Abstract
Communities of arbuscular mycorrhizal (AM) fungi in Mongolian grassland were characterized under gradients of grazing intensity at three study sites of different aridity: mountain forest steppe at Hustai National Park (Hustai), and desert steppe at Mandalgovi and Bulgan. Grazing intensity was classified into three categories: lightly grazed (LG), moderately grazed (MG), and heavily grazed (HG). With regard to floristic composition, grazing decreased the shoot biomass of Poaceae species, especially Stipa spp. Distinctness of the AM fungal communities was observed among the three study sites, but most of the AM fungal operational taxonomic units (OTUs) that comprised over 1.0% of the total reads were ubiquitous. This result indicates that the AM fungal communities may be derived from similar AM fungal floras in correspondence with environmental factors. The composition of AM fungal communities differed significantly among the grazing intensities at all study sites. The relative abundance of the most dominant AM fungal OTU of the LG plots decreased with an increase in grazing intensity at all study sites. The mean proportions of the most dominant AM fungal OTUs also decreased with increased grazing intensity at Hustai. Dominance by a single AM fungal taxon may be a typical ecological feature of the AM fungal symbiosis, and grazing disturbs AM fungal community structure.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Arbuscular mycorrhizal (AM) fungi are considered to be non-specific root colonizers of AM fungal host plants, but some preferences are often observed in the combinations between plants and AM fungi (Smith and Read 2008). Accordingly, as demonstrated by van der Heijden et al. (1998), AM fungal composition may be a determinant of diversity, composition, and productivity of the host plants. The plant growth response is different depending on the AM fungal species colonizing the roots (Klironomos 2003; Pringle and Bever 2008), and a positive correlation between AM fungal species richness and plant productivity has been experimentally demonstrated (Maherali and Klironomos 2007). Furthermore, AM fungal sporulation, which may be an index of AM fungal biomass, has been observed to differ among host plant species in a laboratory microcosm (Bever et al. 1996). These results indicate that plant and AM fungal communities interact, and that AM symbioses can affect the composition of plant communities.
Mongolia, located in East Asia, comprises a vast land area of about 156 million km2. Approximately 70% of the land area is grassland comprising mountain forest steppe, steppe, and desert steppe (Ulziikhutag 1989), which are located around the northeast, central, and southern parts of Mongolia with annual precipitation of about 300–400 mm, 125–250 mm, and 100–130 mm, respectively (Jigjidsuren and Johnson 2003). These areas have been traditionally used as rangeland for livestock, mostly sheep, and the number of livestock has increased after democratization of Mongolia in 1992. Consequently, the vegetation has been seriously degraded as a result of overgrazing (Sasaki et al. 2008).
The majority of plant species in the grassland vegetation, belonging to families such as Poaceae, Fabaceae, Asteraceae, Convolvulaceae, and Amaryllidaceae, are known to have symbioses with AM fungi (Wang and Qiu 2006). Given that AM fungi provide the host plant with a variety of benefits, the symbioses are likely to be indispensable for maintenance of the vegetation, especially under the low-fertility soil conditions (Goomaral et al. 2013a). Nevertheless, it is anticipated that AM fungi also may be affected by vegetation degradation caused by overgrazing, because the fungi are completely dependent on their host plant for a source of carbon (C). Previous field studies in arid ecosystems have reported decreases in AM fungal colonization rate in response to grazing (Bethlenfalvay and Dakessian 1984; Bethlenfalvay et al. 1985; Bai et al. 2013). In contrast, several experimental or field studies have shown that grazing has a limited effect on AM fungal colonization (Allen et al. 1989; Eom et al. 2001; Lugo et al. 2003; Medina-Roldán et al. 2008; Tian et al. 2009). It is likely, therefore, that a variety of factors, such as grazing intensity, plant species, environmental conditions, or a combination of these factors can affect AM fungal colonization.
Diversity of AM fungi also can be affected by grazing in steppe vegetation. Tian et al. (2009) reported that degradation of grassland in Inner Mongolia, China did not induce a significant decline in the colonization rate of AM fungi, but induced distinct lowering of AM fungal spore numbers and species diversity. Ba et al. (2012) observed that AM fungal diversity is higher under light to moderate grazing pressure and declined under intense grazing pressure in a meadow steppe in northeast China. In these studies, the composition of the AM fungal community was investigated based on the number of spores collected in soil samples. However, spore numbers do not usually reflect AM fungal species abundance in roots because of different sporulation abilities among AM fungal species. Instead, characterization of AM fungal communities using molecular techniques for fungal identification can provide a relatively reliable representation of the community in roots (Chagnon and Bainard 2014).
Several recent studies have used molecular methods to investigate the AM fungal communities associated with a number of plant species in Mongolian grassland. Goomaral et al. (2013a) observed that a group of AM fungi belonging to the genus Rhizophagus was dominant in roots of the Poaceae species Stipa krylovii, which is the grass species most preferred as forage by livestock in forest steppe in the Hustai National Park and steppe in Uvurkhangai. However, no studies have previously attempted to determine the effect of plant community degradation on the AM fungal community as a whole in a Mongolian grassland ecosystem.
The objective of the present study was to elucidate the effects of vegetation degradation caused by grazing on the community structure of AM fungi in two types of Mongolian grassland, i.e., mountain forest steppe and desert steppe. Given the correlations between productivity and/or diversity of host plants and AM fungal species richness observed in previous studies (van der Heijden et al. 1998; Maherali and Klironomos 2007; Hiiesalu et al. 2014), we hypothesized that grazing in the Mongolian steppe may induce a decrease in AM fungal diversity. Soil chemical properties, such as soil pH and available phosphorus (P), also may be determinants of AM fungal community structure (An et al. 2008; Yoshimura et al. 2013). Therefore, we analyzed the correlation of soil chemical properties, namely soil pH, available P, soil total C, and soil total nitrogen (N), as well as plant community factors, such as plant biomass, plant diversity, and plant species composition, with AM fungal community structure. Dumbrell et al. (2010) observed that AM fungal communities are typically dominated by a single taxon, representing on average 40% of total abundance within the community, in which the dominant AM fungal type differs among communities and is not a widespread generalist AM fungus. In the present study, we evaluated the proportion of dominant AM fungi at the study sites in relation to the effect of grazing intensity on the grassland ecosystem.
