Abstract
Interaction with orchid mycorrhizal fungi (OMF) is essential to all members of the Orchidaceae, yet we know little about whether or how OMF abundances in substrates shape orchid populations. While root-associated OMF diversity is catalogued frequently, technological constraints have impeded the assessments of OMF communities in substrates until recently, thereby limiting the ability to link OMF communities in a habitat to population responses. Furthermore, there is some evidence that edaphic and microclimatic conditions impact OMF in soil, yet we lack an understanding of the coupled influences of abiotic environment and OMF structure on orchid population dynamics. To discover the linkages between abiotic environment, OMF community structure, and population size, we characterized the microclimatic conditions, soil physicochemistry, and OMF communities hosted by roots and soil across large and small populations of a terrestrial orchid endemic to California Floristic Province in North America. By using high-throughput sequencing of the ITS2 region of nrDNA amplified from root and soil DNAs, we determined that both roots and soil of larger populations, which were high in phosphorus but low in zinc, organic matter, and silt, were dominated by Tulasnellaceae OTUs. In comparison, roots and soil from smaller populations of the orchid hosted higher relative abundances of the Ceratobasidiaceae. In this multiyear, range-wide study that simultaneously measured habitat environmental conditions, and soil and root OMF communities, our results suggest that soil chemistry is clearly linked to soil and root OMF communities, which then likely alter and shape orchid populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ecological mechanisms that govern distributions and population dynamics of plant species are complex (Stanton-Geddes et al. 2012). Plants and populations may respond to a host of abiotic and biotic variables that may also interact with each other. In species with multitrophic interactions, such as symbioses with mycorrhizae, there is an additional layer of complexity that must be factored in when attempting to predict plant or population responses (McCormick et al. 2018). Further, when rarity of the plant host is added into this mix, identifying the ecological drivers of their populations becomes increasingly challenging due to the increasingly narrower fundamental niches (Fiedler et al. 2007; Wamelink et al. 2014; Leitao et al. 2016). The family Orchidaceae is one of the largest plant groups on Earth with an estimated 30,000 species (Dressler 1993) that are generally all considered rare in nature. All members of the Orchidaceae are uniquely characterized by an obligatory dependence on orchid mycorrhizal fungi (OMF) that primarily, but not exclusively, constitute rhizoctonia-type Basidiomycetes for seed germination and early development, and partial or complete dependence in later life stages depending on whether the taxa is photosynthetic or non-photosynthetic in adulthood (Girlanda et al. 2011; McCormick and Jacquemyn 2014). Moreover, many orchid taxa are patchily distributed across space and are influenced by specific preferences towards mycorrhizae and microenvironment (McCormick et al. 2016). Combined together, such trophic dependencies and niche preferences translate into the rarity of a large majority of orchid taxa if precisely suitable habitat with ambient conditions for germination, seedling recruitment, and reproduction is not available (Dressler 1993; Swarts and Dixon 2009a).
Distributions of orchid populations have been explained in the context of specificity and availability of OMF, which mostly belong to the three fungal families (Ceratobasidiaceae, Tulasnellaceae, and Sebacinaceae) routinely and pan-globally detected in orchids (Dearnaley et al. 2012; McCormick and Jacquemyn, 2014). Studies emphasizing the rarity of orchid taxa have revealed their generalist (Pandey et al. 2013; Waud et al. 2017) as well as specialist interactions toward OMF (Swarts et al. 2010; Kaur et al. 2017, 2019). Independent of the nature of host-OMF interactions, the presence of appropriate OMF in soil is clearly essential for the sustainability of an orchid population (Swarts and Dixon 2009b). Yet, it is less clear how the community structure of the preferred OMF in soil shapes the widespread variation in population sizes and demography of species within the Orchidaceae (Coates et al. 2006; Jacquemyn et al. 2010; Shefferson et al. 2014; Rock-Blake et al. 2017).
Prior studies based on automated methods of ribosomal intergenic spacer analysis (ARISA) and real-time polymerase chain reaction (qPCR) have shown that spatial gradients in OMF abundances in soil lead to variation in seed germination or plant emergence (McCormick et al. 2009, 2012, 2016; Rock-Blake et al. 2017). Given these positive effects on plant growth, it is likely that OMF abundances can mediate species demographies by affecting seed germination, plant recruitment, vegetative- or reproductive dormancy, reproduction, and/or mortality. To our knowledge, only one study by Jacquemyn et al. (2015) has simultaneously examined the relationship between population size and OMF communities in roots and associated soil of an orchid. Besides this one study, systematic investigations into the potential linkages between orchid population size and OMF communities within roots and soil are severely lacking (McCormick et al. 2018).
Further, the abiotic environment experienced by an orchid population can also alter its demography (Mattila and Kuitunen 2000; Shefferson et al. 2018). For instance, multiple studies have reported that increase in soil moisture and pH variably increases or decreases seed germination in orchids (Diez 2007; Jacquemyn et al. 2015; Waud et al. 2017). Orchids are also known to switch OMF partners in response to changing abiotic conditions such as drought or edaphic properties (McCormick et al. 2006; Bunch et al. 2013; Mujica et al. 2016). While these few studies address pairwise links between environmental predictors and orchid or OMF response, studies delineating tripartite linkages are rarely available. For instance, in the study by McCormick et al. (2006), it cannot be said whether the host orchid switched OMF within the roots strictly in response to drought or whether this switch was mediated by the altered OMF abundances in soil. Similarly, seed germination, plant growth, dormancy, or reproduction responses in orchid populations could be facilitated via OMF community shifts that are ultimately governed by the environment.
Provided the independent effects of OMF communities and abiotic microenvironment on orchid biology, their coupled dynamics might resolve the ambiguities pertaining to the wide disparities in orchid population dynamics. Yet, there are few studies that have considered integrated and coupled distributions of root OMF, their distributions in soil, and microhabitat conditions concurrently (Jacquemyn et al. 2015; Waud et al. 2017). And, such investigations are especially lacking from continents outside of Europe. Considering the wide global diversity of orchids and their life histories, the question of what explains their complex population ecology remains poorly addressed. To narrow this gap, we studied OMF communities in the context of host population size in a rare terrestrial orchid, Platanthera (Piperia) cooperi (S. Watson) R.M. Bateman (Bateman et al. 2003), that is endemic to the North American California Floristic Province. The orchid taxon is an appropriate model because of the wide spatiotemporal variability in population size coupled with their highly heterogeneous microenvironments. We specifically asked whether OMF communities in roots of P. cooperi and in its soil are uniquely distinct across variably sized populations and whether these differences are tied to the microenvironment (microclimate, and/or soil edaphic characters)? We hypothesized that orchid populations of different sizes will host distinct mycorrhizal communities in roots, and these differences will be tied to the community structure of the same fungi in soil and microenvironment.
