Abstract
We report on a novel graphene-based nanoarchitecture modified with plasma-polymerized propargylamine (G-PpPG) and its application in electrochemical sensors for DNA. Films of G-PpPG were characterized by X-ray photoelectron spectroscopy and electrochemical impedance spectroscopy. The presence of graphene enhances the electrochemical activity of the films, and the high density of amino groups (deposited at a low plasma input power) on their surface assists in the immobilization of probe DNA on the water-swollen polymeric network. By contrast, the degree of hybridization of the total complementary target DNA to the probe DNA remains unchanged when G-PpPG nanofilms prepared at higher input power. No substantial non-specific adsorption of totally mismatched target DNA on the polymer films is observed because of the complete coverage of the probe DNA. The detection limit for total complementary target DNA is approximately 1.84 nmol · L−1. The dynamic range extends from 0.1 to 1,000 nmol · L−1. The new nanocomposite may also be used to immobilize other probe DNA sequences, and this makes the approach potentially applicable to the detection of other oligomers.

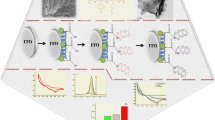
Preparing the DNA sensor made from the graphene-based nanoarchitecture modified by using PpPG (G-PpPG) includes the following processes: (a) Modifying the Au electrode with the graphene nanosheet, (b) depositing the PpPG film onto the Au electrode coated with graphene, (c) immobilizing the probe DNA onto the G-PpPG film, and (d) hybridizing the MM0 target with the G-PpPG film immobilized with P1
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Surface modification through radio frequency (RF) plasma treatment is expected to be suitable for biomedical or biosensor applications because it is controllable and eco-friendly. Moreover, highly cross-linked films can be deposited in such treatment regardless of surface geometries and the formation of multilayer films. Materials with amino functionality have attracted attention because of their application in fabricating biosensors. Amino-functional polymer films prepared through plasma polymerization includes ethylenediamine [1], butylamine [2], heptylamine [3], aniline [4], and allylamine [5]. One of the major conclusions of recent works on plasma-polymerized allylamine (PPAA) films reveals that their amino content of these films has an important role on their biosensing applications. Given that unsaturated monomers can be readily polymerized in RF plasma, the primary amino groups are transformed to have imine or nitrile functions [6]. Consequently, monomers with double or triple bonds are selected during polymerization. For example, PPAA, which is used as the sensitive layer for biosensors in biomedical or protein sensing, has been extensively studied [7]. Foerch et al. [5] used pulsed RF plasma in allylamine polymerization, successful DNA immobilization, and protein adhesion. PPAA was also used to modify the surface of quartz particles to remove humic acid [8]. Rebl et al. [9] reported that the positive charges of PPAA can control cell physiology, which is dominant in the integrin receptor that binds to collagen I. Monomer reorganization always occurs during polymerization because numerous bonds breaks as a result of high electron density and electronic temperature in plasma. This phenomenon forms the highly cross-linked structure and saturated bonds of polymers. Hence, most plasma polymers exhibit poor electrochemical performance and are seldom used as electrochemical biosensors. By contrast, a high degree of conjugation is produced in plasma-polymerized propargylamine (PpPG) because carbon–carbon triple bonds can dissolve into double bonds, thus indicating that PpPG films possibly exhibit electrochemical activity [10].
Graphene has recently attracted significant scientific and technological interest [11]. This material exhibits tremendous potential in numerous applications, including electronics, energy storage, and bioscience/biotechnology because of its unique physicochemical properties such as high surface area (theoretically 2,630 m2 · g for a single-layer graphene), excellent thermal and electric conductivity, and high mechanical strength. However, the bonding interaction of graphene with biomolecules, as well as its adsorptive capability, must be improved because of its poor functionality. Graphene with amino [12], carboxyl [13], and doping functionalizations [14] has good biocompatibility and biological detection characteristics [15].
