Abstract
Arbuscular mycorrhizal fungi (AMF) have been implicated in non-native plant invasion success and persistence. However, few studies have identified the AMF species associating directly with plant invaders, or how these associations differ from those of native plant species. Identifying changes to the AMF community due to plant invasion could yield key plant–AMF interactions necessary for the restoration of native plant communities. This research compared AMF associating with coexisting Bromus tectorum, an invasive annual grass, and Artemisia tridentata, the dominant native shrub in western North America. At three sites, soil and root samples from Bromus and Artemisia were collected. Sporulation was induced using trap cultures, and spores were identified using morphological characteristics. DNA was extracted from root and soil subsamples and amplified. Sequences obtained were aligned and analyzed to compare diversity, composition, and phylogenetic distance between hosts and sites. Richness of AMF species associated with Artemisia in cultures was higher than AMF species associated with Bromus. Gamma diversity was similar and beta diversity was higher in AMF associated with Bromus compared to Artemisia. AMF community composition differed between hosts in both cultures and roots. Two AMF species (Archaeospora trappei and Viscospora viscosum) associated more frequently with Artemisia than Bromus across multiple sites. AMF communities in Bromus roots were more phylogenetically dispersed than in Artemisia roots, indicating a greater competition for resources within the invasive grass. Bromus associated with an AMF community that differed from Artemisia in a number of ways, and these changes could restrict native plant establishment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Interactions with the soil microbial community have been implicated in successful invasions by introduced plant species (Callaway et al. 2004; Reinhart and Callaway 2006). In particular, interactions with arbuscular mycorrhizal fungi (AMF) are thought to be important in invasive plant success through a number of mechanisms (Pringle et al. 2009; Richardson et al. 2000; Shah et al. 2009). These mechanisms include suppression of AMF by non-mycorrhizal invaders (Callaway et al. 2008; Stinson et al. 2006), receipt of greater benefits from associations (Harner et al. 2010; Marler et al. 1999), reduced dependence on AMF (Seifert et al. 2009; Vogelsang and Bever 2009), and alterations to the AMF community (Mummey and Rillig 2006; Shah et al. 2010; Zhang et al. 2010).
However, due to the high variation in interactions between AMF and plant species (Gustafson and Casper 2006; Sanders and Fitter 1992; Wilson and Hartnett 1998), it is not known if many of these observed changes are unique, or if they reflect species-specific variation among plant hosts. Because of the influence of extrinsic factors, such as environment (Allen et al. 1995; Opik et al. 2006), land use intensity (Oehl et al. 2003), grazing (Eom et al. 2001), fire (O’Dea 2007), neighbor identity (Hausmann and Hawkes 2009), etc. on AMF communities, comparing changes to AMF communities due to invaders with AMF communities associated with natives becomes difficult to achieve. To determine differences between introduced and native species, native species need to be carefully selected in order to elucidate invader differences that are ecologically relevant (van Kleunen et al. 2010).
Invasive grasses are particularly destructive, as they are globally invasive and alter ecosystem processes (D’Antonio and Vitousek 1992). Bromus tectorum L. (hereafter Bromus) invasion in the semi-arid sagebrush (Artemisia spp.) rangelands of western North America has caused a catastrophic shift from shrub-dominated plant communities to those dominated by this introduced annual grass. Overgrazing of native perennial grasses by cattle allowed Bromus to gain a foothold (Klemmedson and Smith 1964; Knapp 1996), and it has since invaded 20 million ha in the intermountain west (Mack 1989). Bromus burns readily and has reduced the fire return interval substantially in invaded communities (D’Antonio and Vitousek 1992), maintaining its dominance through this alteration of the fire regime (Klemmedson and Smith 1964). Historic fire return interval estimates range from 60 to 110 years across the sagebrush steppe (Whisenant 1990), to as high as 100 to 240 years for stands of Wyoming big sagebrush (Baker 2006). Bromus invasion has reduced these intervals to every 3 to5 years (D’Antonio and Vitousek 1992). Native species are not adapted to this altered fire cycle and are subsequently replaced by Bromus (Klemmedson and Smith 1964; Young and Evans 1978). Bromus often attains nearly 100 % coverage in invaded areas, and approximately 20 % of the historic sagebrush steppe is dominated by Bromus, which restricts establishment of native vegetation (Knapp 1996). Sagebrush historically occurred on over 60 million ha in western North America, and big sagebrush (Artemisia tridentata Nutt., hereafter Artemisia) was dominant or co-dominant with perennial grasses over much of this area (West 1983).
The impacts of Bromus invasion on the AMF community are not well known. Bromus is considered a facultative AMF host (Allen 1984). Bromus reduces AMF diversity colonizing roots of neighboring grass species (Hawkes et al. 2006), and soils invaded by Bromus have lower AMF density than un-invaded soils (Al-Qawari 2002). While Artemisia associates with a high diversity of AMF (Allen et al. 1995), the identity and diversity of AMF species associating with Bromus are not known.
An important question in understanding replacement of Artemisia dominance by Bromus is how this shift alters the diversity of AMF in invaded sagebrush steppe habitat, thereby impacting future recovery and restoration of invaded communities. It is likely that Bromus invasion results in significant alteration to the AMF community, given the disparity that exists between Bromus and the historic native dominant, Artemisia. The goals of this research were to determine how coexisting Bromus and Artemisia differ in their associations with AMF and identify AMF species that may be important for restoration of Artemisia in Bromus-invaded soils. We tested the hypothesis that alpha diversity of AMF associated with Bromus is lower than alpha diversity of AMF associated with Artemisia. AMF species were identified from three distinct locations in Colorado, Utah, and Wyoming, USA (hereafter the CO, UT, and WY sites, respectively), using trap cultures on field-collected soil and root material and DNA extracted from subsamples of the roots and soils.
