Abstract
We describe here an aptasensor for the ultrasensitive detection of Staphylococcus aureus by electrochemical impedance spectroscopy (EIS). Single-stranded DNA was linked to a nanocomposite prepared from reduced graphene oxide (rGO) and gold nanoparticles (AuNP). Thiolated ssDNA was covalently linked to the AuNPs linked to rGO, and probe DNA was immobilized on the surface of an AuNP-modified glassy carbon electrode to capture and concentrate Staph. aureus. The probe DNA of the aptasensor selectively captures the target bacteria in its three-dimensional space, and these results in a dramatic increase in impedance. Scanning electron microscopy, cyclic voltammetry and EIS were used to monitor the single steps of the electrode assembly process. The effect was utilized to quantify the bacteria in the concentration range from 10 to 106 cfu mL−1 and with a detection limit of 10 cfu mL−1 (S/N = 3). The relative standard deviation of Staphylococcus aureus detection was equal to 4.3 % (105 cfu mL−1, n = 7). In addition to its sensitivity, the biosensor exhibits high selectivity over other pathogens.

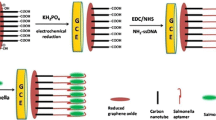
Schematic representation of the GCE surface modification and the detection of S. aureus. Reduced graphene oxide and gold nanoparticle (AuNP) nanocomposite linked by single-stranded DNA was prepared and then used in an aptasensor for the ultrasensitive detection of Staphylococcus aureus through electrochemical impedance spectroscopy. The probe DNA of the aptasensor selectively captures the target bacteria in its three-dimensional space, and these results in a dramatic increase in impedance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Staphylococcus aureus is a Gram-positive, relatively virulent pathogen that causes many types of infections, including dermatitis, gastroenteritis and toxic shock syndrome [1, 2]. S. aureus is temperature (7 to 48 °C) and pH (4.2 to 9.3) tolerant, enabling it to exist in food at high bacterial concentrations and for long periods of time [3, 4]. Improperly treated foods produced in unclean conditions (e.g., raw milk, dairy products and meat products) are easily contaminated with S. aureus, therefore, causes to food borne diseases and threatens human health [5, 6]. Hence, the detection quantification of S. aureus is important to food safety.
Many methods exist for detecting S. aureus in food. The traditional and most common method is based on routine bacterial culture and isolation technology, and this detection method uses standard plate counts after bacterial enrichment in medium [7]. However, the use of this technique is dwindling because it is time consuming (at least 2 days) and has low sensitivity. Recently, bioanalytical methods and mass spectrometry have been used to speed up the detection and ameliorate the sensitivity. Enzyme-linked immunosorbent assay (ELISA) is a common used method based on an antigen-antibody reaction [8]. Although this detection method of S. aureus can improve the sensitivity by two orders of magnitude, the high cost in preparing monoclonal antibodies and the long incubation time (at least 1 day) for ELISA restricted its use. Similar drawbacks were found in the polymerase chain reaction (PCR) technique. Real-time fluorescent quantitative PCR (FQ-PCR) works by detecting the fluorescence of specific nucleic acids in S. aureus and confirming the bacteria’s existence. The required DNA extractions, however, are time consuming, and the results have no direct relationship to the S. aureus concentration [9, 10]. The deficiencies in the current methods create an urgent demand for new, high-stability systems that detect the pathogen in faster and cheaper ways and at lower detection limit [11].
Aptamers are short series of artificial, but functional, single-stranded DNA or RNA oligonucleotides [12] which are obtained via an in vitro evolution process called SELEX (systematic evolution of ligands exponential enrichment) from random-sequence nucleic acid pools (generally, pools of approximately 1015 species) [13]. Aptamer can capture most biomolecules, from small molecules to whole cells, by covalent bond and spatial conformation recognitions. Because of the strong affinity towards targets, aptamers have been extensively used as capture probes in sensors [14]. Currently, aptamers play an important role in biological diagnoses [15], environmental monitoring and food safety because of their high specificity and affinity to target molecular, ability to be synthesized in vitro and ease of functionalization without compromising biological activity [16, 17]. Moreover, compared with protein-binding antibodies, aptamers survive repeated denaturations and renaturations, are stable in storage and are inexpensive [18].