Graphene nanosheets that were modified by using plasma polymerized pyrrole and allylmaine were investigated in a previous work [16]; however, no good electrochemical property was observed. Considering its advantages, developing a new kind of electrochemical biosensor, i.e., a graphene-based nanofilm modified by using PpPG, is significant. In this paper, a novel and highly sensitive DNA sensor based on nanocomposite layers of graphene and PpPG for detecting target DNA in aqueous solutions is reported. Graphene can enhance the electrochemical properties of plasma polymeric films. Although these films have high functionality, they are reserved toward the biocompatibility of the biosensor matrix. In this research, the capabilities of these DNA sensing nanofilms were investigated via electrochemical impedance spectroscopy (EIS), and a high detection limit of 1.82 nmol · L−1 was observed upon target DNA hybridization.
Materials and methods
Chemicals and regents
Propargylamine was supplied by Aladdin Industrial Corporation (www.aladdin-e.com, Shanghai, China). The DNA sequences used were obtained from SBS Genetech Co., Ltd. (www.sbsbio.com, Beijing, China). Graphene was supplied by Nanjing XFNANO Materials Tech Co., Ltd. (www.xfnano.com, Jiangsu, China). All chemicals were used as received, without further purification. The probe oligonucleotide sequences used in this study are listed as follows:
-
P1 (P1, 30 mer): 5’-TTT TTT TTT TTT TTT TGT ACA TCA CAA CTA-3’
-
TMM (Target DNA, 15 mer): 5’-TGT GCC TAA GCC ATA-3’
-
MM0 (Target DNA, 15 mer): 5’-TAG TTG TGA TGT ACA-3’
Pretreating and modifying the Au electrode
Preparing the DNA sensor made from the graphene-based nanoarchitecture modified by using PpPG (G-PpPG) included the following processes: (a) Modifying the Au electrode with the graphene nanosheet, (b) depositing the PpPG film onto the Au film coated with graphene, (c) immobilizing the probe DNA onto the G-PpPG film, and (d) hybridizing the MM0 target with the G-PpPG film immobilized with P1. A schematic representation of the fabrication process of the sensitive DNA sensor is shown in Scheme 1.
Pretreating the Au film was performed according to published procedures [17]. The Au films were successively polished with 0.5 and 0.03 μm alumina slurries made from alumina powder and water on micro cloth pads. After removing the alumina from the surface by using copious amounts of water, the Au films were briefly cleaned in an ultrasonic bath, and then, with ethanol and water between polishing steps. After being washed with ethanol and water, the Au films were cleaned in 0.05 mol · L−1 H2SO4 by cycling them between −0.3 and 1.5 V until a reproducible voltammogram was obtained. Then, the Au films were washed with ultrapure water and dried under a stream of nitrogen.
Modifying the Au films by using graphene nanosheets involved two steps, as shown in Scheme 1: (a) The treated Au films were immersed in 0.01 mol · L−1 octadecanethiol (ODT) solution in ethanol for 0.5 h to form self-assembled ODT layer, and (b) the Au films with a self-assembled ODT layer were incubated in an ethanol suspension with 1 mg · mL−1 graphene for 1 h to ensure that they could combine with the alkyl chains of ODT via intermolecular interaction [18]. Herein, 10 mg graphene was dispersed in 10 mL ethanol and unltra-sonicated for 10 min to form the graphene -ethanol dispersion used.
The polymeric films of PpPG were deposited onto the surface of an Au film, in which propargylamine was used as the monomer during plasma polymerization by employing a hydroquinone-2 plasma-enhanced chemical vapor deposition instrument (Institute of Microelectronics of the Chinese Academy of Sciences, Beijing, China), as illustrated in Scheme 1. The RF generator was operated at 13.56 MHz. Prior to each experiment the samples were activated by argon plasma at a settled flow rate of 5 sccm (standard cubic centimeters per min) for 5 s under a constant procedure pressure of 50 mTorr. The input power was between 5 and 200 W under continuous wave (CW). Propargylamine was frozen by liquid N2 to avoid the quick volatilization. Afterward, it was introduced into the reactor chamber together with argon. The carrier gas was assumed to be saturated with the propargylamine monomer in all cases. The gas flow rate was fixed at 30 sccm. Thicknesses of G-PpPG films were form 24 ± 2 to 78 ± 2 nm for the deposition time within 1 to 5 min at the input power of 5, 50, and 100 W, which was determined by the step apparatus. To decrease the possibility of the samples surface to be oxidized by O2 in air, samples were still in the chamber of the plasma reactor for 10 min after stopping the plasma irradiation.