Materials and methods
Study sites and sampling
Two types of Mongolian grasslands, namely mountain forest steppe in Hustai National Park (Hustai) and desert steppe in Mandalgovi and Bulgan (Undarmaa et al. 2018), were selected for inclusion in this study (Fig. 1, Table S1). The mean temperature in January and July, and mean annual precipitation at Hustai National Park are − 20.8 °C, 24.5 °C, and 207.7 mm, respectively. The corresponding variables recorded at the Mandalgovi meteorological station in Mandalgovi are − 18.2 °C, 20.1 °C, and 128.4 mm, and those recorded at Saikhan meteorological station in Bulgan are − 13.8 °C, 22.7 °C, and 146.9 mm, respectively (averages for 1995–2016; Mongolia National Agency for Meteorology and Environmental Monitoring, Mongolia).
Sampling at Hustai, Mandalgovi, and Bulgan was conducted in early August in 2014, 2015, and 2016, respectively. At each study site, the sampling area was arbitrarily selected to represent different grazing intensities provisionally classified into three categories: lightly grazed (LG), moderately grazed (MG), and heavily grazed (HG), based on visual inspection of the floristic composition. The categorization reflected the approximate relative coverage ratio of Poaceae species (> 50% LG, 20–40% MG, and < 15% HG) based on Sasaki et al. (2005). Hustai is a national park; therefore, the sampling sites were located around the buffer zone of the protected area. Accordingly, the grazing intensity of the sampling sites was considered to be relatively consistent. The sampling sites at Mandalgovi and Bulgan were located in rangeland and the grazing intensity can be changeable because of variable animal activity.
For each grazing intensity, three sampling sites, e.g., designated HL1, HL2, and HL3 for the LG site at Hustai, were arbitrarily selected to represent the typical vegetation of the grazing intensity in the region (Fig. 1). The sampling areas for each grazing category neighbored those for another grazing category, e.g., LG and MG, or MG and HG, and the area differed in size depending on the grazing of each study site. Accordingly, the scale of sampling site separation also differed among the study sites. At each sampling site, three sampling plots (1 m × 1 m) were established with each plot separated by about 10 m, and were designated, for example, HL1–1, HL1–2, and HL1–3 for the HL1 sampling site. In each plot, all shoots were collected for each plant species, and three soil cores containing plant roots (5 cm diameter and 10 cm depth) were arbitrarily collected. The three soil cores were pooled to form one composite sample per plot. The plant roots were separated from the soil by sieving and washed with tap water. For the root sample, fine roots (unlignified and less than 2 mm in diameter) were collected for molecular analysis of AM fungi.
Soil chemical analysis
Soil samples dried at room temperature for 1 week were used for analyses of soil chemical properties. Soil pH (H2O) at the soil/water ratio of 1:2.5 (v/v) and available P (Truog-P) content were measured. Total N and total C contents were analyzed using a CN analyzer (Macro Corder JM1000CN, J-Science Lab Co., Ltd., Kyoto, Japan).
Molecular analysis
Fine roots (about 300 mg fresh weight) were randomly selected from each root sample and stored in 99.5% ethanol until processed. After crushing the roots in liquid nitrogen, total DNA was extracted using the DNeasy® Plant Mini Kit (Qiagen, Tokyo, Japan) following the manufacturer’s instructions. DNA extracts were quantified using the QuantiFluor® dsDNA System with the Quantus™ Fluorometer (Promega, Tokyo, Japan). The DNA extracts were diluted with TE buffer to 1 ng μl−1 for use as DNA templates for PCR amplification.
Partial AM fungal gene sequences for the small subunit of the nuclear ribosomal RNA gene (SSU rDNA) were amplified from the diluted DNA by PCR using Takara Ex Taq™ Hot Start Version (Takara Bio, Otsu, Japan) with the primers AMV4.5NF and AMDGR (Sato et al. 2005). Sequences of Primer A-key and Primer P1-key for sequencing in the Ion Torrent System (Life Technologies, Carlsbad, CA, USA) were linked to the 5′ end of AMV4.5NF and AMDGR, respectively. To identify sequences from different samples, a set of sample-specific 8-bp tag sequences (Hamady et al. 2008) and a common 3-bp key sequence were inserted between Primer A-key and AMV4.5NF. Thus, the forward primer was 5′-Primer A-key +8-bp tag+3-bp key+AMV4.5NF-3′ and the reverse primer was 5′-Primer P1-key +AMDGR-3′. The PCR reaction mixture contained 1.0 μl diluted DNA solution, 0.5 U Taq DNA polymerase, 0.25 μM each primer, 0.2 mM each dNTP, and 2 μl supplied PCR buffer, in a total volume of 20 μl. The PCR amplification was performed on a Thermal Cycler Gene Atlas (ASTEC, Fukuoka, Japan) using the following program: initial denaturation at 94 °C for 2 min, followed by 35 cycles at 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min, then final elongation at 72 °C for 10 min. The PCR products were purified using the HiYield™ Gel/PCR DNA Fragments Extraction Kit (RBC Bioscience, Taipei, Taiwan). Purified PCR products were pooled in equimolar quantities to prepare templates for sequencing.
The size and quality of library DNA were checked using the Agilent High Sensitivity DNA Kit and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). With 25 μl of the 16 pM library diluted, an emulsion PCR was conducted with the Ion OneTouch™ 2 system (composed of the Ion OneTouch™ 2 Instrument and Ion OneTouch™ ES) and Ion PGM™ Hi-Q™ OT2 400 Kit (Life Technologies). Sequencing was conducted using an Ion Torrent™ Personal Genome Machine™ (Life Technologies) with the Ion PGM™ Hi-Q™ Sequencing Kit and the Ion 318™ Chip Kit (Life Technologies). The sequence data were deposited in the DDBJ sequence archive (DRA) under accession number DRA006370.
