Abstract
Arbuscular mycorrhizal (AM) fungal spores were isolated from field transplants and rhizosphere soil of Hedera rhombea (Miq) Bean and Rubus parvifolius L., which form Paris-type and Arum-type AM, respectively. DNA from the spore isolates was used to generate molecular markers based on partial large subunit (LSU) ribosomal RNA (rDNA) sequences to determine AM fungi colonizing field-collected roots of the two plant species. Species that were isolated as spores and identified morphologically and molecularly were Gigaspora rosea and Scutellospora erythropa from H. rhombea, Acaulospora longula and Glomus etunicatum from R. parvifolius, and Glomus claroideum from both plants. The composition of the AM fungal communities with respect to plant trap cultures was highly divergent between plant species. Analysis of partial LSU rDNA sequences amplified from field-collected roots of the two plant species with PCR primers designed for the AM fungi indicated that both plants were colonized by G. claroideum, G. etunicatum, A. longula, and S. erythropa. G. rosea was not detected in the field-collected roots of either plant species. Four other AM fungal genotypes, which were not isolated as spores in trap cultures from the two plant species, were also found in the roots of both plant species; two were closely related to Glomus intraradices and Glomus clarum. One genotype, which was most closely related to Glomus microaggregatum, was confined to R. parvifolius, whereas an uncultured Glomeromycotan fungus occurred only in roots of H. rhombea. S. erythropa was the most dominant fungus found in the roots of H. rhombea. The detection of the same AM fungal species in field-collected roots of H. rhombea and R. parvifolius, which form Paris- and Arum-type AM, respectively, shows that AM morphology in these plants is strongly influenced by the host plant genotypes as appears to be the case in many plant species in natural ecosystems, although there are preferential associations between the hosts and colonizing AM fungi in this study.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Two distinct arbuscular mycorrhizal (AM) morphologies, the Arum and Paris types, have been described on the basis of the presence or absence of intercellular hyphal growth in the root cortex (Gallaud 1905). The factors determining the development of these two morphological types of AM have remained largely unclear. In a survey of Arum- and Paris-type distribution in a Japanese sand-dune ecosystem (Ahulu et al. 2004), it was noted that the majority of pioneer plants, consisting mainly of annual and perennial herbs, formed Arum-type AM, whereas the majority of late succession plants, mostly evergreen shrubs and trees, formed Paris-type AM. Conversely, Yamato and Iwasaki (2002) in a study of AM morphology in some Japanese broad-leaved forests found that the tendency to form Paris-type AM was more prominent in herbaceous plants than in woody plants and suggested that, from an ecological point of view, Paris-type AM may be more advantageous for slow growing herbaceous understory plants. In fact, Brundrett and Kendrick (1990a,b) had previously suggested that slow root growth coupled with long root life span and gradual AM fungal colonization in Paris-type plants may be the best growth strategy under the low-nutrient and high-stress conditions that often prevail in natural ecosystems.
It is generally thought that host plant genotype influences AM morphology because it has been shown that the same AM fungus can form either Arum and Paris type depending on the host plant species (Jacquelinet-Jeanmougin and Gianinazzi-Pearson 1983; Bedini et al. 2000). However, Cavagnaro et al. (2001) provided evidence that in tomato, the colonizing fungus can also control AM morphology. Kubota et al. (2004) reported that tomato and cucumber grown in soil containing the same AM fungal inoculum formed both morphologies, sometimes even in the same root system, while Clethra barbinervis (Sieb. & Zucc.) only harbored the Paris-type AM.
Although AM fungi show no clear specificity toward the plants they colonize, there is evidence for preferential selectivity between host plants and mycorrhizal fungal genotypes in managed or natural ecosystems (Bever et al. 1996; Helgason et al. 2002; Vandenkoornhuyse et al. 2002, 2003; Johnson et al. 2003). However, this is not the rule in all natural ecosystems (Rosendahl and Stukenbrock 2004; Stukenbrock and Rosendahl 2005). Although spore diversity data in soil have given useful information concerning the effect of plant diversity on the AM fungal community (Bever et al. 1996; Eom et al. 2000; Johnson et al. 2003; Landis et al. 2004), it is becoming evident that there are differences between AM fungi colonizing plant roots and those found in soil (Clapp et al. 1995; Renker et al. 2005). Molecular techniques bypass the difficulty of accurately differentiating AM fungi to the species level from hyphal morphology within roots, which is accentuated by the existence of AM fungi that do not stain at all within the roots, or only very weakly using standard dyes (Morton and Redecker 2001). Among the different molecular techniques that have been used to determine AM fungal diversity in plant roots, nested PCR based on the large subunit (LSU) region of ribosomal RNA genes (rDNA) has been successfully applied to the detection of distinct AM fungal species colonizing plant roots in microcosm experiments (van Tuinen et al. 1998; Jacquot et al. 2000; Kjoller and Rosendahl 2001) and to study AM fungal diversity in roots from the field (Jacquot-Plumey et al. 2001; Turnau et al. 2001; Gollotte et al. 2004; Rosendahl and Stukenbrock 2004; Stukenbrock and Rosendahl 2005).