Preparing the phosphate buffer and the DNA solutions
Stock DNA solutions were prepared in phosphate buffered saline (pH 7.4), which was prepared by mixing 8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4, and 0.24 g KH2PO4 in 1 L Milli-Q water. All solutions were prepared immediately before the experiments and were stored at 4 °C until use.
DNA immobilization/hybridization on the G-PpPG nanofilms
DNA was immobilized in 100 nmol · L−1 P1 DNA phosphate buffered saline with an immersion time of 24 h. Before analysis, all samples were thoroughly rinsed with phosphate buffered saline, and then with water. The samples were then dried under flowing nitrogen. DNA was immobilized in 1 μM probe DNA phosphate buffered saline solution for 24 h. Before analysis, each sample was rinsed with phosphate buffered saline and Milli-Q water, and then dried under flowing nitrogen. Finally, the DNA immobilized films were immersed in 1 μmol · L−1 target DNA solution for 24 h, and then, rinsed with phosphate buffered saline and Milli-Q water.
Characterization studies
X-ray photoelectron spectroscopy (XPS) was recorded on an AXIS HIS 165 spectrometer (Kratos Analytical Ltd., Manchester, UK) with a monochromatized Al Kr X-ray source (1486.71 eV photons). The nanofilms were analyzed at a 90° takeoff angle, and the core levels were calibrated based on the first component of the C 1 s core-level peaks, which was set at 284.6 eV. The chemical structure of the film was analyzed by Fourier transform infrared spectroscopy (FTIR) by using a Nicolet 850 spectrometer (Thermo Electron Corporation, Massachusetts, USA). The relative areas of the absorption bands at approximately 3,300 cm−1 (for -NH2 groups) and at 2,900 cm−1 (for CHx groups) were used to estimate and maximize approximate amine functional group densities. EIS was performed by using a CHI660D electrochemical workstation (Shanghai CH Instrument Company, Shanghai, China). A conventional three-electrode cell was used, including an Ag/AgCl (saturated KCl) electrode as the reference electrode, platinum slides as the counter electrode, and Au film substrate as the working electrode. EIS was obtained at a potential of −0.2 VMSE at a frequency range of 106 Hz to 10−2 Hz with AC amplitude of 5 mV. The spectra were then analyzed using the Zview2 software, which uses a non-linear least-squares fit to determine the parameters of the elements in the equivalent circuit. All electrochemical measurements were carried out in 5 mmol · L−1 K3[Fe(CN)6]/K4[Fe(CN)6] (1:1) mixture in 0.1 mol · L−1 KCl.
Results and discussion
Chemical performance of G-PpPG nanocomposites
The chemical compositions of the G-PpPG nanofilms deposited under different plasma conditions were investigated via FTIR and XPS. In the FTIR spectra of the G-PpPG nanofilms prepared under different plasma input powers (Fig. 1), the adsorption band at ~3,360 cm−1 corresponds to amino, imine, and amide bonds. The peak at ~1,650 cm−1 could be attributed to the bending vibrations of N–H or the stretching vibrations of the C = C or C = N groups. In addition, multiple adsorption bands at 2,960, 2,940, and 2,880 cm−1 were assigned to aliphatic alkyl groups. Regarding the sample deposited at 200 W (Fig. 1 curve iv), a new weak adsorption band at 2,180 cm−1 was clearly observed. This band represents the stretching vibrations of the C ≡ N group, which were not present in other samples deposited under plasma input powers of 5, 20, and 100 W. Comparing the FTIR spectra of the four G-PpPG nanofilms deposited at different plasma input powers, the relative intensity of the characteristic N–H peak at ~3,360 cm−1 decreased with increasing input power from 5 to 200 W. The same trend was observed in samples deposited at 200 W for different periods. This result could be explained by the easy decomposition of monomer gas into several smaller species through the irradiation of plasma at high input power or through a long period of irradiation, such as that for the C = N or C ≡ N groups, thus leading to the decrease of N–H content in G-PpPG nanofilms [5].
Comparing Fig. 1a and b, the influence of plasma irradiation time on the chemical structure of the plasma polymerized nanofilms was less than that of the plasma input power used. Hence, the effects of plasma polymerization time on various characteristics of plasma nanocomposite films, such as surface morphology, electrochemical property, and sensing activity, were neglected. All experiments were performed with the objective of warranting similar thicknesses (20 ± 3 nm) of the G-PpPG nanofilms deposited under different plasma input powers [19].