Bioinformatics
Sequence data were generated in a FastQ format for each barcode sample. First, the primer sequence was eliminated using FASTX (http://hannonlab.cshl.edu/fastx_toolkit/index.html). The subsequent sequence analysis was performed with the Quantitative Insight into Microbial Ecology (QIIME) 1.8.0 pipeline (Caporaso et al. 2010). The sequences shorter than 200 bp and the sequences with expected errors predicted by Phred (Q) scores greater than 0.5 were removed. Quality sequences were then clustered into operational taxonomic units (OTUs) with USERCH8.0.1517 (Edgar 2010) at the 97% similarity threshold. For the representative sequences of OTUs, a chimera check was performed using the UCHIME algorithm (Edgar et al. 2011).
For the fungal partial sequence of SSU rDNA, each OTU was classified into a taxonomic group with the MaarjAM database (Öpik et al. 2010) formatted for QIIME (https://github.com/alk224/QIIME_databases/blob/master/MAARJAM.tar.gz). For the representative sequence of each OTU, BLAST searches against MaarjAM were performed to confirm the affiliations with the virtual taxon (VTX). In addition, 20 identified AM fungal sequences within the phylogenetic range of the obtained sequences were downloaded from the GenBank database. Multiple sequence alignment of the representative and downloaded sequences, and neighbor-joining analysis (Saitou and Nei 1987) with a maximum composite likelihood model were carried out using MEGA version 7 (Kumar et al. 2016). A bootstrap analysis with 1000 replications (Felsenstein 1985) was conducted to assess statistical support for the tree topology. The phylogenetic tree obtained was constructed using FigTree version 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Statistical analysis
For the AM fungal sequence data, the sequences were rarefied to 1000 reads per sample using the function “rrarefy” in the vegan package for R software (Oksanen et al. 2017) in order to avoid bias caused by differences among the number of sequences per sample. For the rarefied data set of the AM fungal OTU composition, rarefaction curves were plotted for each sample to confirm the sampling efficacy using the function “rarecurve” in the vegan package.
Dissimilarities in AM fungal OTU composition among the sampling plots were calculated for the rarefied data set using the function “vegdist” in the vegan package for the Bray–Curtis index. Permutational multivariate analysis of variance (PERMANOVA; 10,000 permutations) was subsequently performed using the function “adonis” in the vegan package to analyze the distinction among the study sites, Hustai, Mandalgovi, and Bulgan.
To investigate the effect of grazing on diversities of plants and AM fungi at each study site (Hustai, Mandalgovi, and Bulgan), the respective Shannon diversity indices were calculated, based on the shoot biomass of each plant species for plants and the number of the rarefied AM fungal sequences belonging to each OTU for AM fungi, using the function “diversity” in the vegan package. The Shannon diversity index was also calculated for AM fungal host plants (species of Brassicaceae, Cyperaceae, Amaranthaceae, and Caryophyllaceae were excluded). The means of total shoot dry weight, shoot dry weight of AM fungal host plants, soil chemical properties (soil pH, Truog-P, soil total C, and soil total N), and species richness and Shannon diversity index for plants and AM fungi were compared among the three grazing intensities (LG, MG, and HG) at each study site by ANOVA and post hoc means comparison with the TukeyHSD function using R software.
Dissimilarities in floristic composition among the plots were assessed at each study site using the function “vegdist” in the vegan package to calculate the Bray–Curtis index based on the shoot biomass of each plant species. PERMANOVA was subsequently performed as described above to test the validity of the a priori categorization of grazing intensity. Given that the sampling plots were nested within the sampling sites, Nested PERMANOVA (adonis (Dissimilarities in floristic composition ~ Grazing intensity/Study plot)) was also applied for the comparison. To visualize the dissimilarities among the plots, nonmetric multidimensional scaling (NMDS) was performed using the function “metaMDS” in the vegan package.
For the AM fungal OTU composition, PERMANOVA and Nested PERMANOVA based on Bray–Curtis index were applied to analyze the distinction among the grazing intensity categories as described above. The dissimilarities in AM fungal community composition among the plots were visualized using NMDS with the function “metaMDS” in the vegan package.
Given that the sampling areas for each grazing intensity were unevenly distributed (Fig. 1), the relationship between the geographic distances among the sampling plots and composition of AM fungal OTUs was investigated with the Mantel test using the function “mantel” (with 10,000 permutations) in the vegan package. A significant correlation was observed between the geographic distance and composition of AM fungal OTUs; thus, partial Mantel tests were performed using the function “mantel.partial” (with 10,000 permutations) to dissect the effect of geographical distance as well as Mantel tests (with 10,000 permutations) for the soil chemical properties (soil pH, Truog-P, total soil C, and total soil N) and vegetation variables (floristic composition and shoot biomass of AM fungal host plants, Poaceae biomass, and plant species richness and the Shannon diversity of AM fungal host plants) to identify factors that were correlated with the composition of the AM fungal OTUs. For the analysis of floristic composition, dissimilarities in floristic composition based on shoot biomass, calculated using the function “vegdist” as described above, were used.
To evaluate the effect of grazing on the AM fungal community structure, the proportion of the most dominant OTU in the sampling plot was compared among the grazing intensities at each study site by ANOVA and post hoc means comparison with the TukeyHSD function as described above.