Material and methods
Study species and populations
Platanthera cooperi is a terrestrial, perennial orchid native to California in the USA, and Baja California in Mexico, where its populations occur in scrub, chaparral, and woodland ecosystems (Ackerman and Lauri 2018). In southern California, P. cooperi occurs in the San Gabriel Mountain range and peninsular ranges of Los Angeles, San Bernardino, San Diego, Ventura, Orange, and Riverside Counties, whereas it is limited to a peninsular range in Baja California, although few records have been reported outside its core distribution in southern California and northern Baja California (Ackerman and Lauri 2018). The habitat of P. cooperi is characterized by mean annual precipitation of 25 cm and has mean minimum and maximum temperatures of 13 °C and 21 °C, respectively. Habitat soil can range from sandy clay loam to sandy loam with pH ranging from 5.1 to 8. Summer dormant plants emerge between December and January from perennating tubers and may remain vegetative or become reproductive during the growing season spanning approximately 6 months. Inflorescences of reproductive plants are visible from March to May. Plants enter anthesis in April, while capsules typically mature and dehisce by June.
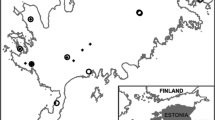
Population sizes vary widely across its range from only tens of individuals to thousands. To capture this wide range and to represent the core geographic range of P. cooperi, we selected two of its largest populations (Point Loma peninsula (PLF) and Santa Catalina island (SCE)) that occur in California and four smaller ones that occur across California and Mexico (Point Loma peninsula (PLE), Santa Catalina island (SCW), Cleveland National Forest (CH), and Baja California, Mexico (MX)) (Fig. 1a). Before finalizing the study locations, we conducted a laborious search at numerous potential P. cooperi sites in Orange, Riverside, and San Diego Counties to locate additional populations. These efforts were based on herbarium records and communication with local and regional orchid experts. While we confirmed one historical location (CH) to find a few individuals in 2017, additional large populations were not found. Populations SCE and PLF were considered large because they can host > 300 and 4000 flowering or > 500 and 10,000 total individuals in a given year, respectively, while each of the smaller populations typically hosts between 1 and 15 flowering or 10 and 30 total individuals per year.
Characterization of orchid mycorrhizal fungal communities within roots and habitat soil of Platanthera cooperi. a A partial map of the USA and Mexico showing the locations of study populations of P. cooperi. Samples were collected from Point Loma peninsula, San Diego County (PLE, PLF), Santa Catalina Island, Los Angeles County (SCE and SCW), Cleveland National Forest, Riverside County (CH), and from one population in Mexico (MX). b Hierarchical clustering of orchid mycorrhizal fungal (OMF) communities recovered from roots (R) and soil (S) across six study populations. Abundances of 112 OTUs belonging to Ceratobasidiaceae and Tulasnellaceae were used, and the heatmap shows the combined abundances of OTUs belonging to the two fungal families. Large populations are marked with an asterisk (*)
Sample collection and DNA extraction
Roots from seedlings (one leaved individual with 6–10 cm leaf length and 0.5–1 cm width), vegetative (individuals with at least two leaves, but lacking an inflorescence), and reproductive (individuals with an inflorescence) individuals from the six study populations were sampled between February and April across 3 years including 2015, 2016 and 2017. At each sampling event, 3 to 5 roots were severed from vegetative and reproductive plants without compromising the whole plant. Seedlings had to be collected whole because of the extremely reduced size of their root systems that typically includes at most 1 to 2 roots ≤ 1 cm in length. Roots were stored at 4 °C until they were shipped overnight to the laboratory. Altogether, 176 individual plants were sampled to represent three phenological stages from six populations across 3 years (Table 1). Once in the laboratory, roots were processed, and DNA was extracted by using the methods described in Pandey et al. (2013). Subsequently, the DNAs from multiple root fragments of a single plant were pooled to obtain an aggregate DNA prep for each sampled plant yielding 176 individual DNA samples. Soil was sampled concurrently with roots. At each population, two replicate soil cores of approximately 20 g were extracted from 2–5 locations within 5–10 cm distance of P. cooperi plants (Table 1). We sampled from 0 to 10 cm of soil profile considering the typical depth to which the roots of the species penetrate. All sample collection surfaces were sterilized with a disinfectant in between the handling of any two samples. Soil samples were placed in dry ice immediately upon collection and shipped overnight to the laboratory where they were transferred to a − 80 °C freezer. Two replicate DNA extractions from each of the 132 individual samples were performed with 250 mg soil per extraction by using MoBio PowerSoil DNA Isolation Kit. The two replicate DNAs per soil sample were subsequently pooled yielding aggregate DNAs for each of the 132 samples.
Fungal library preparation and sequencing
We used the primer pair ITS3 and ITS4-OF (Waud et al. 2014) to amplify the ITS2 region of fungal nrDNA using universal 5′ tail sequences from Alvarado et al. (2018; ITS3-tail: 5′-CCTATGTGGAGAGCCAGTAAGCGATGCTATGGT-GCATCGATGAAGAACGCAGC-3′; ITS4OF-tail: 5′-GTCAACGCTCACTACTGCGATTACCCAAGTCAG-GTTACTAGGGGAATCCTTGTT-3′). Negative controls devoid of any potential source of DNA were included in the sequencing libraries. Fungal amplicons were produced in a two-step protocol as suggested by Berry et al. (2011). A first PCR was carried out for each DNA prep in a 25-μL reaction mixture containing 1× Phusion Green Hot Start II High-Fidelity PCR Master Mix (Thermo Fisher Scientific Inc, MA, USA), an additional 1.5 mM MgCl2 (3.0 mM total), 200 nM of each primer, and 8–10 ng DNA. Thermocycling conditions were the following: 95 °C for 2 min, followed by 30 cycles of 95 °C for 30 s, 55 °C for 30 s, and then 60 °C for 4 min. PCR products were checked on a 1% agarose gel for an amplicon length of 500–550 and purified by bead-prep using carboxylated magnetic beads (GE Healthcare Life Sciences, PA, USA) as described in Rohland and Reich (2012). Two microliters of purified PCR products from the first PCR were used as template to carry out a 25-μL second PCR to add index and flowcell sequences to the amplicons. Thermocycling conditions for the index PCR were the following: 95 °C for 2 min, followed by 15 cycles of 95 °C for 30 s, 60 °C for 30 s, and then 72 °C for 1 min. The indexed amplicons from the second PCR were purified by bead-prep as described above. Products were quantified by using Quantiflour dsDNA System (Promega, WI, USA), and an equal DNA mass of 35 ng from each indexed product was combined to obtain a single pooled library. The pool was purified with bead-prep and sequenced on a MiSeq Desktop Sequencer (Illumina Inc, CA, USA) running in paired end 2 × 300 bp mode at the Center for Biotechnology and Genomics at Texas Tech University.