Materials and methods
Study sites
Three sites were selected that contain coexisting Bromus and Artemisia, are geographically separated, and represent three distinct levels of plant community age (Table 1). By selecting study sites where the target host plant species were interspersed, site effects and previous disturbances that alter the AMF community were minimized. Detailed site descriptions are available in Online Resource 1.
Sample collection
To compare AMF associations of Bromus and Artemisia, 16 individuals of each host at each site were selected and excavated (Fig. A1, Online Resource). Sampling was conducted from May 19 to May 22, 2008 to obtain samples during the estimated peak colonization interval for both hosts (Busby et al. 2012; Trent et al. 1994). Because these plant species differ drastically in their growth strategies, juveniles of Artemisia less than 15 cm in height were selected to more appropriately compare a long-lived shrub to a winter annual grass, as juveniles would be much closer in age, mass, height, root volume, and C fixation capacity to Bromus. To minimize the effects of one host on the other, individuals were selected that were at least 50 cm from individuals of the other target host. Sixteen interspersed individuals of each host plant species fitting the above criteria across each selected study site were identified. Soil and roots were excavated and removed from a depth and diameter of 15 cm immediately around the stem of each individual. Soils were placed in sterile bags and kept cool for transport to the greenhouse. A subsample each of roots and soil was removed from each excavated host sample. Subsamples were placed in 50 mL conical centrifuge tubes and placed on ice for transport to the laboratory. An additional subsample was removed from the roots of five individuals of each host at each site and placed in 70 % ethanol for observation of AMF colonization.
Root colonization
Root subsamples collected for colonization were washed in tap water, cleared to remove pigments in 2.5 % KOH for 30 min at 90 °C, re-rinsed with tap water, and acidified in 1 % HCl for 4 h. Roots were then stained in acid glycerol containing 0.05 % trypan blue for 30 min at 90 °C and destained in acid glycerol for 30 min at 90 °C (Koske and Gemma 1989). Each subsample was observed under 400 × magnification and the presence of hyphae, vesicles, and arbuscules was determined using 100 root intersections per subsample with a crosshair reticle (McGonigle et al. 1990).
Culturing of AMF
Bulk soil samples were used to establish trap cultures for propagation of AMF following Stutz and Morton (1996). Each of the 96 soil samples was mixed 1:1 (v/v) with autoclaved sand, placed in 2.8 L pots, and seeded with 80 to 100 surface-sterilized (5 min in 5 % sodium hypochlorite solution) seeds of Sudangrass (Sorghum bicolor (L.) Moench ssp. drummondii (Nees ex Steud.) de Wet & Harlan). Cultures were placed in a climate-controlled glasshouse at Colorado State University, Fort Collins, CO. The glasshouse was maintained at 20 °C nighttime and 24 °C daytime temperatures and supplemented with sodium vapor lights to maintain 16 h of daylight. Cultures were watered daily and re-randomized on the bench every 2 weeks to minimize microclimate effects. Cultures were grown for 120 days and watering was then stopped to allow soils to dry slowly for induction of sporulation. After 14 days of drying in low-light conditions, Sudangrass shoots were removed and a 250-mL subsample of soil was collected. Cavities remaining from collected soil subsamples were refilled with sterile sand, the cultures were reseeded with 80 to 100 surface-sterilized Sudangrass seeds, and culturing was repeated. This process was repeated for three rounds of culturing to maximize sporulation of AMF species present (Stutz and Morton 1996).
Identification of AMF
AMF spores were isolated from subsamples using sucrose density gradient centrifugation (Daniels and Skipper 1982). A 50-mL subsample of soil removed at each sampling interval was passed through 500- and 38-μm sieves using a water drench. Material retained by the 38-μm sieve was transferred to a 50-mL centrifuge tube containing a 20/60 % sucrose gradient and centrifuged at 1,000 × g for 1 min. The supernatant was poured into a 38-μm sieve and thoroughly rinsed. The rinsed material retained on the 38-μm sieve was transferred to a glass Petri dish, and spores were isolated and removed under a dissecting microscope using manually extended glass pipette tips. Isolated spores were mounted in polyvinyl lacto-glycerol and Meltzer’s reagent (Koske and Tessier 1983; Stutz and Morton 1996), and species were identified using spore wall characteristics (Morton 1988). Mounted spores were compared to voucher specimens at the International Culture Collection of (Vesicular) Arbuscular Mycorrhizal Fungi (INVAM, West Virginia University, Morgantown, WV, USA) and to published descriptions of AMF species.
Molecular analyses
Roots were rinsed in sterile, DNA-free water, and DNA was extracted using the FastDNA Spin Kit (MP Biomedicals, Irvine, CA, USA). DNA was extracted from a maximum of 50 mg of roots, using the entire root subsample for Bromus and only fine roots for Artemisia. For soils, DNA from 400 mg (dry weight) soil was extracted using the FastDNA Spin Kit for Soil (MP Biomedicals). AMF DNA was amplified using the AM1 (5′-GTT TCC CGT AAG GCG CCG AA-3–)–NS31 (5′-TTG GAG GGC AAG TCT GGT GCC-3′) primer pair (Helgason et al. 1998; Simon et al. 1992). PCR amplifications were conducted using the Expand High Fidelity Plus PCR System (Roche, Penzburg, Germany) with 25 μL reaction mixtures containing 2.5 μL of 2 μM of each primer, 1 μL of 100 μM dNTPs, 1 μL Expand High Fidelity Enzyme Mix in 1.25 U/μL, 2.5 μL of 1 mM MgCl2, 5 μL buffer, 1 μL 1 % BSA, 1 μL of 10 ng/μL template DNA, and 8.5 μL H2O. Reaction conditions were as follows: hot start, 94 °C for 2 min, 10 cycles of: 94 °C for 15 s, 58 °C for 30 s, 72 °C for 45 s; 20 cycles of: 94 °C for 15 s, 58 °C for 30 s, 72 °C for 45 s + 5 s per cycle; 72 °C for 5 min. PCR products were purified using the GENECLEAN Turbo Kit (Qbiogene, Montreal, QC, Canada), verified using agarose gel electrophoresis, and quantified on a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Positive PCR products were ligated into a pCR®2.1-TOPO vector using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA, USA). Ligated products were transformed into One Shot® chemically competent Escherichia coli cells (Invitrogen). The transformed cells were spread onto a Luria–Bertani (LB) (Becton Dickinson, Franklin Lakes, NJ, USA) agar plate containing 50 μg/mL ampicillin and were incubated overnight at 37 °C. Fifteen well-isolated ampicillin-resistant colonies were selected randomly from each sample, transferred to 96-well plates, and incubated for 24 h using a shaker incubator at 37 °C and 320 rpm in 2× LB broth containing 50 μg/mL ampicillin. After incubation, the plates were centrifuged at 1,500 × g for 7 min for further plasmid DNA isolation. Plasmid DNA isolation was performed on 15 clones per soil or root sample using the Montage Plasmid MiniprepHTS Kit (Millipore, Billerica, MA, USA) following manufacturer’s instructions. Isolated DNA was sequenced by the Colorado State University Proteomics and Metabolomics Facility using an ABI 3130xL Genetic Analyzer (Applied Biosystems, Carlsbad, CA, USA).
