA sensor for testing real samples, six spiked blank new-born calf serum samples including 0.5 pM, 5.0 pM, 50 pM, 5.0 and 50 nM target DNA were measured with the developed method. According to the obtained SWASV peak current, the level of target DNA in each sample was calculated according to the above-mentioned regression equation: y (μA) = 0.0434 × C [target DNA] + 2.29624. Experimental results indicated that the assayed results by the electrochemical DNA sensor for the above-mentioned six samples were 0.46 pM, 5.6 pM, 56.8 pM, 4.87 and 48.5 nM, respectively. The recoveries were 92, 112, 113.6, 97.4, and 97.4 %, respectively. Therefore, the QD-based electrochemical DNA sensor may be applied to the determination of target DNA in complex matrices.
Conclusions
In summary, we have developed a convenient and feasible DNA sensing strategy with high sensitivity and selectivity by using CdS QD-based amplification protocol accompanying a strand-displacement reaction. The signal was amplified through the labeling quantum dots accompanying the releasing cadmium ions with a high-efficient anodic stripping voltammetric method. Highlight of this work is exploiting a new type of QD-based nanolabel for the signal amplification of electrochemical DNA sensor by coupling with a highly sensitive stripping voltammetric technique. These features, as well as its other advantages, such as convenience of operation, enzyme-free nature, and low cost, make it further applicable for other nucleic acids by controlling the sequence of hairpin-blocker DNA, and thus verify the versatility of the assay. Nevertheless, only one disadvantage of this method is to use an in-situ generated mercury electrode for the detection of target DNA. To intend for routine use, future work should focus on the improvement of the assay system (e.g., directly using the cleaned gold electrode or platinum electrode).
References
Khodakov D, Ellis A (2014) Recent developments in nucleic acid identification using solid-phase enzymatic assays. Microchim Acta 181:1633–1646
Chang K, Deng S, Chen M (2015) Novel biosensing methodologies for improving the detection of single nucleotide polymorphism. Biosens Bioelectron 66:297–307
Ikbal J, Lim G, Gao Z (2015) The hybridization chain reaction in the development of ultrasensitive nucleic acid assays. TrAC Anal Chem 64:86–99
Rodiger S, Liebsch C, Schmidt C, Lehmann W, Resch-Genger U, Schedler U, Schierack P (2014) Nucleic acid detection based on the use of microbeads: a review. Microchim Acta 181:1151–1168
Chen Y, Xiao L, Liu Y, Li X, Zhang J, Shu Y (2014) A lipase-based electrochemical biosensor for target DNA. Microchim Acta 181:615–621
Blackstock D, Chen W (2014) Halo-tag mediated self-labeling of fluorescent to molecular beacons for nucleic acid detection. Chem Commun 50:13735–13738
Gerasimova Y, Kolpashchikov D (2014) Enzyme-assisted target recycling (EATR) for nucleic acid detection. Chem Soc Rev 43:6405–6438
Pu F, Ren J, Ou X (2014) Nucleic acid and smart materials: advanced building blocks for logic systems. Adv Mater 26:5742–5757
Xu M, He Y, Gao Z, Chen G, Tang D (2015) Isothermal cycling and casecade signal amplification strategy for ultrasensitive colorimetric detection of nucleic acid. Microchim Acta 182:449–454
Shen J, Li Y, Gu H, Xia F, Zuo X (2014) Recent development of sandwich assay based on the nanobiotechnologies for proteins, nucleic acids, small molecules, and ions. Chem Rev 114:7631–7677
Wang F, Lu C, Willner I (2014) From casecade catalytic nucleic acids to enzyme-DNA nanostructures: controlling reactivity, sensing, logic operations, and assembly of complex structures. Chem Rev 114:2881–2941
Zhu D, Zhang L, Ma W, Lu S, Xing X (2015) Detection of mircoRNA in clinical tumor samples by isothermal enzyme-free amplification and label-free graphene oxide-based SYBR Green I fluorescence platform. Biosens Bioelectron 65:152–158
Wong J, Yip S, Lee T (2014) Ultrasensitive and closed-tube colorimetric loop-mediated isothermal amplification assay using carboxyl-modified gold nanoparticles. Small 10:1495–1499
Lu W, Qin X, Luo X, Chang G, Sun X (2011) CdS quantum dots as a fluorescent sensing platform for nucleic acid detection. Microchim Acta 175:355–359
Wu Y, Xue P, Kang Y, Hui K (2013) Highly specific and ultrasensitive graphene-enhanced electrochemical detection of low-abundance tumor cells using silica nanoparticles coated with antibody-conjugated quantum dots. Anal Chem 85:3166–3173
Kokkinos C, Economou A, Petrou P, Kakabakos S (2013) Microfabricated tin-film electrodes for protein and DNA sensing based on stripping voltammetric detection of Cd(II) released from quantum dots labels. Anal Chem 85:10686–10691
Zhang B, Tang D, Goryacheva I, Niessner R, Knopp D (2013) Anodic-stripping voltammetric immunoassay for ultrasensitive detection of low-abundance proteins using quantum dot aggregated hollow microspheres. Chem Eur J 19:2496–2503
Lai W, Zhuang J, Tang J, Chen G, Tang D (2012) One-step electrochemical immunosensing for simultaneous detection of two biomarkers using thionine and ferrocene as distinguishable signal tags. Microchim Acta 178:357–365
Sun A, Jia F, Zhang Y, Wang X (2014) Gold nanocluster-encapsulated glucoamylase as a biolabel for sensitive detection of thrombin with glucometer readout. Microchim Acta. doi:10.1007/s00604-014-1440-1
Fu L, Tang D, Zhuang J, Lai W, Que X, Chen G (2013) Hybridization-induced isothermal cycling signal amplification for sensitive electronic detection of nucleic acid. Biosens Bioelectron 47:106–112
Zhu J, Ding Y, Liu X, Wang L, Jiang W (2014) Toehold-mediated strand displacement reaction triggered isothermal DNA amplification for highly sensitive and selective fluorescent detection of single-base mutation. Biosens Bioelectron 59:276–281
Qiu L, Shen Z, Wu Z, Shen G, Yu R (2015) Discovery of the unique self-assembly behavior of terminal suckers-contained dsDNA onto GNP and novel “light-up” colorimetric assay of nucleic acids. Biosens Bioelectron 64:292–299
Luo M, Li N, Liu Y, Chen C, Xiang X, Ji X, He Z (2014) Highly sensitive and multiple DNA biosensor based on isothermal strand-displacement polymerase reaction and functionalized magnetic microparticles. Biosens Bioelectron 55:318–323
Yu Z, Zaitonuna A, Lai R (2014) Effect of redox label tether length and flexibility on sensor performance of displacement-based electrochemical DNA sensor. Anal Chim Acta 812:176–183
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (grant no.: 21405128) and the Research (Initial) Fund for the Doctoral Program of Xinxiang University.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Sun, AL., Zhang, YF. & Wang, XN. Sensitive voltammetric determination of DNA via a target-induced strand-displacement reaction using quantum dot-labeled probe DNA. Microchim Acta 182, 1403–1410 (2015). https://doi.org/10.1007/s00604-015-1467-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-015-1467-y