The precise chemical structures of the G-PpPG nanofilms were also confirmed via XPS, in which the atomic% of the films deposited at different plasma input powers is summarized in Table 1. This table demonstrates that carbon % in the nanofilms increased, whereas nitrogen % and oxygen % decreased with increasing input power used to prepare the G-PpPG nanofilms. As discussed earlier, numerous species were produced because of the decomposition of propargylamine under high plasma input power [5]. Considering the differences in the chemical bonds of the G-PpPG nanofilms deposited under different plasma conditions, the fitted curves of C 1 s and N 1 s core-level XPS spectra were collected and simulated (Fig. 2). As shown in Fig. 2a, the three peaks centered at ~283.9, ~284.6, and ~286.4 eV were assigned to the C = C, C–H/C–C, and C–O/C–N groups, respectively. The relative content of carbon-related and nitrogen-related groups in the G-PpPG nanofilms could be deduced from the ratio of their peak areas to the total peak areas. As shown in the Fig. 2 a, the relative intensity of C–O/C–N in the G-PpPG nanofilms decreased with increasing plasma input power used. This result was mainly attributed to the formation of small molecules, such as CO, CO2, NO, or NO2, at a high plasma input power. These molecules would be pumped out from the plasma reactor, thus resulting in a decrease in the contents of C–O/C–N groups in the samples [20]. In addition, the result demonstrated that the relative intensity of the C = C group in the G-PpPG nanofilms deposited at 200 W is higher than those of the samples prepared at 20 W. This phenomenon was attributed to the equilibrium between the deposition of the polymer film and the etching of the overlayer of the polymer films in the plasma [20]. Aside from the presence of C = C in graphene, the C = C group could also be produced as a result of the polymerization of C ≡ C bonds in the monomer during film deposition and alkyl group decomposition in the polymer chain. Consequently, more C = C groups would be formed at high plasma input power, thus leading to an increase in its relative intensity in the G-PpPG nanofilms.
Similar to the N 1 s core-level XPS spectra Fig. 2b, three peaks were mainly fitted at ~398.8, ~399.3, and ~400.8 eV, which correspond to the functional groups of C = N, C–N/N–H, and C–N = O, respectively. As mentioned earlier, monomer molecules are easily dissociated into smaller species at high input power. Hence, the C = N group could have resulted from the dehydrogenized C–N group, whereas the C–N = O group resulted from the reaction of the active surface of graphene with the irradiated species in plasma. All these processes reduced the relative content of the C–N or N–H groups in the G-PpPG nanofilms deposited at 200 W, as shown in Fig. 2c.
Electrochemical properties of the G-PpPG nanofilms
The semicircle diameter is equal to the electron-transfer resistance, R et . EIS spetra of the samples at each stage were summarized in Fig. 3. These spectra were modeled with the Randles equivalent circuit consisting of solution resistance (R1), charge-transfer resistance (R2), and constant-phase element (CPE) (Fig. S1, Electronic Supplementary Material, ESM). As shown in Fig. 3a, After ODT was self-assembled onto the gold surface, the R et value dramatically increased from 0.26 to 2.79 kΩ, suggesting that ODT acts as an insulating layer and causes the difficulty in the interfacial electron transfer. While graphene was self-assembled with ODT, the R et value decreases gravely from 2.79 to 0.67 kΩ. Thus, the presence of graphene could improve the electron transfer because of the restoration of the graphitic network of sp2 bonds in graphene. For the samples of the pristine PpPG deposited onto the composite electrode of Au-ODT under the plasma input power of 20, 100, and 200 W, their EIS spectra are shown in Fig. 3b. As the plasma input power used in the preparation of the PpPG films increased from 20 to 200 W, the R et value of the composite electrode decreased from 1.91 to 0.67 kΩ. Additionally, The EIS spectra of the G-PpPG nanofilms deposited at 20, 100, and 200 W are shown in Fig. 3c, in which the R et value decreased from 0.43 to 0.24 kΩ, which is smaller than that of the pristine PgPG (Fig. S2, ESM). It clearly demonstrates that graphene enhances the electrical activity of plasma polymers in electrolyte solutions. As discussed earlier, a high intensity of C = C bonds was observed in the G-PpPG nanofilms, which could enhance the conjugation degree of the plasma polymeric film. Subsequently, the electrochemical activity of the highly conjugated structure of the nanocomposite films will be strengthened, thus leading to the rapid transfer of electrons at the interface between the electrode and the electrolyte solution, and the low R et value of the composite electrode [21].