Electrochemical impedance spectroscopy (EIS) is an ultrasensitive analytical technique that detects impedance variations along reaction interfaces. By applying small amplitude sine wave perturbations at different frequencies, the impedance value of a given electrochemical system changes [19]. In 2009, Gamella declared that EIS is likely to obtain higher sensitivities and cause less damage to biomaterials compared with other electrochemical methods of analysis [20], such as cyclic voltammetry (CV) and differential pulse voltammetry (DPV). Tiny changes in reactive surface can be rapidly and accurately detected by a transducer and enlarged by an amplifier, making EIS the most suitable detection method for aptamer-based biosensors. In addition, compared with other quantitative detection approaches (e.g., chemiluminescent methods), EIS is simpler because it requires no attachment of biomarkers to the electrodes and thus allows for label-free detection [21]. A series of label-free strategies were developed based on the assertions of these researchers.
In this work, we developed a cost-effective, sensitive, label-free impedimetric aptasensor for the direct detection of S. aureus. The feasibility of applying the aptasensor was also validated using real food samples. Reduced graphene oxide and gold nanoparticles linked by single-stranded DNA (rGO-ssDNA-AuNPs) was decorated on the electrode and worked as an amplification system to improve the lower limit of detection. The superior electron transporting property of rGO and the surface properties of AuNP enhanced the aptasensor detection capability. The thiol-ssDNA, which was covalently linked to the rGO through π-π stacking as well as the AuNPs by Au-S bonds, was required for stability [22]. Subsequently, the thiol-S. aureus-aptamer was self assembled and served to recognize and concentrate S. aureus. Ultrasensitive impedimetric measurements detected and recorded the current changes in the presence and absence of target bacteria. The D-value resistance obtained from the electrochemical measurements allowed us to calculate the concentration of bacteria colonies easily [23]. Finally, the aptasensor was successfully used to detect S. aureus in a real food sample [24].
Materials and methods
Apparatus
CV and EIS measurements were performed on an AUTOLAB PGSTAT302N (Metrohm, Zofingen, Switzerland, http://www.metrohm.com/). All electrochemical experiments were performed using a conventional three-electrode system where the working electrode was fabricated or bare glassy carbon, the reference electrode was Ag/AgCl (sat. KCl) and the auxiliary electrode was platinum wire. SEM was performed on S-4800 field emission scanning electron microscope (Hitachi, Japan, http://www.hitachi.com/). A KJ-300 ultrasonic bath (Wuxi Kejie Electron Instruments Co. Ltd., China, http://www.wxkjcs.com/) was used to wash the electrodes and disperse the nanoparticles. Ultrapure water used in the experiments was obtained from a Millipore Direct-Q® 3 system (Merck Millipore, Billerica, MA, U.S.A. http://www.emdmillipore.com/) and had a resistivity of 18.2 MΩ⋅cm−1.
Reagents
Graphene oxide (GO) sheets were synthesized from graphite powder by the Hummers method [25]. Typically, 2 g of graphite and 1.6 g of NaNO3 were added to 67.5 mL of concentrated H2SO4 in an ice bath. While stirring the mixture continuously, 9 g of KMnO4 was slowly added. After that, the suspension was maintained at 35 °C for 30 min and was then stirred for 5 days at ambient temperature. Afterwards, the paste was treated with 560 mL of deionized water and 30 mL of 30 % H2O2. Finally, the suspension was centrifuged, washed with water and dried overnight, affording GO.