Soil physicochemical characteristics and climatic parameters
At each of the six study populations, three replicate soil cores representing the soil profile from 0 to 10 cm were collected concurrently with root collection. The individual replicates represented the area where majority of the P. cooperi plants were growing. The three replicates were combined to yield one aggregate samples from all study sites and sampling events. Physicochemical analyses included soil texture, organic matter (OM), pH, cation exchange capacity (CEC), nitrate (NO3), phosphorus (P), potassium (K), magnesium (Mg), sodium (Na), calcium (Ca), zinc (Zn), manganese (Mn), iron (Fe), copper (Cu), boron (B), and soluble salts.
Climatic variables were obtained from two large (PLF, SCE) and two small (PLE, SCW) populations. At PLF and PLE, a ECRN-100 logger (Decagon Devices, WA, USA) was used to record precipitation (mm; ECRN-100) while ibuttons (Maxim Integrated, CA, USA) were used to record air temperature (°C) and relative humidity (%) with a Hygrochron DS1923 and soil temperature (°C) was recorded with a Thermochron DS1921G-F5. All climatic data were recorded continuously at a 4-h frequency. We obtained similar data for the island populations SCE and SCW from Santa Catalina Island Climate stations closest to each population. We extracted the climate data collected between December and April to represent the growing season to include plant emergence, vegetative, and reproductive growth of P. cooperi.
Data analysis
Quality filtering and OTU delineation—Primers were first removed from demultiplexed sequences using Cutadapt version 1.10 (Martin 2011), and then paired end reads were joined with fastq-join using a minimum overlap of 30 bp (-m = 30) and 8% maximum difference in the overlapping region (-p = 8) (Aronesty 2013). Sequences were quality trimmed by setting sum maximum error probability to one (--fastq_maxee = 1) and maximum number of Ns in the sequence to zero (--fastq_maxns = 0) in VSEARCH version 2.4.1 (Rognes et al. 2016). Sequences shorter than 150 bp were discarded, and the remaining sequences were subjected to dereplication (--derep_fulllength) and sorting by length (--sortbylength). Next, chimeric sequences were removed (--uchime_ref) using UNITE fungal reference database (Koljalg et al. 2014) with VSEARCH. Operational taxonomic units (OTUs) were identified by de novo clustering of sequences using a 97% similarity threshold with UCLUST (Edgar 2010) in QIIME version 1.9 (Caporaso et al. 2010). Taxonomy was assigned with SINTAX, a non-Bayesian taxonomy classifier (Edgar 2016) by using UNITE reference database supplemented with sequences belonging to OMF families that were recovered from orchid and non-orchid hosts (popset# = 311306615, 1239395834, 337732522, 1248686462, 38606914, 1304517535, 406591741, 451329646, 1304517590). An OTU table was built in QIIME and subsequently filtered for singletons to remove OTUs represented by a single sequence.
Community analyses—A phyloseq object was first created with phyloseq package (McMurdie and Holmes 2013) in R version 3.5.2 (R Core Team 2019). Next, the decontam package (Davis et al. 2017) was used to filter OTUs based on their frequency and prevalence in negative controls (method = “combined”). We then used the iNEXT package (Hsieh et al. 2016) with 100 bootstraps and first order of hill number (q = 1) to generate rarefaction and extrapolation curves for species accumulation to determine whether fungal OTU diversity had been saturated in root and soil samples, collectively. For the community analyses, fungal communities were filtered to retain Tulasnellaceae and Ceratobasidiaceae families to represent OMF communities in root and soil dataset, and we further filtered OTUs from the two OMF families to represent OMF communities with OTUs that showed an overlap between root and soil datasets.
We estimated both alpha diversity and dissimilarity of root and soil OMF communities hosted by large and small populations. First, Hill diversity was calculated based on Shannon diversity index (H1) to estimate alpha diversity representing the effective number of species, or in this case OTUs, based on their richness and evenness in a community (Hill 1973; Jost 2006). The H1 estimates for large and small populations were compared with a non-parametric Kruskal-Wallis test for roots and soil separately. To compare the OMF communities between large and small populations, we first calculated Bray-Curtis dissimilarities from a Hellinger transformed OTU matrix and then applied permutational multivariate analysis of variance (PERMANOVA) to assess if group size (small/large population size) explained variation in community composition (Anderson 2001). Bray-Curtis dissimilarities of the OMF communities were also used for hierarchical clustering of large and small populations with the stats package in R, based on the OMF OTUs. The analyses described up to here were conducted with two datasets to examine the influence of an uneven n of individuals from large and small populations. The two datasets included (1) the complete dataset and (2) a similar n of individuals from large and small populations. The balanced dataset was generated after conducting 10 simulations of random subsampling of root-OMF communities from 55 individuals (which is equal to the number of individuals collected from small populations) associated with large populations. Subsequently, we used ANCOM (analyses of composition of microbiome) to identify the root and soil OMF OTUs that were differentially abundant between large and small populations by using the most stringent adjustment for multiple comparisons (multcorr = 1) (Mandal et al. 2015). Please note that we use the term “abundance” herein to refer to “relative abundance” to report the proportion of DNA sequences assigned to a particular OTU representing its relative frequency.
Network analyses—To determine the co-occurrence patterns of OMF OTUs in roots and soil within two large and two small populations (PLF, PLE, SCE and SCW) representing peninsular and island populations, we computed SparCC correlations for each of the four populations by using 500 permutations and 100 iterations at each step. The SparCC correlations were computed per sampling event; for instance, data from roots of a given population in February 2015 were combined and then correlated with the similarly combined data from soil sampled during the same month and year at the same population. To test if the correlation networks between root and soil OTU communities of large populations were different from small populations, we merged the correlation matrices obtained from root and soil OMF communities from the four populations in CompNet software (Kuntal et al. 2016) after retaining only strong co-occurrence and co-exclusion (absolute SparCC correlation > 0.6) relationships. Subsequently, CompNet detected “communities” based on the Newman’s spectral community finding algorithm (Newman 2006) from the merged correlation networks of the four populations and provided enrichment scores for these communities to show their exclusive or overlapping distributions across four populations.