The aim of the present study was to use molecular analytical techniques to investigate the composition of AM fungal communities in roots of two sand-dune plant species that form morphologically different AM. Hedera rhombea (Miq) Bean is an evergreen climber that forms Paris-type AM, and Rubus parvifolius L. is a deciduous shrub that forms Arum-type AM (Ahulu et al. 2004). AM fungi trapped from the rhizosphere and field transplants of two plant species were molecularly tracked directly in field-collected roots by nested PCR.
Materials and methods
Study site
The experimental site was the coastal sand-dune ecosystem located in the campus of Niigata University, which was surveyed to determine the distribution of AM morphological types (Ahulu et al. 2004). It is a mixed stand of Japanese black pine (Pinus thunbergii Parl.), deciduous and evergreen broad-leaved trees, as well as annual and perennial herbs and grasses. The top 10 cm has the following characteristics: sandy texture, pH (H2O) 5.9, and available P content (0.002 N H2SO4 extractable) 40 μg P kg-1 soil(Ahulu et al. 2004). Six random 5 × 5 m2 plots approximately 20 m from each other were set up within the site.
Isolation of AM fungal spores
Four replicate field plants of H. rhombea and R. parvifolius were transplanted together with rhizosphere soil from each of the six plots selected within the study site. Each plant sample (shoot and roots together) was carefully excavated to avoid detaching roots from the shoots. The plant and soil trap cultures were used to trap and sporulate AM fungal species associated with the two plant species.
Plant trap culture
Replicate field plants of H. rhombea and R. parvifolius collected in replicates were washed thoroughly to remove all soil and grown in 1-l-capacity pots of sandy soil that had been sieved through a 2-mm mesh and pasteurized twice at 80°C for 45 min.
Soil trap
Rhizosphere soil collected from each replicate plant sample of each species was separately sieved through a 2-mm mesh. One-liter-capacity pots were each filled with 600 ml of twice-pasteurized (80°C for 45 min) sandy soil followed by 200 ml sieved rhizosphere soil from each replicate plant sample and finally with 200 ml of pasteurized sandy soil before sowing with five white clover (Trifolium resupinatum L.) seeds.
A total of 24 pots per plant species per trap culture method were kept in the greenhouse (14 h daylight and temperature at 25 and 20°C day and night, respectively) and watered daily with 50–100 ml tap water for 10 weeks. At the end of the growth period, 200 ml aliquot soil samples were taken from each replicate pot of the different methods, and AM fungal spores were extracted by wet sieving and sucrose centrifugation methods (Daniels and Skipper 1982).
Morphological identification of AM fungal spores
Spore characteristics were distinguished using procedures outlined by Morton (1998) based on morphological characteristics of spore size, wall ornamentation, hyphal attachment, spore wall structure, color and histochemical reactions in PVLG and PVLG + Melzer’s reagent.
Multiplication and maintenance of AM fungal isolates
The fresh AM fungal spores recovered from the trap pots and identified morphologically to genus level were used to produce clean pot cultures as follows. One-liter-capacity pots were filled with the sterilized sandy soil, and holes 3 cm deep and 3 cm wide were made in the center of each pot. Using sterile Pasteur pipettes, 10–30 spores of each isolated AM fungal type were placed in replicate pots, and the holes were covered up before sowing three to five white clover seeds over them. The pots were kept in the greenhouse and watered daily for 12 weeks. At the end of the growth period, 200-ml portions of soil from the trap pots were assessed for AM fungal species purity following the procedures outlined by Morton (1998) based on the spore characteristics to check the uniformity of spores from each pot. The fungal isolates were maintained on the living white clover plants and kept in the greenhouse until molecularly identified.
Field-collected roots
Root samples from three individual plants of H. rhombea and R. parvifolius were collected in July 2004 from each of the six randomly selected plots within the study site. Whole plants with intact root systems were carefully excavated, attached soil was shaken off gently, and roots from other plant species were eliminated before washing thoroughly with running tap water. Each root sample was divided into two parts and 0.5 g per sample cleared in 10% KOH at 95°C for 1 h, acidified for 20 min in 2% HCl, and stained in 0.05% trypan blue (Phillips and Hayman 1970). Root fragments were mounted on slides and examined microscopically to determine AM morphology and frequency of mycorrhiza in the root system (F%) using the method of Trouvelot et al. (1986). The data obtained were analyzed statistically by ANOVA followed by Bonferroni post-tests to make comparisons between means after arcsine transformation using GraphPad Prism version 4.00 for Windows, GraphPad Software (San Diego, CA, USA, http://www.graphpad.com). The remaining roots were dried separately at 50°C for 48 h and kept under airtight conditions until used for molecular analysis of AM fungal diversity.