Results
Floristic composition and plant biomass
Number of species in each plant family at the three grazing intensities, LG, MG, and HG, for the study sites Hustai, Mandalgovi, and Bulgan is shown in Table S2. Among the plant species recorded in this study, those belonging to the Amaranthaceae, Brassicaceae, Cyperaceae, and Caryophyllaceae are considered to be non- or weakly mycorrhizal plants according to Tester et al. (1987). In the LG plots at Hustai and Mandalgovi, 23 and 24 plant species were recorded, respectively, and the species number decreased with increase in grazing intensity at both sites. The mean Shannon diversity values for total plants and AM fungal host plants were not significantly different among the grazing intensities at Hustai and Mandalgovi (Table 1). At Bulgan, only nine species were recorded in the LG plots, and the species number showed little variation with grazing intensity (Table S2).
Effects of grazing on plant shoot biomass are presented in Table S3. At all study sites, plants of Poaceae species were dominant in the LG plots, and the biomass of Poaceae species substantially decreased with increase in grazing intensity. Among the Poaceae species, Stipa spp. were dominant, with specifically S. krylovii at Hustai and Mandalgovi, and S. glareosa at Bulgan. A similar tendency was observed in the Amaryllidaceae for Allium spp. The total plant shoot biomass was high in LG plots at Hustai (120.5 g m−2) and decreased with increase in grazing intensity. At Mandalgovi, biomass of Amaranthaceae species, predominantly Chenopodium spp., increased with grazing and, consequently, biomass of AM fungal host plants significantly decreased in the HG plots. At Bulgan, biomass of Asteraceae species increased with grazing. Consequently, total plant shoot biomass increased significantly with increase in grazing intensity at this study site.
Differences in composition of plant species biomass among the three provisional grazing intensities were statistically confirmed for all study sites by PERMANOVA and nested PERMANOVA considering the nested design of the sampling plots (Table 2). Higher F values, suggesting stronger distinction among the grazing intensities, were obtained for the arid environment at Bulgan. Differences in the composition of plant species biomass were also indicated in NMDS plots (Fig. 2).
Non-metric multidimensional scaling plot of floristic composition at a Hustai, b Mandalgovi, and c Bulgan. Stress value 0.17 for Hustai, 0.18 for Mandalgovi, and 0.08 for Bulgan. PERMANOVA showed significant differences in the floristic composition among the three grazing intensities, lightly grazed (LG), moderately grazed (MG), and heavily grazed (HG), at each study site (Hustai: R2 = 0.42, P < 0.01; Mandalgovi: R2 = 0.56, P < 0.01; Bulgan: R2 = 0.94, P < 0.01)
Soil chemical properties
The mean soil pH in LG plots at Hustai, Mandalgovi, and Bulgan was 6.16, 6.91, and 8.59, respectively, and the pH value increased with increase in grazing intensity at all study sites (Table 3). At Hustai and Mandalgovi, Truog-P (available P) content increased with increase in grazing intensity, although the contents at Mandalgovi were comparatively low. The Truog-P content at Bulgan decreased with increase in grazing intensity. Total C and total N contents were comparatively low at Mandalgovi. Slight increases in total C content with increasing grazing intensity were observed at Mandalgovi and Bulgan.
Classification of the AM fungi detected
Pyrosequencing yielded a total of 685,134 high-quality AM fungal DNA sequences classifiable to 139 OTUs based on a 97% sequence similarity threshold. After rarefaction to 1000 sequences per sample, 130 OTUs were retained. Rarefaction curves of AM fungal OTUs plotted for each sample are shown in Fig. S1. The 130 OTUs included members of the Glomeraceae (108 OTUs), Claroideoglomeraceae (10 OTUs), Diversisporaceae (five OTUs), Paraglomeraceae (five OTUs), Archaeosporaceae (one OTU), and Gigasporaceae (one OTU). At Hustai, we detected 109 OTUs, including members of the Glomeraceae (92 OTUs), Claroideoglomeraceae (nine OTUs), Paraglomeraceae (four OTUs), Diversisporaceae (three OTUs), and Archaeosporaceae (one OTU), and the number of fungal OTUs increased with increase in grazing intensity (Table S4). At Mandalgovi, we detected 87 OTUs, including members of Glomeraceae (73 OTUs), Diversisporaceae (five OTUs), Claroideoglomeraceae (four OTUs), Paraglomeraceae (three OTUs), Archaeosporaceae (one OTU) and Gigasporaceae (one OTU). In contrast to Hustai, at Mandalgovi the observed fungal OTUs decreased with increase in grazing intensity (Table S4). At Bulgan, only members of Glomeraceae (76 OTUs) and Claroideoglomeraceae (four OTUs) were detected (Table S4).
Significant differences among the three study sites, Hustai, Mandalgovi, and Bulgan, in the composition of AM fungal OTUs was shown by PERMANOVA (F = 15.7, p < 0.001).
Effect of grazing intensity on AM fungal community
Significant differences among the three grazing intensities in the composition of AM fungal OTUs was confirmed by PERMANOVA and nested PERMANOVA at all study sites, of which higher F values were observed for Hustai (Table 2). The differences were also indicated in NMDS plots (Fig. 3). Analysis of mean species richness and Shannon diversity confirmed that AM fungal diversity was enhanced with increasing grazing intensity at Hustai, whereas AM fungal species richness decreased with increase in grazing intensity at Mandalgovi (Table 1).
Non-metric multidimensional scaling plot of arbuscular mycorrhizal (AM) fungal community composition at a Hustai, b Mandalgovi, and c Bulgan. Stress value 0.21 for Hustai, 0.20 for Mandalgovi, and 0.22 for Bulgan. PERMANOVA showed significant differences in the AM fungal communities among the three grazing intensities, lightly grazed (LG), moderately grazed (MG), and heavily grazed (HG), at each study site (Hustai: R2 = 0.26, P < 0.01; Mandalgovi: R2 = 0.17, P = 0.01; Bulgan: R2 = 0.14, P = 0.03)
Mean read numbers of each fungal OTU for each grazing intensity at the three study sites, retained after rarefaction to 1000 reads per sample, are shown in Table 4, in which those with over 1.0% of total reads were selected as major OTUs. For the 16 major OTUs, a phylogenetic tree based on partial SSU rDNA sequences was constructed (Fig. 4). Among the 16 OTUs, 15 OTUs were detected at all study sites (Table 4).