The S. aureus aptamer sequence [26] 5′-HS-(CH2)6- GCA ATG GTA CGG TAC TTC CTC GGC ACG TTC TCA GTA GCG CTC GCT GGT CAT CCC ACA GCT ACG TCA AAA GTG CAC GCT ACT TTG CTA A-3′ was synthesized by Shanghai Sangon Biological Science & Technology Company (Shanghai, China, http://www.sangon.com/). The aptamer was diluted with TE buffer (10 mM Tris–HCl buffer containing 1 mM EDTA). Before use, the DNA solution was stored at −20 °C. Analytical reagent grade chloroauric acid (HAuCl4), sulphuric acid (H2SO4), hydrogen peroxide (H2O2), potassium dihydrogen phosphate (KH2PO4), 2-mercaptoethanol, K3[Fe(CN)6] and K4[Fe(CN)6] were used without further purification. All chemicals were purchased from Sinopharm Chemical Reagent Co., (Shanghai, China, http://www.reagent.com.cn/). 0.1 M PBS (pH = 7.4) was used in experiment.
Bacterial strains and culture media
S. aureus ATCC 29213 was obtained from the American Type Culture Collection (ATCC, U.S.A. http://www.atcc.org/). The strain was cultured in LB media for 12 h to reach to the growing stationary phase in an incubator-shaker at 37 °C. Then the cultures were serially diluted (10-fold steps), 10 μL of bacteria solutions were transfer to LB agar plates and incubated overnight in incubator-shaker at 37 °C. The concentrations of bacteria on plates were counted to determine the number of colony-forming units per milliliter (cfu mL−1). Finally, the stationary phase cultures were diluted to 10–108 cfu mL−1 with ultrapure water for the electrochemical measurements.
Attachment and reduction of GO on glassy carbon electrode
Before use, the bare glassy carbon electrode (GCE) (3 mm in diameter, CH Instruments Inc. http://www.chinstr.com/) was polished successively with 1.0, 0.3 and 0.05 μm alumina powder on suede to obtain a mirror-like appearance. The electrode was ultrasonically rinsed for 5 min each in ultrapure water, absolute alcohol and then ultrapure water. Afterwards, the GCE was electrochemically cleaned in 0.5 M H2SO4 using a cyclic potential scan between −0.2 and +1.6 V for 20 cycles at 100 mV s−1 to remove possible impurity. Finally, the electrode was washed with ultrapure water and dried with N2 gas.
To prepare GO-ssDNA, 1 mg of GO in 1 mL of PBS was sonicated for 30 min to obtain a homogeneous solution. Then, 10 μL of thiol-ssDNA was added, and the solution was mixed in a GL-88B vortex shaker (Kylin-Bell Lab Instruments Co., Ltd. China, http://www.ql-lab.com/). To ensure sufficient interaction between the GO and ssDNA, the solution was allowed to stand for 12 h before use. Then, 6 μL of the resultant GO-DNA solution was dropped onto the GCE surface and was dried in the oven at 37 °C, forming a thin film. Afterwards, electrochemical reduction was accomplished by CV with a range from −0.2 to 1.0 V at 100 mV s−1 in 0.1 M KH2PO4 solution. After 20 cycles, GO was reduced, and rGO-ssDNA/GCE was obtained [27].
Fabrication of the aptasensors
After gently washing the rGO-ssDNA/GCE with ultrapure water, Au nanoparticles were electrochemically deposited onto it by submersion for 15 s in 1 % HAuCl4 solution containing 0.1 M KNO3 supporting electrolyte at −200 mV, and a rGO-ssDNA-AuNP modified electrode was thus obtained [28]. Then, 5 μL of S. aureus aptamer was dropped onto the rGO-ssDNA-AuNP/GCE and dried naturally at room temperature. Finally, the fabricated electrode was immersed in 0.1 M 2-MCE solution for 1.5 h to remove any nonspecific adsorptions. Thus, a probe DNA-modified electrode expressed with aptamer/rGO-ssDNA-AuNP/GCE was obtained. The electrode was stored at 4 °C.