Nyquist plots of EIS spectra in 5 mM K3[Fe(CN)6]/K4[Fe(CN)6] (1:1) mixture and phosphate buffered saline (pH 7.4, with 0.1 M KCl) within the frequency range of 0.01 Hz to 100 kHz, with 5 mV as the amplitude of the composite electrode of (a) the bare Au film, Au-ODT, and Au-ODT-G, (b) Au-ODT-PpPG, and (c) Au-ODT-G-PpPG nanocomposite films deposited at 20, 100, and 200 W, respectively
Immobilizing/hybridizing DNA on the G-PpPG nanofilms detected by EIS
DNA immobilization and hybridization were verified via EIS, which is ultra sensitive to changes in interfacial impedance on bio-recognition events occurring at the surface/electrolyte interface [22]. Fig. 4 shows the Nyquist plots, wherein the double-stranded DNA (dsDNA) was immobilized on the surface of the modified Au electrode by using the G-PpPG nanofilms deposited at different P input values. Given that all EIS spectra of the samples exhibited similar trends, Fig. 4a presents only one EIS Fig. of the Au electrode, which was modified by using the G-PpPG films deposited at 200 W in 5 mM K3[Fe(CN)6]/K4[Fe(CN)6] (1:1) mixture and phosphate buffered saline (pH 7.4, with 0.1 M KCl) within the frequency range of 0.01 Hz to 100 kHz with 5 mV as the amplitude. The simulated R et values of all samples at different stages of DNA immobilization/hybridization are summarized in Fig. 4b. After the deposition of the PpPG films onto the surface of the Au electrode modified by using graphene, the semicircle diameter of the EIS spectrum demonstrated an apparent enlargement compared with that of the Au electrode modified by using pristine graphene, thus indicating a decrease in the electrochemical activity of the composite electrode. This result was mainly attributed to the coverage of the plasma polymer films, which always exhibits a weaker electrochemical property than the graphene nanosheets, thus leading to the increase in R et value of up to 0.105 kΩ. Generally, DNA can be immobilized onto plasma polymeric films that contain a large number of amino groups via the static–electronic interaction between them [23]. When P1 DNA bonded to the G-PpPG nanofilm, a dramatic increase in R et value was observed compared with that in the as-prepared G-PpPG film. Typically, plasma polymeric films swell into a 3D network in aqueous solutions, in which biomolecules cannot only bond onto the surface of plasma polymer films, but also penetrate it [24]. Once a considerable part of the DNA chain is immobilized in bulk onto the polymer chains, the negatively charged phosphate groups on the oligonucleotide strands will repel the successive bonding of DNA, thus resulting in significant DNA chain immobilization on the polymer surface. Consequently, approaching the electrode surface will be difficult for [Fe(CN)6]3-/4- because of the presence of a huge amount of negatively charged phosphate groups in the single-stranded DNA (ssDNA) [25]. In all samples Fig. 4b, when the TMM DNA was hybridized with the P1 DNA immobilized onto the G-PpPG nanofilms, no substantial difference was observed between them, thus suggesting that no non-specific adsorption occurred between TMM DNA and the plasma films because of the complete coverage of P1 DNA of the modified electrode [26]. However, after MM0 DNA hybridization with P1 DNA, the R et value of the composite electrode apparently decreased, thus indicating the enhancement of the electrochemical activity of the electrodes in the electrolyte solutions. This trend was also observed in the other system, in which plasma polymer films were used as the sensitive layer for DNA biosensing [27]. However, this trend is different from those reported by other researchers [28]. The actual mechanism behind the improvement in electrochemical property that resulted from dsDNA hybridization upon Au electrode modified with plasma polymeric films needs to be investigated further.