Neighbor-joining phylogenetic tree constructed from partial sequences of the small subunit of nuclear ribosomal RNA gene (SSU rDNA) of arbuscular mycorrhizal (AM) fungi obtained from Mongolian grasslands at Hustai, Mandalgovi, and Bulgan. The selected OTUs sequences with over 1.0% of total reads, obtained with an Ion Torrent PGM for the PCR products amplified by the primers AMV4.5NF and AMDGR, were analyzed with identified AM fungal sequences downloaded from the GenBank database. The tree was rooted with Claroideoglomus etunicatum (AJ852598). Bootstrap values are shown where they exceed 50% (1000 replications). The scale is shown so that evolutionary distances can be inferred
At all study sites, the proportion of the most dominant fungal OTU in the LG plots decreased with increase in grazing intensity. At Hustai, OTU001 was the most dominant AM fungus in the LG plots, and constituted 49.1, 25.0, and 2.7% of the total fungi under LG, MG, and HG, respectively. The AM fungi in OTU001, identified as Rhizophagus fungi, belong to VTX00325 in the MaarjAM database (Fig. 4, Table 4). At Mandalgovi, OTU002 was the most dominant AM fungus in the LG plots, and constituted 45.5, 39.4, and 21.8% of the total fungi under LG, MG, and HG, respectively. The AM fungi in OTU002, identified as Rhizophagus fungi, belong to VTX00295 in the MaarjAM database (Fig. 4, Table 4). At Bulgan, OTU004 was the most dominant AM fungus in the LG plots, and constituted 24.6, 13.9, and 16.5% of the total fungi under LG, MG, and HG, respectively. The AM fungi in OTU004, identified as a member of the Glomeraceae, belong to VTX00156 in the MaarjAM database (Fig. 4, Table 4).
The mean proportion of the most dominant OTUs was comparatively high at about 25–50% (Table 5). The proportion of the most dominant OTUs decreased with increase in grazing intensity at Hustai. In the LG plots, the proportion was 50.1%, of which the majority was composed of the OTU001 fungi as described above. No significant correlations were observed between grazing intensity and proportion of the most dominant OTUs at Mandalgovi and Bulgan (Table 5).
Correlations between AM fungal community and environmental variables
Among the soil chemical properties, significant correlations with the AM fungal communities were observed for Truog-P at all study sites and for soil pH at Hustai and Mandalgovi as indicated by Mantel tests (Table 6). At Hustai and Mandalgovi, results of partial Mantel tests to dissect the effect of geographic distances were also significant for Truog-P and soil pH (Table 6). Among the vegetation variables, significant correlations were observed for floristic composition and shoot biomass of AM host plants, and Poaceae biomass at Hustai and Mandalgovi as indicated by Mantel tests. However, shoot biomass of AM fungal host plants and Poaceae biomass at Hustai, and floristic composition of AM host plants at Mandalgovi were not significant by the partial Mantel tests (Table 6).
Discussion
Floristic composition
Among the three sampling sites, plant biomass was highest at Hustai, which reflected its relatively high annual precipitation. At all study sites, grazing decreased the shoot biomass of Poaceae species, of which the majority were Stipa spp. (Table S3). Stipa spp. are known to be highly preferred forage plants by livestock in the region and the decrease in their biomass in response to grazing in Mongolian grassland was previously reported by Sasaki et al. (2008). Total shoot biomass also decreased with increase in grazing intensity at Hustai, whereas biomass increased at Bulgan on account of the luxuriance of Asteraceae plants. At Mandalgovi, as a result of the decreased abundance of Poaceae and increased abundance of Amaranthaceae plants, the shoot biomass of AM fungal host plants was substantially decreased by grazing (Table 1). Amaranthaceae species are known to be non- or weakly mycorrhizal plants (Tester et al. 1987). A significant effect of grazing on floristic composition at all study sites was confirmed by PERMANOVA and nested PERMANOVA.
Soil chemical properties
The soil pH was relatively high at Mandalgovi and Bulgan, which receive lower precipitation than Hustai (Table 3). The high pH of arid soils is a well-known feature caused by accumulation of alkaline cations (de Caritat et al. 2011). A slight but significant increase in pH with enhanced grazing intensity was observed at all study sites, which may be caused by excreta from livestock as suggested by Haynes and Williams (1992). As summarized by Wang et al. (2009), heavy grazing with reduced ground cover may induce increases in evaporation, soil temperature, and decomposition of soil organic matter, which all promote the processes of soil salinization and alkalization.
With regard to soil nutrient contents, the available P (Truog-P), total C, and total N contents were relatively low at Mandalgovi, which may reflect the sandy soil texture. At Hustai and Mandalgovi, the available P content increased with increase in grazing intensity, which may also be caused by excreta from livestock. By contrast, the available P content decreased with increasing grazing intensity at Bulgan. The reason for the lower soil P content in the heavily grazed area is not clear, but the area may have been avoided by domesticated animals for a long period because of the slow growth of the plants in the arid environment.
Molecular identification of AM fungi
Among the 130 AM fungal OTUs obtained in this study, 108 OTUs were members of the Glomeraceae. Given that the DNA primers used for the SSU rDNA regions are inclined to amplify Glomeraceae sequences (Van Geel et al. 2014), the true frequency of AM fungi in other families may be higher than indicated by this result. In the present study, AM fungal communities were estimated based on the number of reads of the partial sequence of SSU rDNA obtained by PCR and next-generation sequencing with an Ion Torrent™ PGM™. Although the read number does not exactly reflect biological abundance because of the biases introduced by DNA extraction, primer selectivity, and DNA sequencing, as well as variable copy numbers of ribosomal genes (Amend et al. 2010), a strong correlation between read number and genic abundance was confirmed for the PCR products of partial SSU rDNA of AM fungi (Yamato et al. 2016). Considering the semi-quantitative nature of the PCR products for this region of the AM fungal DNA, and the similar bias among samples induced by applying the same PCR procedure, it is reasonable to rely on the semi-quantitative reads for the analysis in this study.