Characterization and electrochemical measurements
SEM, CV and EIS were employed to characterize each step of the GCE modification. A disassembled glassy carbon electrode was decorated with the rGO-ssDNA-AuNPs nanocomposite in the previous steps. The SEM in Fig. 2 was captured with this particular electrode. CV was performed from −0.2 to +0.6 V in a 0.1 M KCl solution containing 5 mM K3[Fe(CN)6] and K4[Fe(CN)6], at a scan rate of 100 mV s−1. EIS was performed from 105 to 10−1 Hz in a 7.4 pH 0.1 M KCl solution containing 5 mM K3[Fe(CN)6] and K4[Fe(CN)6]. The impedance data were analyzed and fitted to equivalent circuits using NOVA 1.8 software. Electron transfer resistance (Ret) and the D-value resistance (ΔRet) were calculated using the following equation:
where Ret(aptamer) is the resistance value of the modified electrode and Ret(aptamer-bacteria) is the resistance after aptamer-bacteria complex formation [29].
Analytical application
Fresh fish obtained from the China Resources Vanguard Supermarket (Wuxi, China) were washed with ultrapure water. All the samples were sterilized before adding different concentrations of S. aureus. Then the back of fish were cut off carefully under aseptic conditions and ground up in sterilization pot. The mince was treated with normal saline and the diluent was suspended by stirring. After letting the solids settle, the supernatant was removed and used as sample solutions.
Water samples obtained from Tai Lake were also used to evaluate the aptasensor. Firstly, the water samples were sterilized and left to stand for 30 min to precipitate macroaggregates. Then S. aureus in different concentrations was added to the aseptic water samples and the sample solutions were obtained [30].
Impedimetric measurements were carried out after incubating in all sample solutions for 30 min at 37 °C. Meanwhile, the samples were also tested with the flat counting method for comparison.
Results and discussion
Detection principle of the aptasensor
The impedimetric aptasensor’s method of detecting S. aureus and each step of GCE modification is depicted in Scheme 1. This biosensor employed a novel nanocomposite as a signal amplifier and a S. aureus aptamer as a capture probe. First, the GO-ssDNA solution was prepared by adding thiol-ssDNA to a GO solution. To ensure sufficient interaction between the GO and thiol, the solution was allowed to stand for overnight. The solution was dropped on the GCE surface to form a thin GO-ssDNA layer. The layer was electrochemically reduced to rGO-ssDNA in a KH2PO4 solution. Afterwards, AuNPs were electrodeposited, forming strong Au-S bonds with thiol-ssDNA. Staphylococcus aptamer was self-assembled onto the rGO-ssDNA-AuNP/GCE surface, and a layer-by-layer structure was thus formed. The electrode was incubated in 2-MCE to cover unoccupied areas [31].
In the presence of S. aureus, the aptamer-based electrode captured the bacteria, and the electric currents in the three-electrode test system were blocked accordingly. Through this experimental setup, the S. aureus concentration was quantified using the D-value resistance [24].
Characterization of the modified electrodes
CV and EIS were used to characterize each step of the biosensor fabrication. Cyclic voltammograms obtained in Fe(CN)6 3-/4- solution are displayed in Fig. 1.1. In CV, bare GCE (line a) normally presents itself as a pair of reversible redox peaks. After attaching GO-ssDNA, we observed a decrease in the redox peak current (line b). GO is a poor electrical conductor, and GO’s covalent conjugation with ssDNA further reduced its conductivity. An increase in current was observed from line b to line c. Because removing oxygenated functional groups from GO results in electrical conductivity similar to graphene, the redox current increase from line b to line c confirmed the electrochemical reduction of GO-ssDNA to rGO-ssDNA in the KH2PO4 solution. A dramatic increase in redox peak current emerged when AuNPs were deposited on the electrode (line d). AuNPs are widely used to enhance the transfer of electrons in biosensors because of its excellent conductivity same as electronic wires. The super high surface area of AuNPs is also helpful to load more capture probe. So the increase occurred because the AuNPs improved the conductivity and increased the effective area of the electrode. Finally, the redox current decreased after attaching aptamer (line e). Consistent with electric charge theory, the negatively charged aptamer impeded electron movement in the Fe(CN)6 3-/4- solution. The CV observations also proved that the aptamer had been successfully attached to the electrode.