Phylogenetic analyses—To examine the placement of Tulasnellaceae and Ceratobasidiaceae OMF OTUs recovered from P. cooperi roots among previously known OMF from the same fungal families in other orchid species, we generated maximum likelihood (ML) phylograms. First, 197 Tulasnellaceae sequences (45 represent P. cooperi OTUs and 152 GenBank reference sequences from other orchids) and 288 Ceratobasidiaceae sequences (67 P. cooperi associated OTUs and 221 GenBank reference sequences) were aligned separately with T-coffee (Di Tommaso et al. 2011). Midpoint rooted ML trees were then generated for each family with RAxML-HPC2 version 7.4.2 (Stamatakis 2006), and the best tree from 1000 random parsimonious trees was assigned clade support values based on 1000 bootstrap replicates. Phylograms were visualized using FigTree version 1.4.2 (Rambaut 2012).
Soil, microclimate, and affinities of root and soil OMF communities for microenvironment—We used the non-parametric Kruskal-Wallis test to detect differences in physical and chemical properties of soil in large and small populations. Variation in microclimate was assessed via a linear mixed-effect model (LMM) on weekly means of the explanatory variables measured between December and April of each of the years between 2014 and 2017. In the LMM, we used population size as the fixed effect with month and year serving as random effects. The influence of soil physicochemical and microclimatic predictors on root and soil OMF communities was tested by performing redundancy analysis (RDA) with the vegan package in R. To improve the fit of the RDA model, soil physicochemical variables were first subjected to forward selection (forward.sel) with adespatial package (Dray et al. 2016).
Results
Our Illumina MiSeq runs yielded > 33 million (33,326,134) sequences across root and soil samples from which 63% were successfully joined to 10,484,997 sequences. Of the joined reads, 10% and 5% were discarded during quality filtering and chimera filtering steps, respectively while 88% of the sequences were dereplicated. A total of 1,055,470 sequences were subsequently used for clustering that yielded 8,936 OTUs. We further removed 2,271 singletons, and 1,870 non-fungal and 308 contaminant OTUs, and thus, the remaining 4,487 OTUs represented the root and soil fungal communities associated with P. cooperi. Rarefaction and extrapolation curves revealed sufficient diversity saturation in both sources (Fig. S1).
Selection and representation of OMF communities
From 4487 fungal OTUs identified in roots and soil, 579 OTUs belonged to the fungal families known to host OMF, hereafter referred to as “OMF OTUs.” Of these 579 OMF OTUs, P. cooperi roots hosted 382 OTUs, whereby Ceratobasidiaceae (206 OTUs) and Tulasnellaceae (101 OTUs) represented 80% of the OMF OTUs and > 99% of OMF sequences (Fig. S2). Soil fungal community from all sites combined, on the other hand, was composed of 359 OTUs from fungal families known to host OMF. In the order of dominance, Thelephoraceae, Tulasnellaceae, Agaricaceae, Ceratobasidiaceae, and Tricholomataceae collectively represented > 75% of the OMF community in soil (Fig. S2). Of these, Ceratobasidiaceae was represented by 89 OTUs while Tulasnellaceae was represented by 54 OTUs. Given the strong dominance of Ceratobasidiaceae and Tulasnellaceae in root OMF communities, we retained these two families in roots and soil to examine the co-distributions of OMF OTUs that are preferred by the orchid.
When we examined the overlap of Ceratobasidiaceae and Tulasnellaceae OTUs recovered from roots and soil, 112 OTUs were common to both. Of these, 67 belonged to the Ceratobasidiaceae and the remaining 45 belonged to Tulasnellaceae. Altogether, the 112 OTUs accounted for 87% of all OMF sequences within roots (Table 1), while they accounted for 23% of the OMF communities in soil. To determine the effect of selection of 112 OTUs on OMF diversity, we calculated and compared the effective number of species by using 112 and 382 OTUs (total number of OTUs belonging to Tulasnellaceae and Ceratobasidiaceae). The effective number of species showed a high correlation (Pearson r2 = 0.80, P < 0.001) between the two datasets, thus we retained the 112 OTUs that were detected both in roots and in soil for all analyses presented below.
OMF communities in the context of host population size
Sampling effort to capture OMF diversity across both population size groups was sufficient (Fig. S3). And, considering that the results from both the complete and the subsampled datasets mirrored each other, we are retaining the results based on the complete dataset here. The α diversity of 112 OMF OTUs in roots was higher (H1 = 4.18; P < 0.001) in larger populations in comparison with small populations (H1 = 2.27). The OMF community structure was also dissimilar between large and small populations (r2 = 0.24, pseudo-F1/174 = 55.35, P = 0.001, Table 2). ANCOM detected 29 OTUs as differentially abundant between large and small populations (false discovery rate (FDR) < 0.05 for all). Nineteen of the 29 differentially abundant OTUs belonged to the Tulasnellaceae and were more abundant in larger populations, representing > 75% of the root OMF communities. These same Tulasnellaceae OTUs only accounted for 11% of the OMF communities in small populations. In contrast, the 10 differentially abundant Ceratobasidiaceae OTUs represented a smaller proportion (< 0.01%) of the root OMF community in larger populations while they represented 64% of the OMF communities in smaller populations.
When examining the community structure of the 112 OMF OTUs in soil, the α diversity in soil was also larger (H1 = 3.16) in larger populations than in smaller populations (H1 = 2.27; P = 0.006). We also detected dissimilarity in the structure of soil-associated 112 OMF OTUs between large and small populations (r2 = 0.08, pseudo-F1/113 = 9.75, P = 0.001; Table 2). ANCOM detected only 2 OTUs as differentially abundant between large and small populations (FDR < 0.05 for both), both belonging to Tulasnellaceae that were also differentially abundant in root OMF communities. Hierarchical clustering with the 112 shared OMF OTUs grouped both root and soil OMF communities of the two large populations (SCE and PLF) together (Fig. 1b).
Network analyses of root and soil OMF OTUs
We used the 112 OMF OTUs shared between roots and soil to create independent OTU networks for root and soil communities of each of the four populations (Fig. 2). After filtering edges with absolute SparCC correlation > 0.6 from each network, a total of 129 nodes and 2372 edges were used to compare the structure of root and soil OTU networks of four populations (Fig S4). Network diameter, density, and clustering coefficient were 5, 0.29, and 0.54, respectively. The most abundant OTU (OTU8712 belonging to Tulasnellaceae) in roots and soil showed highest measure of betweenness (819) and degree (106) centrality in the network (Fig. 2). We identified five communities (identified by union of networks in CompNet that uses the method outlined in “igraph” software (Csardi and Nepusz 2006)) from OTU networks of four populations of which communities 1, 3, 4, and 5 were highly enriched in roots and soil of two large populations (PLF and SCE) with subtle differences between the two (Fig. 3a). In contrast, two small populations (PLE and SCW) showed differential and pronounced enrichment of either community 2 or 5 in roots and soil (Fig. 3a). Large populations also showed a higher number of edges (SCE = 1496, PLF = 530) in comparison with small populations (PLE = 242, SCW = 394). Moreover, edge distribution showed higher overlap between two large populations in comparison with small populations (Fig. 3b). Based on the similarity of neighboring nodes, two large populations clustered more closely to each other in comparison with small populations (Fig. 3c). Overall, the OMF community networks were more similar between large populations in comparison with small populations.