Extraction of DNA from spores and roots
For molecular identification of the AM fungi trapped and maintained in pure culture on white clover, 10–50 identical spores were placed individually in 0.5 ml Eppendorf tubes, crushed with micropestles in 10 μl sterile distilled water, and heated at 95°C for 5 min. The extracts were diluted and used as DNA template for PCR. Similarly, DNA was extracted from spores of a reference INVAM isolate of Scutellospora erythropa (Koske & Walker) (MA453B).
To determine AM fungi colonizing field-collected roots, 1.0 g aliquots of dried samples were ground separately in liquid nitrogen. Twenty-milligram samples were transferred to 1.5 ml Eppendorf tubes, and activated charcoal was added to darkly colored root samples before suspending in 1 ml of DNA extraction buffer [20% TE (10 mM Tris–HCl pH 8, 1 mM EDTA), 80% (10 mM Tris–HCl), and 0.1 mM EDTA]. This is a modification of DNA extraction method described by van Tuinen et al. (1998). Activated charcoal was added to eliminate the pigments from the root extracts, which might be PCR-inhibiting polyphenolic compounds. The suspension was boiled for 10 min and centrifuged at 11,000×g for 10 min. Supernatants were diluted 10 and 100 times with TE buffer for a first PCR.
PCR amplifications of a partial LSU rDNA region
The primer pair LR1 (5′-GCATATCAATAAGCGGAGGA-3′) and FLR2 (5′-GTCGTTTAAAGCCATTACGT-3′) that is specific for fungi in general (van Tuinen et al. 1998; Trouvelot et al. 1999) was used for the first PCR amplification of the 5′-end of LSU rDNA sequences from spores, as described by Gollotte et al. (2004). PCR products were separated in a 1.4% agarose gel in TAE buffer (40 mM Tris, pH 7.8, 20 mM acetic acid, 2 mM EDTA), and DNA was visualized under UV light after staining with ethidium bromide (Sambrook et al. 1989) to confirm the presence and the size of the expected DNA fragment. In the case of the root samples, the first PCR products were diluted 100 times and used as templates for a second PCR with the primers LR1 (van Tuinen et al. 1998; Trouvelot et al. 1999) and FLR4 (Gollotte et al. 2004) under the same PCR conditions to increase the amount of DNA available for cloning.
Cloning and sequencing of PCR products
The PCR products generated from each AM fungus isolated from the trap cultures were cloned separately into the PCR 4 Topo vector (Invitrogen) according to the manufacturers’ recommendations. PCR products obtained from pooled root samples of each plant species were cloned separately. The presence of cloned inserts was checked by PCR, directly on the bacterial colonies diluted 100 times in water, using primers T3 and T7, and rDNA clone libraries were constructed for each AM fungal spore isolate and replicate root sample. Recombinant plasmids from five and ten bacterial colonies, respectively, from rDNA libraries for AM fungal spore isolates and replicate root samples of each plant species from the six plots were extracted and purified using Nucleospin® Plasmid (Macherey-Nagel) following the manufacturers’ recommendations. Each purified DNA sample was sequenced on both strands at MWG Biotech, Germany, for analysis.
Construction of phylogenetic trees
Insert sequences from plasmids were aligned with AM fungal DNA database sequences over 400 bp using ClustalW 1.8.1 and optimized manually using Bioedit version 4.8.4 software (North Carolina State University). Phylogenetic analyses were performed with the neighbor-joining algorithm using Mortierella polycephala Coem., Mortierella multidivaricata RK Benj., and Dissophora decumbens Thaxter as out-group species. Sequences from the AM fungal spore isolates and field-collected roots have been deposited in EMBL under the accession numbers AM040291–AM040435.
Statistics
The effect of location and trap culture method on abundance of different AM fungi isolated from rhizosphere and field transplants of the two plant species was assessed by two-way ANOVA, with the randomly selected plots and trap culture methods as factors, which examined the significance of interaction between the factors. Bonferroni’s multiple comparison test with a 95% confidence interval was then applied to make comparisons between means using GraphPad Prism version 4.00 for Windows, GraphPad Software (San Diego California USA, http://www.graphpad.com).
The effect of location and host plant species on frequency of occurrence of the different AM fungal genotypes in roots of the host plant species was assessed by univariate ANOVA following multivariate analysis of variance (MANOVA) analysis. Statistical analyses were carried out with NCSS and PASS software, Number Cruncher Statistical Systems (J. Hintze, Kaysville, UT, USA. http://www.ncss.com). The acceptable level of significance for this study was set at P < 0.05. LSD multiple range test was performed to test significant differences among the means provided that the previous ANOVA analysis gave a significant result. Both Hotelling–Lawley Trace statistics and Wilks’ lambda statistics were used to estimate the F ratio of the MANOVA analysis (Jongman et al. 1987).