(a) Nyquist plots of the EIS spectra of the 5 mM K3[Fe(CN)6]/K4[Fe(CN)6] (1:1) mixture and phosphate buffered saline (pH 7.4, with 0.1 M KCl) within the frequency range of 0.01 Hz to 100 kHz, with 5 mV as the amplitude of the Au electrodes modified at different stages: (i) G-PpPG, (ii) G-PpPG-P1, (iii) G-PpPG-P1-TMM, and (iv) G-PpPG-P1-MM0. (b) Ret values of the Au electrodes modified by using the G-PpPG nanofilms deposited at 20, 100, and 200 W at different stages before and after DNA immobilization/hybridization
The comparison of R et values deduced from the EIS curves during DNA immobilization of the three samples deposited at 20, 100, and 200 W Fig. 4b obtained a similar trend. Given that the electrochemical activity of the Au electrode depended on the electrode component, variations in R et values could represent the amount of biomolecules bonded onto the electrode surface to a certain extent. It shows that P1 DNA would likely immobilize the Au electrode modified by using the G-PpPG nanofilm deposited at a low P input , i.e., 20 W, with the highest ΔR et value of 1.905 kΩ. As shown in Fig. S 2 (ESM), high-intensity amino groups were reserved in the G-PpPG nanofilm prepared at 20 W, thus releasing additional positively charged amino groups, which subsequently adsorbed more DNA strands than the other G-PpPG films. No clear non-specific adsorption was apparent between TMM and the G-PpPG films in all samples. In addition, nearly the same amount of MM0 DNA hybridized with P1 was immobilized on the three G-PpPG nanofilms, in which the same ΔR et , R et, and MM0-R et, P1, were obtained. This result revealed that MM0 DNA strands hybridized first with P1 DNA immobilized onto the surface of the G-PpPG nanofilms. This situation produced a negatively charged phosphate-rich matter in the cover layer as well as a few MM0 DNA strands that could hybridize with the P1 DNA immobilized in the interior of the G-PpPG bulk because of the repletion interaction among the phosphate groups. All G-PpPG nanofilms could probably be used for DNA sensing in the future.
Detection limit of the developed DNA sensor
To estimate the efficiency of the hybridization of MM0 DNA with the immobilized P1 DNA, MM0 DNA with different concentrations within the range of 0.1 nM to 1,000 nM were incubated into the system after P1 was immobilized onto the PpPG nanofilms deposited at 200 W for 5 min, and were detected via EIS [Fig. 5a]. As MM0 DNA concentration increased, R et value decreased corresponding with the formation of dsDNA. The difference between the R et value of the ssDNA immobilized electrode and that after hybridization with MM0, ΔR et = R et, MM0–R et, P1, was adopted as the measurement signal. The ΔR et value was linear with the logarithm of the concentration of MM0 [Fig. 5b]. The dynamic detection range for MM0 DNA was from 0.1 nM to 1,000 nM, with the regression equation ΔR et = 1.54-0.41 logCMM0, and the regression coefficient (R) = 0.996. The detection limit was estimated to be 1.84 nM, which is comparable with those of previous assay techniques based on EIS, QCM, and SPR (Table 2).
Conclusions
In summary, a new graphene-based nanocomposite film modified by using a conjugated plasma polymer, i.e., G-PpPG, was developed. The new film exhibited high electrochemical performance. The chemical structure of the G-PpPG nanofilms depended on the plasma conditions used during plasma deposition, including plasma input power and polymerization time. At low P input , the G-PpPG nanofilms contained considerable amino groups, and helped DNA immobilization via the electro–static interaction between the negatively charged phosphate groups of DNA and the positively charged amino groups of the films. This finding suggests that the probe DNA did not only immobilize onto the nanofilm surface, but also into the interior of the polymeric network, thus leading to a decrease in the electrochemical activity of the composite electrode. Unlike other systems used in DNA biosensors, the electrochemical activity of the novel system improved through the formation of the double helix dsDNA after the MM0 DNA was hybridized with the probe DNA. Challenges and issues remain at the interface of the dsDNA and the electrolyte in plasma polymeric films. Moreover, the amount of dsDNA exhibited no clear change among the G-PpPG nanofilms deposited at 20, 100, and 200 W. The developed olDNA sensor based on the G-PpPG films presented a detection limit of 1.84 nM, with an MM0 concentration from 0.1 nM to 1000 nM. This fabricated electrochemical biosensor can be used to detect other biomolecules.