At Hustai, 109 OTUs in five AM fungal families were detected, but only 87 OTUs in six families and 80 OTUs in two families were detected at Mandalgovi and Bulgan, respectively. The overall higher taxonomic diversity of AM fungi at Hustai may reflect the higher plant productivity at this site.
Distribution of AM fungi in Mongolian grassland
After rarefaction to 1000 sequences per sample, 130 AM fungal OTUs were retained in total, and distinct AM fungal communities among the study sites, Hustai, Mandalgovi, and Bulgan, were observed. Meanwhile, dominant AM fungi were commonly distributed among the study sites. Actually, among the 16 OTUs that comprised more than 1.0% of the total reads, 15 OTUs were detected at all study sites (Table 4). A search of the virtual taxa (VTX) in the MaarjAM database for the 16 OTUs revealed that 13 of the OTUs, namely OTU001 (VTX00325), OTU002 (VTX00295), OTU003 (VTX00304), OTU004 (VTX00156), OTU005 (VTX00167), OTU006 (VTX00166), OTU008 (VTX00130), OTU010 (VTX00214), OTU011 (VTX00100), OTU 015 (VTX00222), OTU016 (VTX00301), OTU051 (VTX00113), and OTU098 (VTX00156) are represented by sequences previously obtained from Mongolian steppes by Goomaral et al. (2013a, b). This result suggests that these AM fungi are widely distributed in Mongolian grassland, and that some of these AM fungi may have proliferated in congruence with environmental factors.
Effect of grazing on the composition of AM fungi
Significant differences in the composition of AM fungal communities among the grazing intensity categories at all study sites were confirmed by PERMANOVA (Table 2). The Mantel test identified significant correlation between soil pH or soil P and composition of AM fungal communities at all study sites, which indicated that soil chemical properties influence the AM fungal community (Table 6).
The areas for each grazing category neighbored another sampling area on the contiguous plain, but the area of the plots differed depending on the site. Consequently, the scale of plot separation was different in this study. Mantel tests to analyze the relationship between geographic distance and composition of AM fungal communities were significant at all study sites, and the highest R value was obtained at Hustai, to which the greater distance between the plots may have influenced (Table 6). Accordingly, partial Mantel tests to dissect the effect of geographic distance were applied in addition to Mantel tests of the effect of environmental and vegetation variables on composition of AM fungal communities (Table 6). Shoot biomass of AM fungal host plants and Poaceae biomass at Mandalgovi were also significantly correlated with AM fungal community composition by the partial Mantel tests. However, the vegetation variables were not significantly correlated with AM fungal community composition at Bulgan, in contrast to the comparatively high distinctness in floristic composition at this site (Table 2).
Interestingly, correlation between grazing intensity and AM fungal diversity showed conflicting results among the study sites. Grazing increased species richness and Shannon diversity of AM fungi at Hustai, which contradicts the initial hypothesis of this study, whereas grazing decreased species richness and Shannon diversity of AM fungi at Mandalgovi (Table 5) where, although total shoot biomass was not decreased by grazing, the increase in biomass of Amaranthaceae and Cyperaceae, which are non- or weakly mycorrhizal plants (Tester et al. 1987), was responsible for the lower biomass of the AM fungal host plants (Table S3). Given that a positive correlation between plant productivity and AM fungal species richness has been experimentally demonstrated (Maherali and Klironomos 2007), the lower AM fungal host biomass at the grazed sites may be the reason for the lower AM fungal diversity at Mandalgovi.
The effect of grazing on AM fungal symbiosis reported in previous studies is inconsistent. Grazing did not affect AM fungal colonization and/or spore numbers in some studies (Lugo et al. 2003; Medina-Roldán et al. 2008), whereas significant reductions were reported by other authors (Bethlenfalvay and Dakessian 1984; Bethlenfalvay et al. 1985; Su and Guo 2007; Bai et al. 2013). Decline in species richness or shift in species composition in response to grazing has been commonly reported (Eom et al. 2001; Su and Guo 2007; Ba et al. 2012; Bai et al. 2013; Tian et al. 2009). However, to the best of our knowledge, no previous study has considered the effect of grazing on the dominant AM fungi.
Effect of grazing on dominance of single AM fungal taxon
The most dominant fungus OTU001 in LG plots at Hustai accounted for 49.1% of the obtained sequences, which decreased with increasing grazing intensity to only 2.7% in HG plots (Table 4). The dominant AM fungi in LG plots also decreased with grazing at Mandalgovi and Bulgan, but the trends were less marked and the dominant fungi differed, i.e., OTU002 was dominant at Mandalgovi and OTU004 was dominant at Bulgan (Table 4).
At Hustai, the proportion of the most dominant OTUs was high, 50.1% on average in the LG plots, and the proportion decreased with increasing grazing intensity (Table 5), in which the most dominant fungal OTUs were not identical among the sampling plots. The decrease in the proportion of dominant OTUs at Hustai may be one reason for the increase in AM fungal diversity with grazing (Table 1). Dumbrell et al. (2010) observed that AM fungal communities are typically dominated by a single taxon, representing on average 40% of total abundance within the community, and the dominant AM fungal taxa differ among plant communities. Given that the dominance of a single AM fungal taxon in the study plots was also generally observed in the present study, it may be a typical ecological feature of AM fungal symbiosis. As an explanation for the dominance of a single AM fungal taxon, Dumbrell et al. (2010) proposed a stochastic process, that is, an AM fungal mycelium that colonizes new roots can gain additional C, and extra-radical mycelium of this fungus will be able to extend to other uncolonized plant roots. In this manner, a single fungal taxon may achieve dominance. Kiers et al. (2011) observed that plants can detect, discriminate, and reward the optimal fungal partners with more carbohydrates and, in turn, the fungal partners enforce cooperation by increasing nutrient transfer only to those roots providing higher amounts of carbohydrates. This interaction may be one reason for the dominance of a single fungal taxon in the AM symbiosis, and the dominance may be striking in a stable environment with one dominant plant. The OTU001 fungus, the most dominant AM fungal OTU in LG plots at Hustai, may be selected in the stable environment dominated by Stipa krylovii. The OTU001 fungi belong to the dominant AM fungal group that colonize Stipa krylovii at Hustai (Goomaral et al. 2013a).