Union of orchid mycorrhizal fungal (OMF) operational taxonomic unit (OTU) networks hosted by root and soil of four populations of Platanthera cooperi. A total of 112 OTUs belonging to Ceratobasidiaceae and Tulasnellaceae were used. The nodes from each population are color coded where multicolored nodes are shared among populations. The five communities within the networks are represented by the color of node labels. Large populations are marked with an asterisk (*)
Community analyses of orchid mycorrhizal fungal (OMF) operational taxonomic unit (OTU) networks hosted by the root and habitat soil of two large and two small populations of Platanthera cooperi. Total 112 OTUs belonging to Ceratobasidiaceae and Tulasnellaceae were included. a Enrichment scores of five OMF communities across the four populations. b Distribution of edges across the four populations to show exclusive and shared edges. c A dendrogram showing grouping of the four populations based on similarity of neighboring nodes. Large populations are marked with an asterisk (*)
Phylogenetic placement of the 112 OMF OTUs present in roots and soil
Of the 45 Tulasnellaceae OTUs, 44 grouped together in a single clade; within this single clade, a majority clustered exclusively while four OTUs were somewhat segregated and associated with sequences reported from temperate terrestrial orchid genera Cypripedium and Gymnadenia (Fig. S4). Tulasnellaceae OTUs derived from P. cooperi did not show close phylogenetic relationships with sequences derived from other Platanthera species except OTU 7449, which was nested within a clade representing Tulasnellaceae OTUs previously recovered from P. yadonii, P. chlorantha, and P. pollostantha. Almost all of the 67 Ceratobasidiaceae OTUs recovered from P. cooperi also segregated from the reference sequences in the ML tree. Within the separated clade, however, the Ceratobasidiaceae sequences exhibited a wider phylogenetic breadth by segregating in multiple sub-clades. Ceratobasidiaceae OTUs from P. cooperi associated with sequences reported from other terrestrial orchids including Spiranthes, Goodyera, Platanthera, Rhizanthella, Coelogyne, Epipactis, and Limodorum species (Fig. S5).
Affinities of the 112 OMF OTUs toward soil physicochemical analytes and microclimate
Soil: When soil profiles were compared across the six study populations, large and small populations showed differences in soil phosphorus (P) concentrations (large = 181 mg/l, small = 32 mg/l, P = 0.01; Table S1). Based on the four soil analytes (Zn, P, OM, and silt) that were identified by forward selection, the RDA explained variation (F = 2.16, P = 0.011) in OMF communities associated with roots and soil of P. cooperi (Fig. 4). Only RDA1 was identified as the significant axis (F = 4.7, P = 0.029) explaining 30% of the variation in root and soil OMF communities. RDA1 showed highest absolute biplot score for Zn (0.89) followed by P (− 0.59), while the species scores were highest for a Ceratobasidiaceae OTU [OTU7535 (0.65)] and a Tulasnellaceae OTU [OTU8712 (− 0.38)]. Accordingly, we observed higher relative abundance of OTU8712 and concentration of phosphorus (P) and lower relative abundance of OTU7535 and zinc (Zn) content in two large populations (SCE and PLF) that clustered together (Fig. 4).
Redundancy analyses (RDA) with 112 Ceratobasidiaceae and Tulasnellaceae operational taxonomic units (OTUs) identified within roots and habitat soil of Platanthera cooperi across six populations (PLE, PLF, SCE, SCW, CH, MX) with respect to the corresponding soil physicochemical predictors. Root and soil samples were pooled to combine samples collected across multiple years. Of the 18 variables that were measured over multiple years, phosphorus (P), zinc (Zn), organic matter (OM), and silt were identified as most relevant by forward selection. Large populations are marked with an asterisk (*)
Microclimate: Of the four measures of microclimate, soil temperature was higher in small populations (18 °C) when compared with large populations (17 °C; P < 0.001; Table S2). The RDA model composing of air temperature, soil temperature, relative humidity, and precipitation, however, failed to explain (F = 1.85, P = 0.07) the distinct structures of root and soil OMF communities in large and small populations.
Discussion
It has been posited earlier that orchid mycorrhizal communities and microenvironment could shape orchid population dynamics by affecting orchid seed germination, seedling emergence, and flowering (Hutchings 2010; McCormick et al. 2018). However, although previous studies have reported some relationships between orchid abundance and OMF communities at local scales (McCormick et al. 2016; Voyron et al. 2017), the effect of OMF communities on orchid population ecology at regional scales spanning the distribution range of species has not been studied extensively. Moreover, linkages of root OMF communities with their corresponding structure in the substrate or with microenvironment are rarely reported. Herein, we report one of the first few sets of evidence for linkages among population size, root and soil OMF communities, and microenvironment associated with Platanthera cooperi. We show that population size of this rare, terrestrial orchid is clearly linked to the composition of OMF communities in its roots and soil and that the fungal community structures are in turn shaped by soil chemistry.
In this study, the two most widely recognized OMF families, Tulasnellaceae and Ceratobasidiaceae, dominated the roots and soil communities of P. cooperi; however, their distinct distribution patterns were clearly linked to the population size. Specifically, the fungal family Tulasnellaceae was more abundant in roots and soil of large populations while Ceratobasidiaceae was dominant in smaller populations. Further, the structure and co-occurrences of OMF OTUs in roots and soil were more similar for the two large populations compared with small populations. Our findings support the observations by Jacquemyn et al. (2015) who reported concordance between root and soil OMF communities of a rare orchid Neottia ovata, but in contrast to our results, mycorrhizal communities of N. ovata were independent of the size of its populations. It should be noted, though, that N. ovata displays a more generalist approach in its selection of OMF compared with P. cooperi, which might explain its lack of OMF specificity in the context of its population size. Our results also support the previously reported local scale role of irregularly distributed OMF in soil in giving rise to patchy distributions of orchids (McCormick et al. 2016; Oja et al. 2017). With the increasing evidence for the latter pattern, McCormick et al. (2018) suggested a potential positive effect of OMF abundances on population size of orchid species. Our results clearly support the hypothesized link, while also considering the factors that might govern OMF abundances in soil.