The CVs (1) and Nyquist plots (2) of different modified electrodes in 0.1 M KCl solution containing 5 mM K3[Fe(CN)6] and K4[Fe(CN)6] (pH 7.4): (a) bare GCE; (b) GO-ssDNA modified GCE; (c) rGO-ssDNA modified GCE; (d) rGO-ssDNA modified GCE combined with AuNPs; (e) aptamer modified rGO-ssDNA-AuNPs/GCE
EIS measurements led to similar conclusions. As can be observed in the Nyquist plot (Fig. 1.2), the electron transfer resistance (Ret) was proportional to semicircular diameter at high frequencies [32]. The Nyquist plot of bare GCE was a typically scatter diagram with a Ret of 412 Ω. Due to the big resistance of GO, the Ret were dramatically increase to 1.60 KΩ after dropping GO-ssDNA on the electrode. After that, the electrochemical reduction of GO-ssDNA to rGO-ssDNA leads to a decrease in Ret from 1.60 KΩ to about 1.03 KΩ. Obviously, after deposition of AuNPs, the Ret of the electrode was sharply decreased. Owing to the super conductive ability of AuNPs, it was about 200 Ω lower of the Ret compared with bare GCE. In the end, a slightly growth from 196 to 351 Ω was obtained after attaching aptamer. As expected, each assembly step characterized by EIS afforded results consistent with the corresponding CV data. In summary, characterization of the designed aptasensor was performed using parallel sets of CV and EIS measurements, rGO-ssDNA-AuNP nanocomposites, enhanced the electronic transmission capacity and increased the amount of immobilized probe.
A comparable experiment to a system that does not contain an rGO was also developed to investigate the effects of carbon nanomaterial. We can see from Fig. 2, the current of rGO–AuNPs (line c) is bigger than AuNPs (line b). The excellent electron transporting property of rGO improved the electric conductivity. The other reason may be that the amount of AuNPs deposited on the electrode surface increased because of the large rGO surface area. Besides, the binding affinity between GCE and rGO are higher than it was between GCE and AuNPs. Therefore, the sensitivity and stability of the biosensor is promoted by the carbon nanomaterial.
Optimization of the experiment conditions
The distribution of AuNPs on the electrode surface had a large impact on the biosensor’s performance. We reasoned that increasing the amount of AuNPs to immobilize more aptamer would not cause a significant increase in the resistance of the biosensor. A deposition time of 15 s in 1 % HAuCl4 solution afforded the best morphology. SEM was used to examine the rGO-ssDNA-AuNPs nanomaterial (Fig. 3). The base layer exhibited a typical rGO ribbon-shaped and crumpled structure. The AuNPs were regularly sized and of uniform morphology, they were adsorbed to the surface in a thin layer. The introduction of rGO-ssDNA to AuNPs created a strong and durable Au-S attachment to the aptamer. The SEM image clearly revealed each layer of the layer-by layer structure.
The incubation time, which is another important factor in biosensor ability, was also investigated. Using the sensor created from the optimized AuNP deposition conditions, we recorded the aptasensor’s EIS response to 105 cfu mL−1 S. aureus at different incubation times. The D-value resistance increased sharply in the first 30 min and changed little subsequently, even after an extended period of time. We concluded that the relatively constant D-value resistance resulted from saturation of the aptamer with bacterium.
Analytical performance
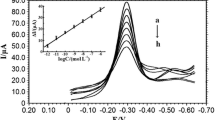
In this work, we correlated resistance values with concentrations of S. aureus. Our designed aptasensor was used to evaluate S. aureus concentrations of 101 to 106 cfu mL−1. The impedance spectra (Fig. 4) demonstrated that the semicircular diameters in the Nyquist plots, which are proportional to the resistance of the work electrode, increased with increased bacterial density. We further investigated the relationship between resistance and S. aureus concentration by calculating the resistance value of each parallel determination through a fitting procedure. The Randles equivalent circuit mode employed is illustrated in Fig. 3, where Rs is the resistance of the solution. The impedance of the faradaic reaction, which consisted of the electron transfer resistance (Ret) and a Warburg element (W), was connected to a capacitor CPE in a parallel circuit. By reading the Ret value after fitting, we observed that increasing the S. aureus 10-fold resulted an approximate 45 Ω increase in Ret.