Results
Community differentiation and spore abundance of AM fungi isolated in trap cultures
Five AM fungal spore types belonging to four described AM fungal genera (Glomus, Acaulospora, Gigaspora, and Scutellospora) were found in the soil and plant trap cultures of the selected plant species (Table 1). Glomus species were found in association with either plant species; however, Gigasporaceae was found only in trap cultures of H. rhombea, whereas Acaulosporaceae was limited to R. parvifolius. Spore morphological characters used to identify the isolates to genus level are shown in Table 2. The AM fungal spore isolates were referred to at the generic level until the molecular characterization was done due to the inability to confirm their species identities using only physical characteristics of the spores. All the isolates from trap cultures of R. parvifolius and H. rhombea were successfully multiplied and maintained on living white clover plants except spores of Glomus-like sp. 5 (Tables 1 and 2) isolated from soil trap culture of H. rhombea and Glomus-like species 6 (Tables 1 and 2) obtained from plant trap culture of R. parvifolius.
The composition of the AM fungal communities with respect to plant trap cultures was highly divergent between plant species, as revealed by the different spore types isolated from the plant species (Tables 1 and 2). It was not possible to statistically analyze the abundance of Scutellospora sp. and Gigaspora sp. on one hand and Glomus sp. 5 on the other hand, which were found only in the plant trap and soil trap cultures of H. rhombea, respectively (Fig. 1a,b). Similarly, the abundance of the undescribed species 6 that sporulated only in the soil trap cultures of R. parvifolius (Fig. 2b) could not be statistically analyzed. Glomus sp. 1 sporulated most abundantly in both soil and plant trap cultures of H. rhombea with significant differences in spore numbers between trap culture methods (P < 0.01) as well as plots (P < 0.001) (Fig. 1 and Table 3). Spore abundance of Glomus sp. 2 was significantly different between trap methods (P < 0.001) but not between plots (Fig. 1 and Table 3). Spore numbers of Acaulospora sp. and Glomus sp. 4 were significantly higher in soil trap culture with R. parvifolius (P < 0.01) but showed no significant difference between plots for either trap method (P > 0.1) (Fig. 2a,b and Table 4). Glomus sp. GE showed high significant differences in spore abundance between trap methods (P < 0.01) and plots (P < 0.01) alike (Fig. 2a,b and Table 4).
Molecular identification of AM fungal spores isolated in soil trap cultures
LSU rDNA sequences obtained from the AM fungal spore isolates Glomus sp. 1 and 2 from H. rhombea and isolates Glomus sp. 3 and 4 from R. parvifolius (Table 1) formed three fairly distinct phylogenetic clusters (A, B, and C) within the phylogenetic group formed by Glomus claroideum (Schenck & Smith) and Glomus etunicatum (Becker & Gerd.) (Fig. 3). For the isolates unique to R. parvifolius, sequences from Glomus sp. GE matched most closely with G. etunicatum, while those from the Acaulospora sp. grouped with Acaulospora longula (Spain & Schenck) (Fig. 3). For the Gigaspora and Scutellospora sp. recovered only from the plant trap cultures of H. rhombea (Table 1), sequences obtained from the Gigaspora sp. clustered with database sequences of Gigaspora rosea (Nicol. & Schenck) (Fig. 3), while spores of the Scutellospora sp. shared morphological features with spores of S. erythropa (Koske & Walker) (MA453B). Partial LSU rDNA sequences from Scutellospora sp. and S. erythropa grouped together and formed a distinct cluster with Scutellospora heterogama (Nicol. & Gerd.) (Fig. 3).
Root colonization and AM morphology
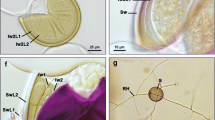
Percent AM fungal colonization of roots of H. rhombea (44.1 ± 4.6) and R. parvifolius (59.6 ± 5.8), collected from the six randomly selected plots, was significantly different (P < 0.05). Hyphal coils characteristic of Paris-type AM were observed within the root cortex of H. rhombea. Extensive longitudinal intercellular hyphae, typical arbuscules, and many vesicles were apparent in the root cortex of R. parvifolius, indicating an Arum-type AM.