Editing, alignment, and matching of sequence data
Vectors were trimmed and raw sequences were edited using Geneious Pro version 5.0.4 (Biomatters Ltd., Auckland, New Zealand) with the following criteria: minimum sequence length of 300 bp and high quality DNA greater than 80 %. Sequences resulting from editing were analyzed for chimeric sequences by the Bellerophon server (http://comp-bio.anu.edu.au/bellerophon/bellerophon.pl) using Huber–Hugenholtz correction and a window size of 200 bp (Huber et al. 2004). Sequences passing the above screening were assembled using custom assembly parameters: 50 bp word length with an index word length of 13 bp, maximum gap size of 1, 2 % maximum gaps per read, and 2 % maximum mismatches. Assembled sequences were then compared to the BLAST database to identify AMF sequences on February 6, 2012. All AMF sequences were submitted to GenBank under accession numbers JF683552–JF683578.
Phylogenetic analyses of sequence data
Assembled sequences from Geneious were imported into MegAlign v.8.1.3 (Lasergene, DNASTAR, Inc., Madison, WI, USA) and aligned using ClustalW with a gap penalty of 15, a gap length penalty of 6.66, divergent sequence delay of 30 %, and a DNA transition weight of 0.5. A phylogenetic tree was constructed from the alignment using Geosiphon pyriformis as outgroup. Branch divergence was calculated by MegAlign.
Mean divergence (D) was calculated for each root and soil sample using the following equation:
Where N s = number of unique AMF sequences in sample, and D ij = divergence between AMF sequences i and j.
Statistical analyses of data
Individual plant hosts sampled from each site were treated as samples, with 16 replicate samples taken from each host population at each site, with three replicate sites. Mean AMF species richness in trap cultures, root DNA samples, and soil DNA samples was used as an index of AMF diversity to compare alpha diversity between sample sets within and across sites and hosts. Gamma diversity was calculated as the total species richness for a set of samples within and across sites and hosts. The alpha and gamma diversity calculations were then used to calculate respective beta diversities using the equation β = (γ / α) − 1 (Whittaker 1972). Mean AMF alpha diversity was compared between hosts using t tests with α = 0.05.
Site and host effects and their interaction on AMF composition in cultures, roots, and soils were analyzed using multivariate analyses of variance with α = 0.1 using SAS version 9.1 (The SAS Institute, Cary, NC, USA). To identify potential AMF species associations with a particular host, indicator species analysis was conducted across sites using PC-ORD version 5.31 (MjM Software, Gleneden Beach, OR, USA) with 4,999 permutations.
Results
All root subsamples observed for AMF colonization contained AMF (Table A1, Online Resource). Mean AMF colonization for Artemisia was 28.5 %, while mean colonization for Bromus was 10.6 %. For both host plant species, AMF percent colonization was lowest in samples from the CO site while samples were highest at the WY site.
All 96 trap cultures produced AMF spores, with a range from 1 to 12 species. A total of 32 AMF species were isolated from the 96 trap cultures, including seven unidentified species that were isolated across all three sites (Fig. 1). Funneliformis mosseae was the most common species observed in the culture study, occurring in 83 of the 96 cultures across all sites (Fig. 1). Other common species associating with both hosts and occurring at all three sites were Glomus aggregatum, Claroideoglomus claroideum, Diversispora eburnea, Rhizophagus intraradices, Diversipora versiforme, Viscospora viscosum, Diversispora spurca, Paraglomus occultum, and Entrophospora infrequens (Fig. 1). Two species were common in only two of the three sites: Archaeaospora trappei and Septoglomus constrictum (Fig. 1). Acaulospora delicata (WY site) and Pacispora scintillans (UT site) were moderately common at only one site and not isolated elsewhere (Fig. 1). Many isolated AMF species (16 of the 32 total species isolated) occurred in less than 5 % of the cultures.
Total frequency of arbuscular mycorrhizal fungi species across and within three study sites (a total, b Colorado, c Utah, and d Wyoming), based on identification of spores produced in 16 trap cultures for each host plant species (Bromus tectorum or Artemisia tridentata) at each site using soil samples for each host plant species (N = 96 cultures total). Black bars indicate B. tectorum associations, white bars indicate Artemisia tridentata associations. Ac. Acaulospora, Ar. Archaeospora, C. Claroideoglomus, D. Diversispora, E. Entrophospora, F. Funneliformis, G. Glomus, Pac. Pacispora, Par. Paraglomus, R. Rhizophagus, Sc. Scutellospora, Se. Septoglomus, V. Viscospora
Of the 96 samples collected for roots and soils, usable DNA was extracted from a total of 63 root and 82 soil samples. Failed samples were not randomly distributed, with 24 of the 33 failed root samples being Artemisia roots (11 of 16 Artemisia root samples from the UT site failed). Failed soil samples were also clustered, with 6 of the 14 samples occurring from Artemisia soils at the CO site. Of the samples yielding extractable DNA, one Artemisia and six Bromus root samples contained no AMF DNA, while six Artemisia and eight Bromus soil samples contained no AMF DNA.