Higher relative abundances of Tulasnellaceae associated with large populations of P. cooperi suggest their ecological advantage over Ceratobasidiaceae. Provided that OMF partners differ in their efficiencies in inducing seed germination (Sharma et al. 2003; Otero et al. 2005; Swarts et al. 2010) and accessing nutrients (Nurfadilah et al. 2013), the differences in soil fungal profiles, combined with preferential selection by the orchid, are likely to influence population size by regulating germination, vegetative, and/or reproductive success of orchid individuals (McCormick et al. 2018)(McCormick et al. 2018). In fact, we also observed that regardless of their size, populations that hosted flowering individuals also hosted higher relative abundances of Tulasnellaceae in roots and soil, suggesting a role of mycorrhizal community structure in reproductive fitness of individuals (Fig. S6). Although experimental evidence for phenology related fungal selection or succession is currently pending for P. cooperi, Bidartondo and Read (2008) have previously reported in situ germination of Epipactis and Cephalanthera seeds with a wider range of OMF while higher specificity was implicated in the transition of a protocorm to seedling stage. To date, studies of orchid-OMF interactions have heavily, and understandably, emphasized the role of fungi in inducing seed germination (McCormick et al. 2018) while fungal regulation, via specificity or abundance, of phenological progression and reproductive fitness is poorly addressed. Simultaneously, orchid populations might persist weakly at sites where the preferred fungal OTUs are scarce by associating with secondary OMF partners. This might also be a result of low OMF diversity and higher heterogeneity in OMF community composition among small populations in comparison with the large ones. The specificity of P. cooperi toward Tulasnellaceae in the context of population size in our study parallels the patterns observed in another terrestrial orchid, Isotria medeoloides, where higher abundances of preferred OMF in soil increased the post-dormancy emergence of plants (Rock-Blake et al. 2017).
The structure of OMF communities could be influenced by the chemical and physical characteristics of a substrate (Bunch et al. 2013). For instance, McCormick et al. (2012) reported higher abundances of OMF taxa in soil amended with organic materials in comparison with un-amended soil. Furthermore, both substrate type and environmental conditions are implicated in influencing OMF selection by orchid roots (McCormick et al. 2006), whereby Goodyera pubescens switched its OMF partner during drought conditions although the study did not test whether the switch occurred due to any simultaneous changes in soil OMF abundances. Ideally, establishing empirical or observational links between abiotic environment, OMF community structure in substrates, and orchid response would help detect independent and codependent mechanisms that alter mycorrhizal behavior of orchids. In our study, Zn, P, OM, and silt explained the dissimilar community structures of OMF in roots and soil across variably sized populations of P. cooperi. OMF communities in larger populations were operating under increased phosphorus in soil, whereas the OMF communities in small populations were exposed to high organic matter, zinc, and silt in its soil. In contrast, a study by Mujica et al. (2016) from central Chile observed a higher abundance of Ceratobasidiaceae (as opposed to Tulasnellaceae) in roots of Bipinnula fimbriata at locations with high phosphorus.
While the coupling of OMF communities and soil physical and chemical environment associated with P. cooperi was evident, we did not observe an influence of microclimate on OMF communities. Although larger populations experienced lower soil temperature in comparison with the smaller populations, an undetected link between microclimate and OMF communities might surface if sample size could be increased. Provided that cooler air temperature can improve population fitness in orchids (Hutchings 2010; Shefferson et al. 2018), cooler soil temperature may also either directly increase plant fitness in terrestrial orchids, or may improve access to nutrients by increasing OMF abundances in soil. Lower soil temperature could also increase soil moisture, which is known to increase OMF abundances in soil and orchid seed germination (Diez 2007; Jacquemyn et al. 2015; Waud et al. 2017).
To conclude, we show that the demography of a rare temperate terrestrial orchid is linked to a combination of its biotic and abiotic niche preferences. Together, cooler soil, higher phosphorus, and lower zinc appear to facilitate higher abundances of Tulasnellaceae OTUs that are preferred by P. cooperi across its phenology (seedlings, vegetative, and reproductive plants). On the other hand, sites that are dominated by the Ceratobasidiaceae under an environment that is warmer and high in zinc and organic matter may only support smaller populations of the orchid with noticeably fewer reproductive individuals. Our study adds support for the hypothesis that fitness of orchid species is linked to abundances of preferred OMF in the substrate and that the spatial variation in OMF abundances in soil is likely governed by the edaphic and microclimatic characteristics of the soil. To the best of our knowledge, this is only a second study after Jacquemyn et al. (2015) that links orchid demography with both biotic (fungal community structure) and abiotic (soil physical, chemical, and climatic environment) predictors simultaneously. Surely, additional studies are warranted to understand the effect of soil environment and fungal communities on orchid demographies, especially considering the wide diversity of orchid lifestyles ranging from subterranean achlorophyllous species relying completely on their fungal partners to epiphytic mixotrophs residing in high canopies of tropical trees, distribution patterns ranging from wide to extremely narrow, and fungal preferences ranging from wide to extremely specific for fungal taxa that are rare in substrates. The direct effects of soil physicochemistry and microclimate on orchid populations also warrant attention to determine the relative contributions of biotic and abiotic environments in orchid niches. It is possible that while distributions of orchids with narrow and specific fungal niches depend on fungal distributions, which may or may not be governed by microclimate, demographies of widely distributed orchids with wider OMF preferences may be more directly regulated by microclimate instead.
Data accessibility
The raw sequences from root and soil samples were deposited in the NCBI SRA database under BioProject PRJNA609930.
References
Ackerman JD, Lauri RK (2018) Piperia cooperi, in Jepson Flora Project (eds.) Jepson eFlora
Alvarado P, Teixeira MDM, Andrews L, Fernandez A, Santander G, Doyle A, Perez M, Yegres F, Barker BM (2018) Detection of Coccidioides posadasii from xerophytic environments in Venezuela reveals risk of naturally acquired coccidioidomycosis infections. Emerg Microbes Infect 7:1–13.