Under the optimized conditions, the calibration curves corresponding to the EIS detection of S. aureus at different concentrations were created and analyzed. A linear relationship existed between the D-value of Ret and the logarithm of the S. aureus concentration. The linear fit obtained was Y = 35.6X + 10.733, and the fit had a 0.9933 correlation coefficient. Our S. aureus biosensor possessed a wide linear range (101 to 106 cfu mL−1) and a remarkably low LOD (10 cfu mL−1). Because this simple electrochemical method of detecting S. aureus did not require pre-enrichment, the experiment was completed within 60 min under the optimized conditions. This time is much shorter than other comparable biochemical analyses, including ELISA and PCR. The specific features of this method are shown in Table 1. Compared with the aptamer recognizing based method reported previously, the method proposed in this work is preferable in the determination of Staphylococcus aureus.
Repeatability and selectivity of the aptasensor
The repeatability of the aptasensor was evaluated by a set of parallel experiments. Under the same conditions, we used the same materials and reagents to make seven rGO-ssDNA-AuNPs modified electrodes to guarantee their consistency in electrochemical performance. Afterwards, the EIS response of the aptasensor to S. aureus in concentrations of 105 cfu mL−1 was checked, respectively. The RSD of 4.3 % indicated that the sensor has excellent repeatability.
To test the specificity of the S. aureus aptasensor, we immobilized the capture probe as before but changed the pathogen to E. coli and Salmonella. Parallel experiments using the 3 different bacteria were performed under the same conditions and bacterial concentrations of 105 cfu mL−1. E. coli and Salmonella exhibited tremendous differences in aptasensor response compared with S. aureus. The data indicated a ΔRet value of nearly 200 Ω for S. aureus, whereas E. coli and Salmonella exhibited ΔRet values of only approximately 20 Ω. These results indicated that the developed aptasensor possessed excellent selectivity.
Testing on a real food sample
The ability of our impedance-based, label-free aptasensor to detect S. aureus in fish and water samples were investigated in a recovery experiment [33]. After incubating at 37 °C for 30 min, a EIS measurement was carried from 105 to 10−1 Hz in a 7.4 pH 0.1 M KCl solution containing 5 mM K3[Fe(CN)6] and K4[Fe(CN)6]. The recovery rates of different samples, which were obtained through standard addition method, varied from 92 to 114 % (Table 2). This result indicated that the aptasensor for S. aureus is suitable for use in real food samples.
Conclusions
A sensitive and selective label-free impedimetric aptasensor for the rapid detection and quantification of S. aureus was successfully developed. The novel nanocomposite, which increased electron transfer and electrochemical signal, was synthesized by introducing ssDNA to rGO and AuNPs successively. By attaching S. aureus aptamer to the surface of rGO-ssDNA-AuNPs/GCE, we achieved the direct detection of S. aureus in food samples without using bio-labels. Under normal circumstances, the detection can be completed within 60 min. Additionally, the use of less expensive reagents reduce the assay costs. These advantages make the designed aptasensor promising for wide use in the rapid and sensitive detection of pathogens in food samples.