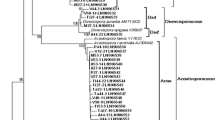
A total of 1,612 sequences were assembled and aligned, resulting in identification of 27 distinct AMF sequences (Fig. 2). Eleven sequences clustered closely (<2 bp substitutions per 100 residues) with a known species isolate in GenBank (Fig. 2). The most common sequence, JF683571, which occurred in root and soil samples from both hosts at all sites, aligned closely with Glomus iranicum (Figs. 1 and 2). Of the 27 AMF sequences identified, 17 occurred in root samples and 21 occurred in soil samples (Fig. 3). Six sequences were unique to roots, while 10 were unique to soils (Fig. 3). The number of sequences isolated from each sample ranged from 1 to 6 for Bromus roots (mean 2.15), 1 to 5 for Artemisia roots (mean 2.17), 1 to 5 for Bromus soils (mean 1.41), and 1 to 4 for Artemisia soils (mean 1.61). Across all sites, 14 AMF sequences were isolated from Bromus roots (four unique) and 11 AMF sequences were isolated from Artemisia roots (two unique). The most common sequence, JF683571, occurred in 76 % of root samples and 55 % of soil samples (Fig. 3). Nine sequences were common across all three study sites. One sequence (JF683568) occurred at only two sites (UT and WY), and 17 sequences were unique to only one of the sites (Fig. A2, Online Resource).
Neighbor-joining tree showing phylogeny of AMF DNA sequences isolated from Bromus tectorum and Artemisia tridentata roots and soils. Fungal sequences from this study are identified by their GenBank sequences beginning with JF683. Closely aligned sequences from GenBank are in bold and are identified by the species name and accession number
Frequency of AMF DNA sequences isolated from Artemisia tridentata (a) and Bromus tectorum (b) roots and soils. Total frequency of AMF sequences across all sample sites, based on occurrence in either roots or soils from Bromus or Artemisia host plants. Black bars indicate root isolates, white bars indicate soil isolates. Naming convention for the sequences are numbered based on the last three digits of the accession number assigned by GenBank
AMF species richness
AMF diversity measures are presented in Table 2, using AMF species richness for diversity measures. Diversity measures are as follows: α diversity is the mean species richness per sample grouping (host and site as groups); γ diversity is the total AMF species richness across sample groups (host and site as groups); and β diversity is the rate of change (i.e., number of unique communities) between sample groups (host and site as groups).
Alpha diversity of AMF species was higher in cultures associated with Artemisia within and among all sites compared to Bromus (Table 2). Gamma diversity was similar among cultures associated with the respective host plant species within and across sites. Beta diversity was higher in cultures with Bromus as host compared to Artemisia within and across all sites (Table 2). Alpha, beta, and gamma diversity indices for AMF DNA sequences in the soil and root samples varied inconsistently between hosts within and between sites. Consistent with these results, t tests indicated that the mean alpha diversity of AMF species isolated from Artemisia cultures was higher than Bromus (t = 2.46, df = 94, P = 0.02), while no host plant difference was observed for root (t = 0.4, df = 61, P = 0.69) or soil DNA (t = 0.56, df = 48.75, P = 0.58) sequences.
AMF species composition
Site had a significant effect on AMF community composition in cultures and root and soil DNA sequences (Table 3). Host identity had a significant effect on AMF community composition in cultures and root DNA sequences (Table 3). The interaction between site and host had a significant effect on AMF community composition only in cultures (Table 3).
Indicator species analysis was used to determine if any of the AMF species were significantly more likely to associate with one host over another. An indicator species is ideal if it occurs in all samples of one group, and none of another group. Indicator species are calculated by multiplying the proportional abundance of a species in a group relative to its abundance in the other group by the proportional frequency of the species in each group. An indicator value (I.V.) of 100 equals perfect indication of maximum indicator group. Across all sites, two AMF species (Archaeospora trappei, I.V. = 20, P = 0.02 and V. viscosum, I.V. = 26.9, P = 0.04) were more frequently associated with Artemisia compared to Bromus in cultures (Table A2, Online Resource), while no indicator species were identified in root or soil DNA sequences.
Phylogenetic distance
Phylogenetic distance analyses produced 351 divergence values between all combinations of paired DNA sequences, with a range from 1.3 to 134.5, and a mean of 57.3. Mean divergence of AMF DNA samples in Bromus roots (40.3 ± 7.8 standard error) was higher (P = 0.04) than in Artemisia roots (16.9 ± 6.2). There was no difference (P = 0.26) between divergence of AMF DNA in soil samples associated with Bromus (24.7 ± 9.1) and Artemisia (39.6 ± 9.5).
Discussion
The results indicate that AMF communities differ between coexisting Artemisia and Bromus. AMF species richness was lower in trap cultures associated with Bromus than in cultures associated with Artemisia. Host had a significant effect on AMF composition in cultures and host roots, and the interaction between site and host had a significant effect on AMF composition in cultures. Two AMF species isolated from trap cultures associated more often with Artemisia than Bromus. DNA sequences isolated from host roots indicated that communities of AMF colonizing Bromus are more phylogenetically distant than communities colonizing Artemisia.
The two methods used to compare AMF communities between hosts differed considerably. Because the trap culture source material comprised soil from underneath host plants, it is possible that the AMF community in each sample was not only influenced by the target host, but by neighboring vegetation and the legacy effects of prior inhabitants of that microsite as well (Hausmann and Hawkes 2009; Hausmann and Hawkes 2010). Trap cultures also introduce cultivation bias due to the varying growth strategies of the different AMF species and vastly different field and culture environmental conditions (Sýkorová et al. 2007). Similar to Sýkorová et al. (2007), F. mosseae (Glomus mosseae) occurred in 86 % of trap cultures, but only 17 % of roots and 10 % of soils. Thus, trap cultures provide little more than a gross assessment of potential associations with the target host plant with which to identify important trends. However, each trap culture provided a sample with approximately 80,000 times the mass of the largest root samples and 10,000 times the mass of the soil samples from which DNA was extracted, allowing each culture to encompass the entire region of host root interaction with the soil.