Anderson MJ (2001) A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46. https://doi.org/10.1046/j.1442-9993.2001.01070.x
Aronesty E (2013) Comparison of sequencing utility programs. Open Bioinforma J 7:1–8. https://doi.org/10.2174/1875036201307010001
Bateman RM, Hollingsworth PM, Preston J, Yi-Bo L, Pridgeon AM, Chase MW (2003) Molecular phylogenetics and evolution of Orchidinae and selected Habenariinae (Orchidaceae). Bot J Linn Soc 142:1–40. https://doi.org/10.1046/j.1095-8339.2003.00157.x
Berry D, Ben Mahfoudh K, Wagner M, Loy A, Ben MK (2011) Barcoded primers used in multiplex amplicon pyrosequencing bias amplification. Appl Environ Microbiol 77:612–612. https://doi.org/10.1128/AEM.05220-11
Bidartondo MI, Read DJ (2008) Fungal specificity bottlenecks during orchid germination and development. Mol Ecol 17:3707–3716. https://doi.org/10.1111/j.1365-294X.2008.03848.x
Bunch WD, Cowden CC, Wurzburger N, Shefferson RP (2013) Geography and soil chemistry drive the distribution of fungal associations in lady’s slipper orchid, Cypripedium acaule. Botany 91:850–856. https://doi.org/10.1139/cjb-2013-0079
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pẽa AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335. https://doi.org/10.1038/nmeth.f.303
Coates F, Lunt ID, Tremblay RL (2006) Effects of disturbance on population dynamics of the threatened orchid Prasophyllum correctum D.L. Jones and implications for grassland management in south-eastern Australia. Biol Conserv 129:59–69. https://doi.org/10.1016/j.biocon.2005.06.037
Csardi G, Nepusz T (2006) The igraph software package for complex network research. InterJournal Complex Syst 1695:1–9
Davis NM, Proctor D, Holmes SP, Relman DA, Callahan BJ (2017) Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6:226. https://doi.org/10.1101/221499
Dearnaley JDW, Martos F, Selosse MA (2012) Orchid mycorrhizas: molecular ecology, physiology, evolution and conservation aspects. In: Fungal Associations, 2nd Edition
Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang JM, Taly JF, Notredame C (2011) T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res 39(suppl_2:W13–W17. https://doi.org/10.1093/nar/gkr245
Diez JM (2007) Hierarchical patterns of symbiotic orchid germination linked to adult proximity and environmental gradients. J Ecol 95:159–170. https://doi.org/10.1111/j.1365-2745.2006.01194.x
Dray S, Blanchet G, Borcard D, Guenard G, Jombart T, Larocque G, Legendre P, Madi N, Wagne H (2016) Adespatial: multivariate multiscale spatial analysis. R Packag version 00–9
Dressler RL (1993) Phylogeny and classification of the orchid family. Cambridge University Press
Edgar R (2016) SINTAX: a simple non-Bayesian taxonomy classifier for 16S and ITS sequences. bioRxiv 074161. https://doi.org/10.1101/074161
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Fiedler PL, Keever ME, Grewell BJ, Partridge DJ (2007) Rare plants in the Golden Gate Estuary (California): the relationship between scale and understanding. Aust J Bot 55:206–220. https://doi.org/10.1071/BT06069
Girlanda M, Segreto R, Cafasso D, Liebel HT, Rodda M, Ercole E, Cozzolino S, Gebauer G, Perotto S (2011) Photosynthetic Mediterranean meadow orchids feature partial mycoheterotrophy and specific mycorrhizal associations1. Am J Bot 98:1148–1163. https://doi.org/10.3732/ajb.1000486
Hill MO (1973) Diversity and evenness: a unifying notation and its consequences. Ecology 54:427–432. https://doi.org/10.2307/1934352
Hsieh TC, Ma KH, Chao A (2016) iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol Evol 7:1451–1456. https://doi.org/10.1111/2041-210X.12613
Hutchings MJ (2010) The population biology of the early spider orchid Ophrys sphegodes Mill. III. Demography over three decades J Ecol 98:867–878. https://doi.org/10.1111/j.1365-2745.2010.01661.x
Jacquemyn H, Brys R, Jongejans E (2010) Size-dependent flowering and costs of reproduction affect population dynamics in a tuberous perennial woodland orchid. J Ecol 98:1204–1215. https://doi.org/10.1111/j.1365-2745.2010.01697.x
Jacquemyn H, Waud M, Merckx VSFT, Lievens B, Brys R (2015) Mycorrhizal diversity, seed germination and long-term changes in population size across nine populations of the terrestrial orchid Neottia ovata. Mol Ecol 24:3269–3280. https://doi.org/10.1111/mec.13236
Jost L (2006) Entropy and diversity. Oikos 113:363–375. https://doi.org/10.1111/j.2006.0030-1299.14714.x
Kaur J, Andrews L, Sharma J (2019) High specificity of a rare terrestrial orchid toward a rare fungus within the North American tallgrass prairie. Fungal Biol 123:895–904. https://doi.org/10.1016/j.funbio.2019.09.010
Kaur J, Poff KE, Sharma J (2017) A rare temperate terrestrial orchid selects similar Tulasnella taxa in ex situ and in situ environments. Plant Ecol 219:1–11. https://doi.org/10.1007/s11258-017-0776-0
Koljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M (2014) Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22:5271–5277. https://doi.org/10.1111/mec.12481
Kuntal BK, Dutta A, Mande SS (2016) CompNet: A GUI based tool for comparison of multiple biological interaction networks. BMC Bioinformatics 17:185. https://doi.org/10.1186/s12859-016-1013-x
Leitao RP, Zuanon J, Williams SE, Baraloto C, Fortunel C, Mendonc FP, Mouillot D (2016) Rare species contribute disproportionately to the functional structure of species assemblages. Proc R Soc B 283:20160084
Mandal S, Van Treuren W, White RA, Eggesbø M, Knight R, Peddada SD (2015) Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis 26:27663. https://doi.org/10.3402/mehd.v26.27663
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17:10–12
Mattila E, Kuitunen MT (2000) Nutrient versus pollination limitation in Platanthera bifolia and Dactylorhiza incarnata (Orchidaceae). Oikos 89:360–366. https://doi.org/10.1034/j.1600-0706.2000.890217.x
McCormick MK, Jacquemyn H (2014) What constrains the distribution of orchid populations? New Phytol 202:392–400. https://doi.org/10.1111/nph.12639
McCormick MK, Lee Taylor D, Juhaszova K, Burnett RK, Whigham DF, O’Neill JP (2012) Limitations on orchid recruitment: not a simple picture. Mol Ecol 21:1511–1523. https://doi.org/10.1111/j.1365-294X.2012.05468.x
McCormick MK, Taylor DL, Whigham DF, Burnett RK (2016) Germination patterns in three terrestrial orchids relate to abundance of mycorrhizal fungi. J Ecol 104:744–754. https://doi.org/10.1111/1365-2745.12556