References
Kim J, Jung D, Park Y, Kim Y, Moon DW, Lee TG (2007) Quantitative analysis of surface amine groups on plasma-polymerized ethylenediamine films using UV–visible spectroscopy compared to chemical derivatization with FT-IR spectroscopy, XPS and TOF-SIMS. Appl Surf Sci 253:4112–4118
Wu Z, Yan Y, Shen G, Yu R (2000) A novel approach of antibody immobilization based on n-butyl amine plasma-polymerized films for immunosensors. Anal Chim Acta 412:29–35
Vasilev K, Poulter N, Martinek P, Griesser HJ (2011) Controlled release of levofloxacin sandwiched between Two plasma polymerized layers on a solid carrier. ACS Appl Mater 3:4831–4836
Agarwala M, Barman T, Gogoi D, Choudhury B, Pal AR, Yadav RNS (2014) Highly effective antibiofilm coating of silver–polymer nanocomposite on polymeric medical devices deposited by one step plasma process. J Biomed Mater Res B 1:33106
Zhang Z, Knoll W, Foerch R, Holcomb R, Roitman D (2005) DNA hybridization on plasma-polymerized allylamine. Macromolecules 38:1271–1276
Fally F, Doneux C, Riga J, Verbist JJ (1995) Quantification of the functional groups present at the surface of plasma polymers deposited from propylamine, allylamine, and propargylamine. J Appl Polym Sci 56:597–614
Mangindaan D, Kuo WH, Chang CC, Wang SL, Liu HC, Wang MJ (2011) Plasma polymerization of amine-containing thin films and the studies on the deposition kinetics. Surf Coat Tech 206:1299–1306
Jarvis KL, Majewski P (2012) Plasma polymerized allylamine coated quartz particles for humic acid removal. J Colloid Interf Sci 380:150–158
Rebl H, Finke B, Ihrke R, Rothe H, Rychly J, Schroeder K, Nebe BJ (2010) Positively charged material surfaces generated by plasma polymerized allylamine enhance vinculin mobility in vital human osteoblastss. Adv Eng Mater 12:356–364
Qiu S, Gao S, Liu Q, Lin Z, Qiu B, Chen G (2011) Electrochemical impedance spectroscopy sensor for ascorbic acid based on copper(I) catalyzed click chemistry. Biosens Bioelectron 26:4326–4330
Shao Y, Wang J, Wu H, Liu J, Aksay IA, Lin Y (2010) Graphene based electrochemical sensors and biosensors: a review. Electroanalysis 22:1027–1036
Huang KJ, Niu DJ, Liu X, Wu ZW, Fan Y, Chang YF, Wu YY (2011) Direct electrochemistry of catalase at amine-functionalized graphene/gold nanoparticles composite film for hydrogen peroxide sensor. Electrochim Acta 56:2947–2953
OuYang F, Huang B, Li Z, Xiao J, Wang H, Xu H (2008) Chemical functionalization of graphene nanoribbons by carboxyl groups on stone-wales defects. J Phys Chem C 112:12003–12007
Zhang LS, Liang XQ, Song WG, Wu ZY (2010) Identification of the nitrogen species on N-doped graphene layers and Pt/NG composite catalyst for direct methanol fuel cell. Phys Chem Chem Phys 12:12055–12059
Yin H, Zhou Y, Ma Q, Ai S, Chen Q, Zhu L (2010) Electrocatalytic oxidation behavior of guanosine at graphene, chitosan and Fe3O4 nanoparticles modified glassy carbon electrode and its determination. Talanta 82:1193–1199
Zhang Z, Liu S, Shi Y, Zhang Y, Peacock D, Yan F, Wang P, He L, Feng X, Fang S (2014) Label-free aptamer biosensor for thrombin detection on nanocomposite of graphene and plasma polymerized allylamine. J Mater Chem B 2:1530–1538
Liu G, Bocking T, Gooding JJ (2007) Diazonium salts: stable monolayers on gold electrodes for sensing applications. J Electroanal Chem 600:335–344
Chen Q, Yan H, Yan C, Pan G, Wan L, Wen G, Zhang D (2008) STM investigation of the dependence of alkane and alkane (C18H38, C19H40) derivatives self-assembly on molecular chemical structure on HOPG surface. Surf Sci 602:1256–1266