On the other hand, sequencing AMF DNA isolated from roots of target hosts provides a specific comparison between hosts that is not limited to the presence of spores, but is limited by sample size. Extraction of DNA from roots is only an accurate measure of the root subsample, while the minute mass of soils extracted likely excludes numerous species that are present in the composite soil sample from which the subsample was removed. Further, many methodological biases can be introduced in DNA isolation and amplification from root and soil samples (Renker et al. 2006). To minimize these biases, we chose to focus on isolating DNA from multiple individual host plants (one PCR amplification per DNA extraction), rather than pooling multiple PCR amplifications from fewer replicate samples. Given that AMF species richness was quite similar between morphological and molecular methods, potential biases were assumed to be minimized.
Most of the discrepancies between species in the trap culture and DNA data sets are due to either gross differences in relative frequencies or the occurrence of unique species or sequences. However, the largest discrepancy was the presence of G. aggregatum in most cultures but not in DNA samples, while G. iranicum and G. indicum occurred in numerous DNA samples but not in cultures. Because of the lack of DNA sequences specific or closely aligned to G. aggregatum, it is likely that this species is synonymous with one or both of G. indicum and G. iranicum, or was misidentified in cultures. All three of these species have similar morphological descriptions (hyaline to yellow in color, spores formed in loose clusters lacking a peridium), reactions to Meltzer’s reagent, and overlapping sizes (Blaszkowski et al. 2010a, b; Schenk and Smith 1982), with the major difference being three wall layers in G. iranicum and two layers in each of G. aggregatum and G. indicum).
Another important difference between trap cultures and DNA sequences was the presence of P. occultum and A. trappei in trap cultures and absence in sequence analyses. Neither Paraglomus nor Archaeospora are amplified by the primer set used for DNA amplification (Lee et al. 2003). Although only one species from each genus was identified in trap cultures, A. trappei was identified as an indicator species for Artemisia. This non-amplification could partially explain why no indicator species were identified from molecular data.
Mean AMF species richness per sample in Artemisia cultures was close to one AMF species more than in Bromus cultures, about a 15 % reduction in species richness associated with Bromus across all sites. The calculated AMF beta diversities from cultures indicate a greater variability of AMF communities associated with Bromus than Artemisia, while the gamma diversities of AMF are about equal between Bromus and Artemisia. So, while either host associates with about as many AMF species at the individual site level and across sites, Artemisia individuals associate with more AMF species than Bromus individuals, and these sagebrush-associated AMF communities are more similar from one individual host to the next when compared to Bromus. Thus, AMF communities associated with Artemisia at a given site are analogous diversity hotspots. The higher concentration of AMF species under Artemisia is supported by the resource island effect of shrubs in arid and semi-arid ecosystems (Reynolds et al. 1999). These shrub resource islands are associated with altered soil microbial diversity (Mummey and Stahl 2003) and harbor higher AMF propagules (Azcón-Aguilar et al. 2003) than shrub interspaces.
The alpha, beta, and gamma diversity measures from DNA sequence analyses did not indicate a consistent trend between hosts. However, the differences observed could be due to the non-random loss of samples concentrated in the Artemisia roots and soils. Failure to extract DNA from sagebrush root and soil samples reduced the overall power of the statistical comparisons between hosts and led to comparisons of unequal sample sizes for all statistical analyses. The large differences between Bromus and Artemisia roots at the UT site undoubtedly skewed the gamma and beta diversity calculations, at the very least. Artemisia produces numerous phenolic and terpenoid compounds (Brown et al. 1975; Jassbi et al. 2010). Secondary compounds such as these can contaminate DNA and interfere with DNA extraction (Friar 2005). Thus, the difficulty in extracting DNA from these samples, given the high concentration in sagebrush samples, was most likely a result of secondary compounds present in the roots that were also exuded into the soil.
Host had a significant effect on AMF community composition in both cultures and roots, most likely due to more than half of the AMF species colonizing roots and almost half of the AMF species isolated from cultures only occurring in one of the two host species. AMF composition in trap cultures indicated a site × host interaction effect, likely due to certain AMF species preferentially associating with one host over another from one site to the next (Fig. 1). While the indicator species preferentially associated with Artemisia across sites, other associations did not produce a consistent trend across sites (for example, D. spurca).
Two AMF species, A. trappei and V. viscosum, were more prevalent in cultures from Artemisia compared to Bromus across multiple sites. These indicator species for Artemisia may be important to Artemisia juveniles, but the present work did not separate the effects of individual AMF species on host growth and survival. Further, while V. viscosum occurred at all three sites, A. trappei was only found at two. So, site effects may also be an important consideration for which AMF species have a strong association with Artemisia. A previous AMF diversity study focused solely on Artemisia across multiple sites found that environmental conditions were more important than host in shaping the Artemisia AMF community (Allen et al. 1995). Thus, the differences across sites not due to host are most likely a reflection of the sites themselves.