References
Ceylan KH, Kulah H, Ozgen C, Alp A, Hascelik G (2011) MEMS biosensors for detection of methicillin resistant Staphylococcus aureus. Biosens Bioelectron 29(1):1–12
Boujday S, Briandet R, Salmain M, Herry JM, Marnet PG, Gautier M (2008) Detection of pathogenic Staphylococcus aureus bacteria by gold based immunosensors. Microchim Acta 163:203–209
Zelada G, Guatavo A, Sebastian JL, Blondeau P, Riu J, Rius FX (2012) Label-free detection of Staphylococcus aureus in skin using real-time potentiometric biosensors based on carbon nanotubes and aptamers. Biosens Bioelectron 31(1):226–232
Pividori MI, Alegret S (2010) Micro and nanoparticles in biosensing systems for food safety and environmental monitoring. An example of converging technologies. Microchim Acta 170:227–242
Naomi B, Avraham R (2000) Staphylococcal enterotoxins. Int J Food Microbiol 61:1–10
Miao TG, Wang ZP, Li S, Wang X (2011) Sensitive fluorescent detection of Staphylococcus aureus using nanogold linked CdTe nanocrystals as signal amplification labels. Microchim Acta 172:431–437
Tsung C, Steven D (1992) Rapid detection of staphylococcus aureus in food by flow cytometry. J Rapid Meth Autom Microbiol 1:133–147
Tang Q, Yuan LL, Xiao XL, Guo P, Hu JB, Ma DD, Gao YY (2013) DNAzyme based electrochemical sensors for trace uranium. Microchim Acta 180:1059–1064
Ding X, Li H, Deng L, Peng Z, Chen H, Wang D (2011) A novel homogenous detection method based on the self-assembled DNAzyme labeled DNA probes with SWNT conjugates and its application in detecting pathogen. Biosens Bioelectron 26(11):4596–4600
Wang HY, Zhang CS, Xing D (2011) Simultaneous detection of Salmonella enterica, Escherichia coli O157:H7, and Listeria monocytogenes using oscillatory-flow multiplex PCR. Microchim Acta 173:503–512
Wang ZJ, Zhang J, Chen P, Zhou XZ, Yang YL, Wu SX, Niu L, Han Y, Wang LH, Boey F, Zhang QC, Liedberg B, Zhang H (2011) Label-free, electrochemical detection of methicillin-resistant staphylococcus aureus DNA with reduced graphene oxide-modified electrodes. Biosens Bioelectron 26(9):3881–3886
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505–510
Ellington AD, Szostak JW, Green R (1990) In vitro genetic analysis of the Tetrahymena self-splicing intron. Nature 346:818–822
Qi HL, Wang C, Cheng N (2010) Label-free electrochemical impedance spectroscopy biosensor for the determination of human immunoglobulin G. Microchim Acta 170:33–38
Xia YF, Gan SW, Xu QH, Qiu XW, Gao PY, Huang SS (2013) A three-way junction aptasensor for lysozyme detection. Biosens Bioelectron 39(1):250–254
Chen ZB, Li LD, Tian Y, Mu XJ, Guo L (2012) Signal amplification architecture for electrochemical aptasensor based on network-like thiocyanuric acid/gold nanoparticle/ssDNA. Biosens Bioelectron 38(1):37–42
Zhao HM, Gao S, Liu M, Chang YY, Fan XF, Quan X (2013) Fluorescent assay for oxytetracycline based on a long-chain aptamer assembled onto reduced graphene oxide. Microchim Acta 180:829–835
Wu SJ, Duan N, Ma XY, Xia Y, Wang HX, Wang ZP, Zhang Q (2012) Multiplexed fluorescence resonance energy transfer aptasensor between upconversion nanoparticles and graphene oxide for the simultaneous determination of mycotoxins. Anal Chem 84(14):6263–6270
Shervedani RK, Akrami Z (2013) Gold–deferrioxamine nanometric interface for selective recognition of Fe(III) using square wave voltammetry and electrochemical impedance spectroscopy methods. Biosens Bioelectron 39(1):31–36
Gamella M, Campuzano S, Parrado C, Reviejo AJ, Pingarron JM (2009) Microorganisms recognition and quantification by lectinadsorptive affinity impedance. Talanta 78(4–5):1303–1309