A significant decrease in the proportion of dominant OTUs in response to grazing was not observed at Mandalgovi and Bulgan. These contrasting results may be caused by a difference in the stability of grazing intensities among the study sites. Hustai is a national park, and the sampling sites are located around the buffer zone of the protected area; therefore, the grazing intensities of the sampling plots are likely to be consistent. In contrast, the sampling sites at Mandalgovi and Bulgan are situated in rangeland, and the grazing intensity can be changeable. Accordingly, the effect of grazing on the AM fungal community structure may be most consistently expressed at Hustai in this study.
Conclusion
Distinct AM fungal communities that reflect the geographical location and different grazing intensities were observed at all study sites. However, the AM fungal OTUs with over 1.0% of the total reads were commonly detected among all the sites. This result indicates that these AM fungi are widely distributed in Mongolian grassland, and some have specifically proliferated from similar AM fungal floras in accordance with environmental and vegetation factors.
Significant differences in floristic and AM fungal compositions among the grazing intensities were confirmed at all study sites. Grazing decreased the shoot biomass of Poaceae species, especially Stipa spp. With regard to the AM fungal composition, although the effects of grazing on AM fungal diversity were inconsistent, the relative abundance of the most dominant OTU fungus in the LG plots decreased with increased grazing intensity at all study sites. The mean proportion of the most dominant OTUs also decreased with increasing grazing intensity at Hustai. As suggested by previous studies, the dominance of a single AM fungal taxon may be a typical ecological feature of AM fungal symbiosis, and grazing may disturb the AM fungal community structure.
References
Allen MF, Richards JH, Busso CA (1989) Influence of clipping and soil water status on vesicular-arbuscular mycorrhizae of twosemi-arid tussock grasses. Biol Fertil Soils 8:285–289
Amend AS, Seifert KA, Bruns TD (2010) Quantifying microbial communities with 454 pyrosequencing: does read abundance count? Mol Ecol 19:5555–5556
An GH, Miyakawa S, Kawahara A, Osaki M, Ezawa T (2008) Community structure of arbuscular mycorrhizal fungi associated with pioneer grass species Miscanthus sinensis in acid sulfate soils: habitat segregation along pH gradients. Soil Sci Plant Nutr 54:517–528
Ba L, Ning J, Wang D, Facelli E, Facelli JM, Yang Y, Zhang L (2012) The relationship between the diversity of arbuscular mycorrhizal fungi and grazing in a meadow steppe. Plant Soil 352:143–156
Bai G, Bao Y, Du G, Qi Y (2013) Arbuscular mycorrhizal fungi associated with vegetation and soil parameters under rest grazing management in a desert steppe ecosystem. Mycorrhiza 23:289–301
Bethlenfalvay GJ, Dakessian S (1984) Grazing effects on mycorrhizal colonization and floristic composition of the vegetation on semiarid range in Northern Nevada. J Range Manag 37:312–316
Bethlenfalvay GJ, Evans RA, Lesperance AL (1985) Mycorrhizal colonization of crested wheatgrass as influenced by grazing. Agron J 77:233–236
Bever JD, Morton JB, Antonovics J, Schultz PA (1996) Host-dependent sporulation and species diversity of arbuscular mycorrhizal fungi in a mown grassland. J Ecol 84:71–82
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Gonzalez Pena A, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Chagnon PL, Bainard LD (2014) Is root DNA a reliable proxy to assess arbuscular mycorrhizal community structure? Can J Microbiol 60:619–624
de Caritat P, Cooper M, Wilford J (2011) The pH of Australian soils: field results from a national survey. Soil Res 49:173–182
Dumbrell AJ, Nelson M, Helgason T, Dytham C, Fiter AH (2010) Idiosyncrasy and overdominance in the structure of natural communities of arbuscular mycorrhizal fungi: is there a role for stochastic process? J Ecol 98:419–428
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Eom AH, Wilson GWT, Hartnett DC (2001) Effects of ungulate grazers on arbuscular mycorrhizal symbiosis and fungal community structure in tallgrass prairie. Mycologia 93:233–242
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Goomaral A, Iwase K, Undarmaa J, Matsumoto T, Yamato M (2013a) Communities of arbuscular mycorrhizal fungi in Stipa krylovii (Poaceae) in the Mongolian steppe. Mycoscience 54:122–129
Goomaral A, Undarmaa J, Matsumoto T, Yamato M (2013b) Effect of plant species on communities of arbuscular mycorrhizal fungi in the Mongolian steppe. Mycoscience 54:362–367
Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R (2008) Error-correcting barcoded primers allow hundreds of samples to be pyrosequenced in multiplex. Nat Methods 5:235–237
Haynes RJ, Williams PH (1992) Changes in soil solution composition and pH in urine-affected areas of pasture. Eur J Soil Sci 43:323–324
Hiiesalu I, Pärtel M, Davison J, Gerhold P, Metsis M, Moora M, Öpik M, Vasar M, Zobel M, Wilson SD (2014) Species richness of arbuscular mycorrhizal fungi: associations with grassland plant richness and biomass. New Phytol 203:233–244
Jigjidsuren S, Johnson DA (2003) Forage plants of Mongolia. Admon Publishing, Ulaanbaatar