Phylotyping of AM fungi in field-collected roots
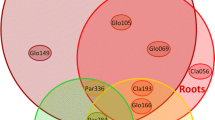
Phylogenetic analysis of rDNA sequences from the selected host plant species showed the presence of both culturable and unculturable species of AM fungi belonging to the genera Glomus, Acaulospora, and Scutellospora as well as an unculturable Glomeromycetes that did not group with any of the identified genera (Fig. 4). A total of 11 distinct phylogenetic clusters were formed (Fig. 4). Only sequences belonging to cluster D—Glomus microaggregatum 1 (most closely related to G. microaggregatum in the databases) and cluster J (an unculturable Glomeromycetes) were found to be in separated R. parvifolius and H. rhombea, respectively (Fig. 4). Clusters E, F, G, H, I, K, M, L, and N (Fig. 4) consisted of sequences obtained from roots of both plant species. With the exception of Gigaspora sp. isolated from spores H. rhombea (Fig. 3), sequences from the isolated AM fungal spores (Fig. 3) matched closely with sequences originating from roots of the two plant species within clusters I, K, M, L, and N which identified with S. erythropa, A. longula, G. etunicatum, G. claroideum 1, and G. claroideum 2, respectively (Fig. 4). On the other hand, sequences from clusters E, F, H, and G obtained from the roots of the two plant species showed close relationships with AM fungal species G. microaggregatum, Glomus intraradices, Glomus clarum, and an unculturable Glomus sp., respectively (Fig. 4).
Phylogenetic analysis to check the origin of the sequences obtained from field-collected roots of Hedera rhombea (Hedera) and Rubus parvifolius (Rubus) (boldface) in comparison to sequences obtained from AM fungal spores isolated from field plants and rhizosphere soil of H. rhombea and R. parvifolius (shaded) and sequences of identified AM fungi in the databases. Bootstrap values were estimated from 100 replicates. Numbers in parentheses indicate the experimental plots and beside them are the numbers of sequences of each AM fungal genotype obtained from the plot
Effect of location and host plant species on frequency of occurrence of different AM fungal genotypes in the roots of the two cooccurring plant species
The frequencies of occurrence of different AM fungal genotypes in roots of R. parvifolius and H. rhombea from the field confirmed differences in community structure depending on the location (Hotelling–Lawley Trace statistic F 55, 62 = 4.01, P < 0.0001; Wilks’ lambda F 55, 62 = 3.70, P < 0.0001) and host plant species (Hotelling–Lawley Trace statistic F 11, 14 = 83.6, P < 0.0001; Wilks’ lambda F 11, 14 = 83.6, P < 0.0001) (Table 5). Colonization of roots of R. parvifolius by AM fungal genotypes, G. intraradices, G. clarum, unculturable Glomus sp., and G. etunicatum, clusters F, H, G, and M, respectively, was strongly affected by location (Table 5 and Fig. 5a). Similarly, the frequency of occurrence of genotypes, G. microaggregatum 1 and G. claroideum 1, clusters D and L, respectively, in roots of R. parvifolius was affected by location but only to a lower extent (Table 5 and Fig. 5a).
Distribution of sequences in phylogenetic clusters present in the roots of AM host plant species H. rhombea and R. parvifolius. Sequence numbers of different AM fungal genotypes isolated in roots of host plants Rubus parvifolius (a) and Hedera rhombea (b) collected from six randomly selected plots within the study site
The host plant strongly affected the frequency of sequences identifying with G. intraradices, S. erythropa, and G. claroideum 2 (clusters F, I, and L) (P < 0.001) (Table 5, Fig. 5a,b). Significant differences with respect to host plant species were also observed in the frequency of sequences from clusters H and N, closely related to G. clarum and G. claroideum, respectively (P < 0.01), and cluster G, an unculturable Glomus sp. (P < 0.05) found in the roots of field-collected R. parvifolius and H. rhombea (Table 5, Fig. 5a,b).
Generally, sequences belonging to phylogenetic clusters D, F, and H corresponding to AM fungal species G. microaggregatum 1, G. intraradices, and G. clarum, respectively, occurred most frequently in the roots of R. parvifolius (Fig. 5a,b). In contrast, sequences of cluster I most closely related to Scutellospora erythropa were dominant in the roots of H. rhombea (Fig. 5a,b)
Discussions
The data obtained in the present work provide evidence that under natural conditions, R. parvifolius and H. rhombea not only accommodate the same AM fungal taxa in their root systems but also show preferential association with different taxa. A total of 11 different phylogenetic clusters of AM fungi belonging to the genera Glomus, Acaulospora, and Scutellospora as well as an unculturable Glomeromycotan fungus were molecularly identified in the roots of the two plant species. The species G. etunicatum, G. claroideum, A. longula, and S. erythropa, also identified from spores in trap cultures, and four unculturable Glomus phylotypes were present in field-collected roots of both plant species. G. claroideum was isolated from field transplants and rhizosphere soil, with both plants showing that G. claroideum is capable of colonizing the plant species for both the Paris and Arum types. This shows that AM morphology depends on the genotype of the host plants used in this study rather than the colonizing fungi. On the other hand, we also found significant differences in the composition of AM fungal species associated with the two host plant species R. parvifolius and H. rhombea, indicating that there is preferential selectivity between colonizing fungi and hosts. Because R. parvifolius and H. rhombea consistently form the Arum- and Paris-type morphologies, respectively, under natural growth conditions (Ahulu et al. 2004), these results reinforce the suggestion that AM morphology is highly influenced by the genotype of the plants used in this study.