Phylogenetic overdispersion is thought to signify competition for limited resources, while phylogenetic clustering is evidence of habitat filters selecting for certain traits possessed by closely related taxa (see Tofts and Silvertown 2000). Closely related AMF species are also believed less likely to coexist due to competitive exclusion as a result of niche overlap (Maherali and Klironomos 2007; Powell et al. 2009). Because sampling in the present study was focused on the two plant hosts and not on the entire AMF species pool existing at each site, comparing divergence in observed AMF assemblages against those occurring by chance from the broader community was not possible. Thus, clustering and overdispersion were not tested. However, AMF sequences isolated from Bromus roots were more phylogenetically dispersed than sequences isolated from Artemisia roots. This indicates that competition between AMF species for resources provided by Bromus is stronger than for Artemisia symbionts. If Bromus is a poor AMF host, it would be expected that the AMF community associating with Bromus would encounter competition for scarce resources, and thus be overdispersed. Alternatively, a host that provides greater resources would host an AMF community less dispersed. Bromus is considered a facultative AMF associate and does not receive significant benefits from nor provides significant benefits to AMF (Allen 1984; Busby et al. 2011). Artemisia is considered AMF dependent and exhibits high biomass responses to certain AMF species (Lindsey 1984). The possibility also exists that Artemisia exhibits greater specificity and may filter certain AMF species to create a clustered AMF community, as evidenced by the presence of associated indicator species.
Future work needs to determine what the observed AMF community changes mean to Artemisia in terms of AMF species loss, as AMF diversity and identity have a strong influence on plant community dynamics (van der Heijden et al. 1998). Further, diversity measures of AMF in trap cultures are only presence/absence data, as sporulation rates associated with a trap culture may not be reflective of relative densities of AMF associating with the initial plant host in the field. A measure of relative densities between AMF species associating with specific hosts would be useful to compare individuals, populations, and communities of plants.
Invasion of shrublands by invasive grasses is a global ecological concern, causing significant alterations to native vegetation on multiple continents (D’Antonio and Vitousek 1992). One important consideration in the restoration of invasive grass-dominated shrublands is the length of time that remnant resource islands, including AMF diversity hotspots, remain intact when invasive grasses are the sole host. Soil fertility differences associated with sagebrush resource islands have been shown to persist for 6 years (Bechtold and Inouye 2007), but island effects on nutrient cycling were reduced within 14 years after shrub removal (Burke et al. 1987). The differences in AMF species associations observed in this study may provide an initial assessment of the changes that might occur as Bromus replaces Artemisia in more heavily invaded rangelands.
Despite the presence of Bromus at the study sites, the plant community was still dominated by native vegetation and was not a Bromus monoculture that is typical of heavily invaded Artemisia shrublands. Changes to the AMF community associated with Bromus that were observed in what could be considered light invasions might become more pronounced as the native vegetation is wholly replaced by dense stands of Bromus that persist over a long period of time. This vegetation shift has already occurred on over 20 % of the historic sagebrush steppe plant community (Knapp 1996). However, comparing severe Bromus infestations with uninvaded Artemisia shrublands is difficult due to variance of sites and the importance of site history on both Bromus invasion and the AMF community. Overgrazing of native perennial grasses often allows Bromus to gain a foothold, and subsequent fires may remove remaining native vegetation (Klemmedson and Smith 1964; Knapp 1996; Young and Evans 1978). Both grazing (Eom et al. 2001) and fire (O’Dea 2007) alter the AMF community. Due to these confounding effects, this study chose to focus on coexisting populations of Bromus and Artemisia so that site differences between the populations could be removed. However, focusing on sites where Bromus coexists with native vegetation likely minimized the differences that might be observed when comparing Bromus monocultures to uninvaded Artemisia shrublands.
References
Allen EB (1984) VA mycorrhizae and colonizing annuals: implications for growth, competition, and succession. In: Williams SE, Allen MF (eds) VA mycorrhizae and reclamation of arid and semi-arid lands. Wyoming Agricultural Experiment Station Scientific Report SA1261. University of Wyoming, Laramie, pp 42–52
Allen EB, Allen MF, Helm DJ, Trappe JM, Molina R, Rincon E (1995) Patterns and regulation of mycorrhizal plant and fungal diversity. Plant Soil 170:47–62
Al-Qawari AA (2002) Relationships among nitrogen availability, vesicular-arbuscular mycorrhizae, and Bromus tectorum in disturbed rangeland sites in Colorado. Colorado State University, Dissertation
Azcón-Aguilar C, Palenzuela J, Roldán A, Bautista S, Vallejo R, Barea JM (2003) Analysis of the mycorrhizal potential in the rhizosphere of representative plant species from desertification-threatened Mediterranean shrublands. Appl Soil Ecol 22(2):9–37
Baker WL (2006) Fire and restoration of sagebrush ecosystems. Wildlife Soc B 34:177–185
Bechtold HA, Inouye RS (2007) Distribution of carbon and nitrogen in sagebrush steppe after six years of nitrogen addition and shrub removal. J Arid Environ 71:122–132
Blaszkowski J, Kovacs GM, Balazs TK, Orlowska E, Sadravi M, Wubet T, Buscot F (2010a) Glomus africanum and G. iranicum, two new species of arbuscular mycorrhizal fungi (Glomeromycota). Mycologia 102:1450–1462