Escamilla GV, Campuzano S, Pedrero M, Pingarron JM (2009) Gold screen-printed-based impedimetric immunobiosensors for direct and sensitive Escherichia coli quantisation. Biosens Bioelectron 24(11):3365–3371
Varghese N, Mogera U, Govindaraj A, Das A, Maiti AK, Rao CNR (2008) Binding of DNA nucleobases and nucleosides with graphene. J Am Chem Soc 130:10876–10877
Kuila T, Mishra AK, Khanra P, Kim NH, Lee JH (2013) Recent advances on the efficient reduction of raphene oxide and its application as energy storage electrode materials. Nanoscale 5(1):52–71
Kochmann S, Hirsch T, Wolfbeis OS (2012) Graphenes in chemical sensors and biosensors. TrAC Trends Anal Chem 39:87–113
Hummers W, Offeman R (1958) Preparation of graphitic oxide. J Am Chem Soc 80:1339
Shao NS, Cao XX, Li SH, Chen LC, Ding HM, Xu H, Huang YP, Li J, Liu NL, Cao WL, Zhu YJ, Shen BF (2009) Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus. Nucleic Acids Res 37(14):4621–4628
Chen D, Feng HB, Li JH (2012) Graphene oxide: preparation, functionalization, and electrochemical applications. Chem Rev 112(11):6027–6053
Wang Y, Zhang S, Du D, Shao YY, Li ZH, Wang J, Engelhard MH, Li JH, Lin YH (2011) Self assembly of acetylcholinesterase on a gold nanoparticles–graphene nanosheet hybrid for organophosphate pesticide detection using polyelectrolyte as a linker. J Mater Chem 21(14):5319
Wang HJ, Yuan R, Chai YQ, Cao YL, Gan XX, Chen YF, Wang Y (2013) An ultrasensitive peroxydisulfate electrochemiluminescence immunosensor for Streptococcus suis serotype 2 based on L-cysteine combined with mimicking bi-enzyme synergetic catalysis to in situgenerate coreactant. Biosens Bioelectron 43:63–68
Yuan JL, Yu Y, Li C, Ma XY, Chen J, Wang ZP (2014) Visual detection and microplate assay for Staphylococcus aureus based on aptamer recognition coupled to tyramine signal amplification. Microchim Acta. doi:10.1007/s00604-013-1120-6
Chen GN, Lu CH, Yang HH, Zhu CL, Chen X (2009) A graphene platform for sensing biomolecules. Angew Chem Int Ed 48:4785–4787
Lu L, Chee G, Yamada K, Jun S (2013) Electrochemical impedance spectroscopic technique with a functionalizedmicrowire sensor for rapid detection of foodborne pathogen. Biosens Bioelectron 42:492–495
Ruan M, Niu CG, Zeng GM (2011) Rapid detection of Staphylococcus aureus via a sensitive DNA hybridization assay based on a long-lifetime luminescent europium marker. Microchim Acta 175:105–112
Duan N, Wu SJ, Zhu CQ, Ma XY, Wang ZP, Yu Y, Jiang Y (2011) Dual-color upconversion flurescence and aptamer-functionalized magnetic nanoparticles-based bioassay for the simultaneous detecion of Salmonella Typhimurium and Staphylococcus aureus. Anal Chim Acta 723:1–6
Sung YJ, Suk HJ, Sung HY, Li T, Poo H, Kim MG (2013) Novel antibody/gold nanoparticles/magnetic nanoparticle nanocomposites for immunomagnetic separation and rapid colorimetric detecion of Staphylococcus aureus in milk. Biosens Bioelectron 43:432–439
Acknowledgments
This work was partly supported by NSFC (21375049), the S&T Supporting Project of Jiangsu Province (BE2011621), National Science and Technology Support Program of China (2012BAK08B01), JSCIQ_2012IK166, Research Fund for the Doctoral Program of Higher Education (20110093110002), NCET-11-0663, and JUSRP51309A.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jia, F., Duan, N., Wu, S. et al. Impedimetric aptasensor for Staphylococcus aureus based on nanocomposite prepared from reduced graphene oxide and gold nanoparticles. Microchim Acta 181, 967–974 (2014). https://doi.org/10.1007/s00604-014-1195-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-014-1195-8