The majority of AM fungal spores which were trap-cultured from H. rhombea and R. parvifolius were also molecularly identified in field-collected roots of the two plant species. However G. rosea, which was isolated from plant trap cultures of H. rhombea, was absent from field-collected root samples, while several AM fungal species, which did not sporulate in trap cultures, were detected in the field-collected roots of H. rhombea and R. parvifolius. These observations underline the bias imposed by trap culture techniques to identify root colonizers, and they strengthen previous conclusions of differences between AM fungal spores in soil and those actually within roots of colonizing individual plant species (Clapp et al. 1995; Kjoller and Rosendahl 2001; Renker et al. 2005). These differences found in trap cultures of the host plant species that form distinct AM morphologies further support the existence of host-specific differences in plant response to AM fungi and in fungal response to plants. Growth of AM fungal species has been found to depend on the associated host plant species (Johnson et al. 1992; Sanders and Fitter 1992; Eom et al. 2000). The relatively high diversity of Glomus species in the field-collected roots of both H. rhombea and R. parvifolius, as compared to the species of Acaulosporaceae and Gigasporaceae, is similar to other data on AM fungi naturally colonizing roots, as for example in the case of Agrostis capillaris L. and Lolium perenne L. in managed grassland (Vandenkoornhuyse et al. 2002; Gollotte et al. 2004).
Differences observed in AM fungal communities in the field-collected roots and trap cultures of the two host plant species with respect to location are in agreement with previous studies, which have reported variations in AM fungal composition and diversity in different ecosystems (Cuenca et al. 1998; Helgason et al. 2002). AM fungal taxa have a specific multidimensional niche that is determined by the plant species that are present at the site and by edaphic factors such as pH, moisture content, and phosphorus (P) and nitrogen (N) availability. As a result, there is a large between- and within-site variation in the composition of AM fungal taxa (Burrows and Pfleger 2002; Hart and Klironomos 2002).
Phylogenetic analysis of the LSU rDNA sequences from the AM fungal spores in this study clearly indicates intraspecific variability as well as overlaps among sequences obtained from morphologically variable spore isolates of G. claroideum from the two plant species. This supports the results of spore morphological and molecular analysis of isolates of G. etunicatum and G. claroideum isolated in pure culture from around the world (Rodriguez et al. 2005). The high levels of sequence diversity first noted in isolates of G. claroideum and G. etunicatum analyzed by multispore extractions have been confirmed by single spore DNA analysis, further supporting reports of high levels of polymorphisms present in the rRNA genes of AM fungal spores (Sanders et al. 1995; Clapp et al. 1995; Rodriguez et al. 2001). The results of the current study are therefore not fully in agreement with the earlier studies, which state that contamination of cultures such as several AM fungi present together in supposedly single species pot cultures on such high variability in sequences was obtained from one fungus (Jansa et al. 2002; Schüßler et al. 2003). However, that may explain the overlaps between sequences from the different isolates of G. claroideum in this study, which might be due to spore similarities, which make accurate visual differentiation of AM fungal spores difficult. Regarding molecular identification of AM fungi in field-collected roots, it is worth noting that a large degree of molecular variability has been reported even within a single individual AM fungus (a single spore), as discussed, for example, by Kuhn et al. (2001) and Sanders (2004). This makes it difficult to know if two or more different AM fungal sequences obtained from a root system originate from one, two, or several different AM fungal individuals.
In conclusion, although environmental biotic and abiotic factors might influence AM morphology in the root and even cause the formation of both Arum- and Paris-type AM in the roots of an individual host plant species, the data presented in this study indicate that under the natural conditions, the host plant genotype is the crucial factor for the plants studied. However, in spite of the occurrence of the same AM fungal genotypes in either plant species, preferential selectivity existed between species of family Glomaceae and Arum-type R. parvifolius on one hand and between S. erythropa belonging to the family Gigasporaceae and Paris-type H. rhombea on the other hand. A comparison of AM morphology of the plant species H. rhombea and R. parvifolius inoculated under pure culture conditions with the AM fungal species isolated in this study and grown under different environmental conditions may be helpful in the clarification of factors that affect AM morphology.