Kiers ET, Duhamel M, Beesetty Y, Mensah JA, Franken O, Verbruggen E, Fellbaum CR, Kowalchuk GA, Hart MM, Bago A, Palmer TM, West SA, Vandenkoornhuyse P, Jansa J, Bücking H (2011) Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 333:880–882
Klironomos JN (2003) Variation in plant response to native and exotic arbuscular mycorrhizal fungi. Ecology 84:2292–2301
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lugo MA, Maza MWG, Cabello MN (2003) Arbuscular mycorrhizal fungi in a mountain grassland II: seasonal variation of colonization studied, along with its relation to grazing and metabolic host type. Mycologia 95:407–415
Maherali H, Klironomos JN (2007) Influence of phylogeny on fungal community assembly and ecosystem functioning. Science 316:1746–1748
Medina-Roldán E, Arredondo JT, Huber-Sannwald E, Chapa-Vargas L, Olalde-Portugal V (2008) Grazing effects on fungal root symbionts and carbon and nitrogen storage in a shortgrass steppe in Central Mexico. J Arid Environ 72:546–556
Oksanen J, Blanachet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens HH, Szoecs E, Wagner H (2017) Package ‘vegan’ community ecology package version 2.4–4 (https://cran.ism.ac.jp/web/packages/vegan/vegan.pdf)
Öpik M, Vanatoa A, Vanatoa E, Moora M, Davison J, Kalwij JM, Rejer Ü, Zobel M (2010) The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytol 188:223–241
Pringle A, Bever JD (2008) Analogous effects of arbuscular mycorrhizal fungi in the laboratory and a North Carolina field. New Phytol 180:162–175
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:404–425
Sasaki T, Okayasu T, Takeuchi K, Undarmaa J, Jadambaa S (2005) Patterns of floristic composition under different grazing intensities in Bulgan, South Gobi, Mongolia. Grassl Sci 51:235–241
Sasaki T, Okayasu T, Shirato Y, Undarmaa J, Okubo S, Takeuchi K (2008) Can edaphic factors demonstrate landscape-scale differences in vegetation responses to grazing? Plant Ecol 194:51–66
Sato K, Suyama Y, Saito M, Sugawara K (2005) A new primer for discrimination of arbuscular mycorrhizal fungi with polymerase chain reaction-denature gradient gel electrophorsis. Grassl Sci 51:179–181
Smith SE, Read DJ (2008) Mycorrhizal symbiosis, 3rd edn. Academic Press, New York
Su YY, Guo LD (2007) Arbuscular mycorrhizal fungi in non-grazed, restored and over-graed grassland in the Inner Mongolia steppe. Mycorrhiza 17:689–693
Tester M, Smith SE, Smith FA (1987) The phenomenon of “nonmycorrhizal” plants. Can J Bot 65:419–431
Tian H, Gai JP, Zhang JL, Christie P, Li XL (2009) Arbuscular mycorrhizal fungi in degraded typical steppe of Inner Mongolia. Land Degrad Dev 20:41–54
Ulziikhutag N (1989) Review of flora of Mongolia, Ulaanbaatar, Ulaanbaatar
Undarmaa J, Tamura K, Luvsan N, Yamanaka N (2018) Rangeland ecosystems of Mongolia. Munkhiin Useg, Ulaanbaartar
van der Heijden MGA, Klironomos JN, Ursic M, Moutoglis P, Streitwolf-Engel R, Boller T, Wiemken A, Sanders IR (1998) Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396:69–72
Van Geel M, Busschaert P, Honnay O, Lievens B (2014) Evaluation of six primer pairs targeting the nuclear rRNA operon for characterization of arbuscular mycorrhizal fungal (AMF) communities using 454 pyrosequencing. J Microbiol Methods 106:93–100
Wang B, Qiu YL (2006) Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16:299–363
Wang L, Seki K, Miyazaki T, Ishihama Y (2009) The causes of soil alkalinization in the Songnen plain of Northeast China. Paddy Water Environ 7:259–270
Yamato M, Takahashi H, Shimono A, Kusakabe R, Yukawa T (2016) Distribution of Petrosavia sakuraii (Petrosaviaceae), a rare mycoheterotrophic plant, may be determined by the abundance of its mycobionts. Mycorrhiza 26:417–427
Yoshimura Y, Ido A, Iwase K, Matsumoto T, Yamato M (2013) Communities of arbuscular mycorrhizal fungi in the roots of Pyrus pyrifolia var. culta (Japanese pear) in orchards with variable amount of soil-available phosphorus. Microbes Environ 28:105–111
Acknowledgements
Mongolian soil and plant samples used for the analysis were transported to Japan with the permission of the General Agency for Specialized Inspection, Mineral Resource Authority, Ministry of Environment and Tourism in Mongolia, and the Plant Protection Station, Ministry of Agriculture, Forestry and Fisheries in Japan. This study was supported by a Grant-in-Aid for Scientific Research (26304046) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, and the Joint Research Program of Arid Land Research Center, Tottori University (2014-2015). We thank Robert McKenzie, PhD, from Edanz Group (www.edanzediting.com/ac), for editing a draft of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Fig. S1
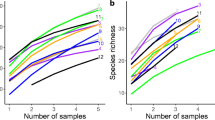
Rarefaction curves of arbuscular mycorrhizal fungal OTUs against the number of reads after rarefication to 1000 reads per sample in each root sample of the three grazing intensities, lightly grazed (LG; Green), moderately grazed (MG; Blue), and heavily grazed (HG; Red) at three study site, a) Hustai, b) Mandalgovi, and c) Bulgan (PNG 96 kb)
Table S1
(XLSX 50 kb)
Table S2
(XLSX 40 kb)
Table S3
(XLSX 42 kb)
Table S4
(XLSX 37 kb)
Rights and permissions
About this article
Cite this article
Kusakabe, R., Taniguchi, T., Goomaral, A. et al. Arbuscular mycorrhizal fungal communities under gradients of grazing in Mongolian grasslands of different aridity. Mycorrhiza 28, 621–634 (2018). https://doi.org/10.1007/s00572-018-0855-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-018-0855-7