McCormick MK, Whigham DF, Canchani-Viruet A (2018) Mycorrhizal fungi affect orchid distribution and population dynamics. New Phytol 219:1207–1215. https://doi.org/10.1111/nph.15223
Mccormick MK, Whigham DF, O’Neill JP, Becker JJ, Sarah W, Rasmussen HN, Bruns And TD, Taylor DL (2009) Abundance and distribution of Corallorhiza odontorhiza reflect variations in climate and ectomycorrhizae. Ecol Monogr. https://doi.org/10.1890/08-0729.1
McCormick MK, Whigham DF, Sloan D, O’Malley K, Hodkinson B (2006) Orchid-fungus fidelity: a marriage meant to last? Ecology 87:903–911
McMurdie PJ, Holmes S (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. https://doi.org/10.1371/journal.pone.0061217
Mujica MI, Saez N, Cisternas M, Manzano M, Armesto JJ, Pérez F (2016) Relationship between soil nutrients and mycorrhizal associations of two Bipinnula species (Orchidaceae) from central Chile. Ann Bot 118:149–158. https://doi.org/10.1093/aob/mcw082
Newman MEJ (2006) Modularity and community structure in networks. Proc Natl Acad Sci 103:8577–8582. https://doi.org/10.1073/pnas.0601602103
Nurfadilah S, Swarts ND, Dixon KW, Lambers H, Merritt DJ (2013) Variation in nutrient-acquisition patterns by mycorrhizal fungi of rare and common orchids explains diversification in a global biodiversity hotspot. Ann Bot 111:1233–1241. https://doi.org/10.1093/aob/mct064
Oja J, Vahtra J, Bahram M, Kohout P, Kull T, Rannap R, Kõljalg U, Tedersoo L (2017) Local-scale spatial structure and community composition of orchid mycorrhizal fungi in semi-natural grasslands. Mycorrhiza 27:355–367. https://doi.org/10.1007/s00572-016-0755-7
Otero JT, Bayman P, Ackerman JD (2005) Variation in mycorrhizal performance in the epiphytic orchid Tolumnia variegata in vitro: the potential for natural selection. Evol Ecol 19:29–43. https://doi.org/10.1007/s10682-004-5441-0
Pandey M, Sharma J, Taylor DL, Yadon VL (2013) A narrowly endemic photosynthetic orchid is non-specific in its mycorrhizal associations. Mol Ecol 22:2341–2354. https://doi.org/10.1111/mec.12249
R Core Team (2019) R: a language and environment for statistical computing. R Found Stat Comput Vienna, Austria URL https://wwwR-project.org/
Rambaut A (2012) FigTree v1.4. https://tree.bio.ed.ac.uk/software/figtree/
Rock-Blake R, McCormick MK, Brooks HEA, Jones CS, Whigham DF (2017) Symbiont abundance can affect host plant population dynamics. Am J Bot 104:72–82. https://doi.org/10.3732/ajb.1600334
Rognes T, Flouri T, Nichols B, Quince C, Mahé F (2016) VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. https://doi.org/10.7717/peerj.2584
Rohland N, Reich D (2012) Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res 22:939–946. https://doi.org/10.1101/gr.128124.111
Sharma J, Zettler L, Sambeek J (2003) A survey of mycobionts of federally threatened Platanthera praeclara (Orchidaceae). Symbiosis 34:145–155
Shefferson RP, Kull T, Hutchings MJ, Selosse MA, Jacquemyn H, Kellett KM, Menges ES, Primack RB, Tuomi J, Alahuhta K, Hurskainen S, Alexander HM, Anderson DS, Brys R, Brzosko E, Dostálik S, Gregg K, Ipser Z, Jäkäläniemi A, Jersáková J, Dean Kettle W, Mccormick MK, Mendoza A, Miller MT, Moen A, Øien DI, Püttsepp Ü, Roy M, Sather N, Sletvold N, Štípková Z, Tali K, Warren RJ, Whigham DF (2018) Drivers of vegetative dormancy across herbaceous perennial plant species. Ecol Lett 21:724–733. https://doi.org/10.1111/ele.12940
Shefferson RP, Warren RJ, Pulliam RH (2014) Life-history costs make perfect sprouting maladaptive in two herbaceous perennials. J Ecol 102:1318–1328. https://doi.org/10.1111/1365-2745.12281
Stamatakis A (2006) RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. https://doi.org/10.1093/bioinformatics/btl446
Stanton-Geddes J, Tiffin P, Shaw RG (2012) Role of climate and competitors in limiting fitness across range edges of an annual plant. Ecology 93:1604–1613
Swarts ND, Dixon KW (2009a) Perspectives on orchid conservation in botanic gardens. Trends Plant Sci 14:590–598. https://doi.org/10.1016/j.tplants.2009.07.008
Swarts ND, Dixon KW (2009b) Terrestrial orchid conservation in the age of extinction. Ann Bot 104:543–556. https://doi.org/10.1093/aob/mcp025
Swarts ND, Sinclair EA, Francis A, Dixon KW (2010) Ecological specialization in mycorrhizal symbiosis leads to rarity in an endangered orchid. Mol Ecol 19:3226–3242. https://doi.org/10.1111/j.1365-294X.2010.04736.x
Taylor DL, Walters WA, Lennon NJ, Bochicchio J, Krohn A, Caporaso JG, Pennanen T (2016) Accurate estimation of fungal diversity and abundance through improved lineage-specific primers optimized for Illumina amplicon sequencing. Appl Environ Microbiol 82:7217–7226. https://doi.org/10.1128/AEM.02576-16
Voyron S, Ercole E, Ghignone S, Perotto S, Girlanda M (2017) Fine-scale spatial distribution of orchid mycorrhizal fungi in the soil of host-rich grasslands. New Phytol 213:1428–1439. https://doi.org/10.1111/nph.14286
Wamelink GWW, Goedhart PW, Frissel JY (2014) Why some plant species are rare. PLoS One 9. https://doi.org/10.1371/journal.pone.0102674
Waud M, Brys R, Van Landuyt W, Lievens B, Jacquemyn H (2017) Mycorrhizal specificity does not limit the distribution of an endangered orchid species. Mol Ecol 26:1687–1701. https://doi.org/10.1111/mec.14014
Waud M, Busschaert P, Ruyters S, Jacquemyn H, Lievens B (2014) Impact of primer choice on characterization of orchid mycorrhizal communities using 454 pyrosequencing. Mol Ecol Resour 14:679–699. https://doi.org/10.1111/1755-0998.12229
Acknowledgments
We specifically thank Lisa Markovchick, Andrew Wastell, Michelle Maley, Chris Gillespie, and Jose Roldan for their assistance with project management at NBPL. We also thank San Diego County Orchid Society and California Native Plant Society for providing additional research funding, and Jardin Botanico San Quintin (Josue Campos), Santa Catalina Island Conservancy, and Cleveland National Forest for providing research permits and for assisting in the field.
Funding
We received support from the Naval Base Point Loma (NBPL; Office of Naval Research, U.S. Department of Defense; Cooperative Agreement Nos. N62473-13-2-4907 and N62473-15-2-0011).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kaur, J., Phillips, C. & Sharma, J. Host population size is linked to orchid mycorrhizal fungal communities in roots and soil, which are shaped by microenvironment. Mycorrhiza 31, 17–30 (2021). https://doi.org/10.1007/s00572-020-00993-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-020-00993-5