References
Ahulu EM, Nakata M, Nonaka M (2004) Arum- and Paris-type arbuscular mycorrhizas in a mixed pine forest on sand dune soil in Niigata Prefecture, central Honshu, Japan. Mycorrhiza 15:129–136
Bedini S, Maremmani A, Giovannetti M (2000) Paris-type mycorrhizas in Smilax aspera L. growing in a Mediterranean sclerophyllous wood. Mycorrhiza 10:9–13
Bever JD, Morton JB, Antonovics A, Schultz PA (1996) Host-dependent sporulation and species diversity of arbuscular mycorrhizal fungi in mown grassland. J Ecol 84:71–82
Burrows L, Pfleger L (2002) Arbuscular mycorrhizal fungi respond to increasing plant diversity. Can J Bot 80:120–130
Brundrett MC, Kendrick WB (1990a) The roots and mycorrhizae of herbaceous woodland plants. I. Quantitative aspects of morphology. New Phytol 114:457–468
Brundrett MC, Kendrick WB (1990b) The roots and mycorrhizae of herbaceous woodland plants. II. Structural aspects of morphology. New Phytol 114:469–479
Cavagnaro TR, Gao L-L, Smith FA, Smith SE (2001) Morphology of arbuscular mycorrhiza is influenced by fungal identity. New Phytol 151:469–475
Clapp JP, Young JPW, Merryweather JW, Fitter AH (1995) Diversity of fungal symbionts in arbuscular mycorrhizas from natural community. New Phytol 130:259–265
Cuenca G, Andrade Z, Escalante G (1998) Diversity of Glomalean spores from natural, disturbed and revegetated communities growing on nutrient-poor tropical soils. Soil Biol Biochem 30:119–125
Daniels BA, Skipper HD (1982) Methods for the recovery and quantitative estimation of propagules from soil. In: Schenck NC (ed) Methods and principles of mycorrhizal research. American Phytopathological Society Press, St. Paul, MN, USA, pp 29–35
Eom AH, Hartnet DC, Wilson GWT (2000) Host plant species effects on arbuscular mycorrhizal fungal communities in tallgrass prairie. Oecologia 122:435–444
Gallaud I (1905) Études sur les mycorrhizes endophytes. Revue General de Botanique 17:5–48, 66–83, 123–136, 223–239, 313–325, 425–433, 479–500
Gollotte A, van Tuinen D, Atkinson D (2004) Diversity of mycorrhizal fungi colonizing roots of grass species Agrostis capillaries and Lolium perenne in a field experiment. Mycorrhiza 14:111–117
Hart M, Klironomos JN (2002) Diversity of arbuscular mycorrhizal fungi and ecosystem functioning. In: van der Heijden MGA, Sanders IR (eds) Mycorrhizal ecology. Springer, Berlin Heidelberg New York, pp 225–242
Helgason T, Merryweather JW, Denison J, Wilson P, Young JPW, Fitter AH (2002) Selectivity and functional diversity in arbuscular mycorrhizas of co-occurring fungi and plants from a temperate deciduous woodland. J Ecol 90:371–384
Jacquelinet-Jeanmougin S, Gianinazzi-Pearson V (1983) Endomycorrhizas in Gentianaceae. I. The fungus associated with Gentiana lutea L. New Phytol 95:663–666
Jacquot E, van Tuinen D, Gianinazzi S, Gianinazzi-Pearson V (2000) Monitoring species of arbuscular mycorrhizal fungi in planta and in soil by nested PCR: application to the study of the impact of sewage sludge. Plant Soil 226:179–188
Jacquot-Plumey E, van Tuinen D, Chatagnier O, Gianinazzi S, Gianinazzi-Pearson V (2001) 25S rDNA-based molecular monitoring of Glomalean fungi in sewage sludge-treated field plots. Environ Microbiol 3:525–531
Jansa J, Mozafar A, Anken T, Ruh R, Sanders I R, Frossard E (2002) Diversity and structure of AMF communities as affected by tillage in a temperate soil. Mycorrhiza 12:225–234
Johnson NC, Tilman D, Wedin D (1992) Plant and soil controls on mycorrhizal fungal communities. Ecology 73:2034–2042
Johnson D, Vandenkoornhuyse P, Leake JR, Gilbert L, Booth RE, Grime JP, Young JPW, Read DJ (2003) Plant communities affect arbuscular mycorrhizal fungal diversity and community composition in grassland microcosms. New Phytol 161:503–515
Jongman RH, Ter Braak CLF, van Tongeren OFR (1987) Data analysis in community and landscape ecology. PUDOC, Wageningen, The Netherlands
Kjoller R, Rosendahl S (2001) Molecular diversity of glomalean (arbuscular mycorrhizal) fungi determined as distinct Glomus specific DNA sequences from roots of field grown peas. Mycol Res 105:1027–1032
Kubota M, McGonigle TP, Hyakumachi M (2004) Co-occurrence of Arum-and Paris-type morphologies of arbuscular mycorrhizae in cucumber and tomato. Mycorrhiza 15:73–77
Kuhn G, Hijiri M, Sanders IR (2001) Evidence for the evolution of multiple genomes in arbuscular mycorrhizal fungi. Nature 414:745–748
Landis FC, Gargas A, Givnish TJ (2004) Relationships among arbuscular mycorrhizal fungi, vascular plants and environmental conditions in oak savannas. New Phytol 164:493–504