Blaszkowski J, Wubet T, Harikumar VS, Ryszka P, Buscot F (2010b) Glomus indicum, a new arbuscular mycorrhizal fungus. Botany 88:132–143
Brown D, Asplund O, McMahon VA (1975) Phenolic constituents of Artemisia tridentata ssp. vaseyana. Phytochemistry 14:1083–1084
Burke IC, Reiners WA, Sturges DL, Matson PA (1987) Herbicide treatment effects on properties of mountain Artemisia soils after fourteen years. Soil Sci Soc Am J 51:1337–1343
Busby RR, Gebhart DL, Stromberger ME, Meiman PJ, Paschke MW (2011) Early seral plant species’ interactions with an arbuscular mycorrhizal fungi community are highly variable. Appl Soil Ecol 48:257–262
Busby RR, Paschke MW, Stromberger ME, Gebhart DL (2012) Seasonal variation in arbuscular mycorrhizal fungi root colonization of cheatgrass (Bromus tectorum), an invasive winter annual. J Ecosyst Ecography. doi:10.4172/2157-7625.S8-001
Callaway RM, Thelen GC, Rodriguez A, Holben WE (2004) Soil biota and exotic plant invasion. Nature 427:731–733
Callaway RM, Cipollini D, Barto K, Thelen GC, Hallett SG, Prati D, Stinson K, Klironomos J (2008) Novel weapons: invasive plant suppresses fungal mutualists in America but not in its native Europe. Ecology 89:1043–1055
D’Antonio CM, Vitousek PM (1992) Biological invasions by exotic grasses, the grass/fire cycle, and global change. Ann Rev Ecol Syst 23:63–87
Daniels BA, Skipper HD (1982) Methods for the recovery and quantitative estimation of propagules from soil. In: Schenk NC (ed) Methods and principles of mycorrhizal research. American Phytopathological Society Press, St. Paul, pp 29–35
Eom AH, Wilson GWT, Hartnett DC (2001) Effects of ungulate grazers on arbuscular mycorrhizal symbioses and fungal community structure in tallgrass prairie. Mycologia 93:233–242
Friar EA (2005) Isolation of DNA from plants with large amounts of secondary metabolites. Method Enzymol 395:3–14
Gustafson DJ, Casper BB (2006) Differential host plant performance as a function of soil arbuscular mycorrhizal fungal communities: experimentally manipulating co-occurring Glomus species. Plant Ecol 183:257–263
Harner MJ, Mummey DL, Stanford JA, Rillig MC (2010) Arbuscular mycorrhizal fungi enhance spotted knapweed growth across a riparian chronosequence. Biol Invasions 12:1481–1490
Hausmann NT, Hawkes CV (2009) Plant neighborhood control of arbuscular mycorrhizal community composition. New Phytol 183:1188–1200
Hausmann NT, Hawkes CV (2010) Order of plant host establishment alters composition of arbuscular mycorrhizal communities. Ecology 91:2333–2343
Hawkes CV, Belnap J, D’Antonio C, Firestone MK (2006) Arbuscular mycorrhizal assemblages in native plant roots change in the presence of invasive exotic grasses. Plant Soil 281:369–380
Helgason T, Daniell TJ, Husband R, Fitter AH, Young JPW (1998) Ploughing up the wood-wide web? Nature 394:431
Huber T, Faulkner G, Hugenholtz P (2004) Bellerophon; a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319
Jassbi AR, Zamanizadehnajari S, Baldwin IT (2010) Phytotoxic volatiles in the roots and shoots of Artemisia tridentata as detected by headspace solid-phase microextraction and gas chromatographic-mass spectrometry analysis. J Chem Ecol 36:1398–1407
Klemmedson JO, Smith JG (1964) Bromus (Bromus tectorum L.). Bot Rev 30:226–262
Knapp PA (1996) Bromus (Bromus tectorum L.) dominance in the Great Basin Desert: history, persistence, and influences to human activities. Global Env Change 6:37–52
Koske RE, Gemma JN (1989) A modified procedure for staining roots to detect VA mycorrhizas. Mycol Res 92:486–488
Koske RE, Tessier B (1983) A convenient, permanent slide mounting medium. Mycol Soc Am Newsl 34:59
Lee J-K, Tae M-S, Eom A-H, Lee SS (2003) Restriction analyses of PCR amplified partial SSU ribosomal DNA to distinguish arbuscular mycorrhizal fungi from other fungi colonizing plant roots. Mycobiology 31:68–73
Lindsey DL (1984) The role of vesicular-arbuscular mycorrhizae in shrub establishment. In: Williams SE, Allen MF (eds) VA mycorrhizae and reclamation of arid and semi-arid lands. Wyoming Agricultural Experiment Station Scientific Report SA1261. University of Wyoming, Laramie, pp 53–68
Mack RN (1989) Temperate grasslands vulnerable to plant invasions: characteristics and consequences. In: Drake JA (ed) Biological invasions: a global perspective. Wiley, London, pp 155–179
Maherali Z, Klironomos JN (2007) Influence of phylogeny on fungal community assembly and ecosystem functioning. Science 316:1746–1748
Marler MJ, Zabinski CA, Callaway RM (1999) Mycorrhizae indirectly enhance competitive effects of an invasive forb on a native bunchgrass. Ecology 1180–1186
McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA (1990) A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol 115:495–501
Morton JB (1988) Taxonomy of VA mycorrhizal fungi: classification, nomenclature, and identification. Mycotaxon 32:267–324
Mummey DL, Rillig MC (2006) The invasive plant species Centaurea maculosa alters arbuscular mycorrhizal fungal communities in the field. Plant Soil 288:81–90
Mummey DL, Stahl PD (2003) Spatial and temporal variability of bacterial 16 S rDNA-based T-RFLP patterns derived from soil of two Wyoming grassland ecosystems. FEMS Microbiol Ecol 46:113–120
O’Dea ME (2007) Influence of mycotrophy on native and introduced grass regeneration in a semiarid grassland following burning. Restor Ecol 15:149–155
Oehl F, Sieverding E, Ineichen K, Mäder P, Boller T, Wiemken A (2003) Impact of land use intensity on the species diversity of arbuscular mycorrhizal fungi in agroecosystems of central Europe. Appl Environ Microb 69:2816–2824
Opik M, Moora M, Liira J, Zobel M (2006) Composition of root-colonizing arbuscular mycorrhizal fungal communities in different ecosystems around the globe. J Ecol 94:778–790
Powell JR, Parrent JL, Hart MM, Klironomos JN, Rillig MC, Maherali H (2009) Phylogenetic trait conservatism and the evolution of functional trade-offs in arbuscular mycorrhizal fungi. P Roy Soc B-Biol Sci 276:4237–4245