Zhang Z, Chen Q, Knoll W, Foerch R, Holcomb R, Roitman D (2003) Plasma polymer film structure and DNA probe immobilization. Macromolecules 36:7689–7694
Zhang Z, Liang P, Zheng X, Peng D, Yan F, Zhao R, Feng C (2008) DNA immobilization/hybridization on plasma-polymerized pyrrole. Biomacromolecules 9:1613–1617
Mani V, Dinesh B, Chen SM, Saraswathi R (2004) Direct electrochemistry of myoglobin at reduced graphene oxide-multiwalled carbon nanotubes-platinum nanoparticles nanocomposite and biosensing towards hydrogen peroxide and nitrite. Biosens Bioelectron 53:420–427
Hu Y, Li F, Bai X, Li D, Hua S, Wang K, Niu L (2011) Label-free electrochemical impedance sensing of DNa hybridization based on functionalized grapheme sheets. Chem Commun 47:1743–1745
Yang Z, Wang X, Wang J, Yao Y, Sun H, Huang N (2009) Pulsed-plasma polymeric allylamine thin films. Plasma Process Polym 6:498–505
Dong H, Zhu Z, Ju H, Yan F (2012) Triplex signal amplification for electrochemical DNA biosensing by coupling probe-gold nanoparticles-graphene modified electrode with enzyme functionalized carbon sphere as tracer. Biosens Bioelectron 33:228–232
Tang KH, Tsai MD (2008) Structure and function of 2: 1 DNA polymerase · DNA complexes. J Cell Physiol 216:315–320
Turcu F, Schulte A, Hartwich G, Schuhmann W (2004) Imaging immobilised ssDNA and detecting DNA hybridisation by means of the repelling mode of scanning electrochemical microscopy (SECM). Biosens Bioelectron 20:925–932
Ma C, Contento NM, Gibson LR, Bohn PW (2013) Redox cycling in nanoscale-recessed ring-disk electrode arrays for enhanced electrochemical sensitivity. ACS Nano 7:5483–5490
Akhavan O, Ghaderi E, Rahighi R (2012) Toward single-DNA electrochemical biosensing by graphene nanowalls. ACS Nano 6:2904–2916
Gu H, Su X, Loh K (2005) Electrochemical impedance sensing of DNA hybridization on conducting polymer film-modified diamond. J Phys Chem B 109:13611–13618
Fu Y, Yuan R, Xu L, Chai Y, Zhong X, Tang D (2005) Indicator free DNA hybridization detection via EIS based on self-assembled gold nanoparticles and bilayer two-dimensional 3-mercaptopropyltrimethoxysilane onto a gold substrate. Biochem Eng J 23:37–44
Peng H, Soeller C, Vigar NA, Caprio V, Travas-Sejdic J (2007) Label-free detection of DNA hybridization based on a novel functionalized conducting polymer. Biosens Bioelectron 22:1868–1873
Qi H, Li X, Chen P, Zhang C (2007) Electrochemical detection of DNA hybridization based on polypyrrole/ss-DNA/multi-wall carbon nanotubes paste electrode. Talanta 72:1030–1035
Chen K, Chen S, Lin H, Yan T, Tseng C (2007) A novel technique to immobilize DNA on surface of a quartz crystal microbalance by plasma treatment and graft polymerization. Mat Sci Eng C 27:716–724
Wu Z, Jiang J, Fu L, Shen G, Yu R (2006) Optical detection of DNA hybridization based on Xuorescence quenching of tagged oligonucleotideprobes by gold nanoparticles. Anal Biochem 353:22–29
Livache T, Maillart E, Lassalle N, Mailley P, Corso B, Guedon P, Roget A, Levy Y (2003) Polypyrrole based DNA hybridization assays: study of label free detection processes versus fluorescence on microchips. J Pharmaceut Biomed 32:687–696
Acknowledgments
This work was supported by the Program for the National Natural Science Foundation of China (NSFC Account Nos. 51173172 and 21104070).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 43 kb)
Rights and permissions
About this article
Cite this article
He, L., Zhang, Y., Liu, S. et al. A nanocomposite consisting of plasma-polymerized propargylamine and graphene for use in DNA sensing. Microchim Acta 181, 1981–1989 (2014). https://doi.org/10.1007/s00604-014-1300-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-014-1300-z