Morton JB (1998) Taxonomy of VA mycorrhizal fungi: classification, nomenclature and identification. Mycotaxon 32:267–324
Morton JB, Redecker D (2001) Two new families of Glomales, Archaeosporaceae and Paraglomaceae, with two new genera Archaeospora and Paraglomus, based on concordant molecular and morphological characters. Mycologia 93:181–195
Phillips JM, Hayman DS (1970) Improved procedures for cleaning roots and staining parasitic and vesicular-arbuscular mycorrhial fungi rapid assesment of infection. Trans Mycol Soc 55:158–160
Renker C, Blanke V, Buscot F (2005) Diversity of arbuscular mycorrhizal fungi in grassland spontaneously developed on area polluted by a fertilizer plant. Environ Pollut 135:255–266
Rodriguez A, Dougall T, Dodd JC, Clapp (2001) The large subunit ribosomal RNA genes of Entrophospora infrequens comprise sequences related to two different glomalean families. New Phytol 152:159–167
Rodriguez A, Clapp JC, Robinson L, Dodd JC (2005) Studies on the diversity of the distinct phylogenetic lineage encompassing Glomus claroideum and Glomus etunicatum. Mycorrhiza 15:33–46
Rosendahl S, Stukenbrock EH (2004) Community structure of arbuscular mycorrhizal fungi in undisturbed vegetation revealed by analysis of LSU rDNA sequences. Mol Ecol 13:3179–3186
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York
Sanders IR (2004) Plant and arbuscular mycorrhizal fungal diversity—are we looking at the relevant levels of diversity and are we using the right techniques? New Phytol 164:415–418
Sanders IR, Fitter AH (1992) Evidence for differential responses between host–fungus combinations of vesicular–arbuscular mycorrhizas from grassland. Mycol Res 96:415–419
Sanders IR, Alt M, Groppe K, Boller T, Wiemken A (1995) Identification of ribosomal DNA polymorphisms among and within spores of Glomales—application to studies on the genetic diversity of arbuscular mycorrhizal fungal communities. New Phytol 130:419–427
Schüßler A, Schwarzott D, Walker C (2003) Glomeromycota rRNA genes—the diversity of myths? Mycorrhiza 13:233–236
Stukenbrock EH, Rosendahl S (2005) Clonal diversity and population genetic structure of arbuscular mycorrhizal fungi (Glomus spp.) studied by multilocus genotyping of single spores. Mol Ecol 14:743–752
Trouvelot A, Kough JL, Gianinazzi-Pearson V (1986) Mesure du taux de mycorhization VA d’un système radiculaire. Recherche de méthodes d’estimation ayant une signification fonctionnelle. In: Gianinazzi-Pearson V, Gianinazzi S (eds) Physiological and genetical aspects of mycorrhizae. INRA, Paris, pp 217–221
Trouvelot S, van Tuinen D, Hijiri M, Gianinazzi-Pearson V (1999) Visualization of ribosomal DNA loci in spore interphasic nuclei of Glomalean fungi by fluorescence in situ hybridization. Mycorrhiza 8:203–206
Turnau K, Ryszka P, Gianinazzi-Pearson V, van Tuinen D (2001) Identification of arbuscular mycorrhizal fungi in soils and roots of plants colonizing zinc wastes in Southern Poland. Mycorrhiza 10:169–174
Vandenkoornhuyse P, Husband R, Daniel TJ, Watson IJ, Duck JM, Fitter AH, Young PW (2002) Arbuscular mycorrhizal community composition associated with two plant species in a grassland ecosystem. Mol Ecol 11:1555–1564
Vandenkoornhuyse P, Ridgway KP, Watson IJ, Fitter AH, Young JPW (2003) Co-existing grass species have distinctive arbuscular mycorrhizal communities. Mol Ecol 12:3085–3095
van Tuinen D, Jacquot E, Zhao B, Gollotte A, Gianinazzi-Pearson V (1998) Characterization of root colonization profiles by a microcosm community of arbuscular mycorrhizal fungi using 25S rDNA-targeted nested PCR. Mol Ecol 7:879–887
Yamato M, Iwasaki M (2002) Morphological types of arbuscular mycorrhizal fungi in roots of understorey plants in Japanese deciduous broadleaved forests. Mycorrhiza 12:291–296
Acknowledgments
This work was partly supported by the International Tropical Timber Organization (ITTO, travel grant to EMA). The authors are grateful to Diederik van Tuinen (INRA) for the helpful discussion and advice, to Odile Chatagnier and Valerie Monfort (INRA) for their technical assistance, and to Joe Morton (INVAM) for providing the culture of S. erythropa.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Matekwor Ahulu, E., Gollotte, A., Gianinazzi-Pearson, V. et al. Cooccurring plants forming distinct arbuscular mycorrhizal morphologies harbor similar AM fungal species. Mycorrhiza 17, 37–49 (2006). https://doi.org/10.1007/s00572-006-0079-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-006-0079-0