Pringle A, Bever JD, Gardes M, Parrent JL, Rillig MC, Klironomos JN (2009) Mycorrhizal symbioses and plant invasions. Annu Rev Ecol Evol S 40:699–715
Reinhart KO, Callaway RM (2006) Soil biota and invasive plants. New Phytol 170:445–457
Renker C, Weibhun K, Kellner H, Buscot F (2006) Rationalizing molecular analysis of field-collected roots for assessing diversity of arbuscular mycorrhizal fungi: to pool, or not to pool, that is the question. Mycorrhiza 16:525–531
Reynolds JF, Virginia RA, Kemp PR, de Soyza AG, Tremmel DC (1999) Impact of drought on desert shrubs: effects of seasonality and degree of resource island development. Ecol Monogr 69:69–106
Richardson DM, Allsopp N, D’Antonio CM, Milton SJ, Rejmanek M (2000) Plant invasions—the role of mutualisms. Biol Rev 75:65–93
Sanders IR, Fitter AH (1992) Evidence for differential responses between host–fungus combinations of vesicular-arbuscular mycorrhizas from a grassland. Mycol Res 96:415–419
Schenk NC, Smith GS (1982) Additional new and unreported species of mycorrhizal fungi (Endogonaceae) from Florida. Mycologia 77:566–574
Seifert EK, Bever JD, Maron JL (2009) Evidence for the evolution of reduced mycorrhizal dependence during plant invasion. Ecology 90:1055–1062
Shah MA, Reshi ZA, Khasa DP (2009) Arbuscular mycorrhizas: drivers or passengers of alien plant invasion. Bot Rev 75:397–417
Shah MA, Reshi ZA, Rasool N (2010) Plant invasions induce a shift in Glomalean spore diversity. Tropic Ecol 51:317–323
SimonL Lalonde M, Bruns TD (1992) Specific amplification of 18 S fungal ribosomal genes from vesicular-arbuscular mycorrhizal fungal communities. App Env Microbiol 58:291–295
Stinson KA, Campbell SA, Powell JR, Wolfe BE, Callaway RM, Thelen GC, Hallett SG, Prati D, Klironomos JN (2006) Invasive plant suppresses the growth of native tree seedlings by disrupting belowground mutualisms. PLoS Biol 4(5):e140. doi:10.1371/journal.pbio.0040140
Stutz JC, Morton JB (1996) Successive pot cultures reveal high species richness of arbuscular endomycorrhizal fungi in arid ecosystems. Can J Bot 74:1883–1889
Sýkorová Z, Ineichen K, Wiemken A, Redecker D (2007) The cultivation bias: different communities of arbuscular mycorrhizal fungi detected in roots from the field, from bait plants transplanted to the field, and from a greenhouse trap experiment. Mycorrhiza 18:1–14
Tofts R, Silvertown J (2000) A phylogenetic approach to community assembly from a local species pool. P Roy Soc B-Biol Sci 267:363–369
Trent TD, Svejcar TJ, Blank RR (1994) Mycorrhizal colonization, hyphal lengths, and soil moisture associated with two Artemisia tridentata subspecies. Great Basin Nat 54:291–300
van der Heijden MGA, Boller T, Wiemken A, Sanders IR (1998) Different arbuscular mycorrhizal fungal species are potential determinants of plant community structure. Ecology 79:2082–2091
van Kleunen M, Dawson W, Schlaepfer D, Jeschke JM, Fischer M (2010) Are invaders different? A conceptual framework of comparative approaches for assessing determinants of invasiveness. Ecol Lett 13:947–958
Vogelsang KM, Bever JD (2009) Mycorrhizal densities decline in association with nonnative plants and contribute to plant invasion. Ecology 90:399–407
West NE (1983) Temperate deserts and semi-deserts, vol 5, ecosystems of the world. Elsevier, Amsterdam
Whisenant SG (1990) Changing fire frequencies on Idaho’s Snake River plains: ecological and management implications. In: McArthur ED, Romney EM, Smith SD, Tueller PT (eds) Proceedings—Symposium on Bromus Invasion, Shrub Die-off, and Other Aspects of Shrub Biology and Management. USDA Forest Service Intermountain Research Station General Technical Report INT-276, Ogden, pp 4–10
Whittaker RH (1972) Evolution and measurement of species diversity. Taxon 21:213–251
Wilson GWT, Hartnett DC (1998) Interspecific variation in plant responses to mycorrhizal colonization in tallgrass prairie. Am J Bot 85:1732–1738
Young JA, Evans RA (1978) Population dynamics after wildfires in sagebrush grasslands. J Range Manage 31:283–289
Zhang Q, Yang R, Tang J, Yang H, Hu S, Chen X (2010) Positive feedback between mycorrhizal fungi and plants influences plant invasion success and resistance to invasion. PLoS One 5(8):e12380. doi:10.1371/journal.pone.0012380
Acknowledgments
The authors thank H. Varani, B. Wolk, L. Bodistow, A. Broz, and the CSU Sequencing Lab (Colorado State University); J. Morton, R. Bills, S. Purin, and B. Wheeler (INVAM); J. Bever and W. Kaonongbua (Indiana University); and N. Raizen (University of Illinois). This research was funded by the United States Army A896 Direct Funded Research Program.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Materials
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 14 kb)
ESM 2
(DOCX 15 kb)
ESM 3
(DOCX 17 kb)
Fig. A1
Spatial distribution of host subsamples at each of the 3 study sites. A) CO site, B) UT site, C) WY site. Study sites are approximately 1,000 m2. Individual host subsamples do not add to 16 in most panes due to samples in close proximity to one another (< 1 m) with overlapping pins. (JPEG 88 kb)
Fig. A2
Fig. A2a1 Frequency of arbuscular mycorrhizal fungi DNA sequences isolated from Bromus tectorum (a) and Artemisia tridentata (b) roots and soils across three study sites: Colorado (1), Utah (2), and Wyoming (3). Black bars indicate root associations, white bars indicate soil associations. Naming convention for the sequences are numbered based on the last 3 digits of the accession number assigned by GenBank. (JPEG 154 kb)
Rights and permissions
About this article
Cite this article
Busby, R.R., Stromberger, M.E., Rodriguez, G. et al. Arbuscular mycorrhizal fungal community differs between a coexisting native shrub and introduced annual grass. Mycorrhiza 23, 129–141 (2013). https://doi.org/10.1007/s00572-012-0455-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-012-0455-x