Abstract
Background
Gastric cancer (GC) is one of the most common causes of cancer deaths worldwide; however, reliable and non-invasive screening methods for GC are not established. Therefore, we conducted this study to develop a biomarker for GC detection, consisting of urinary microRNAs (miRNAs).
Methods
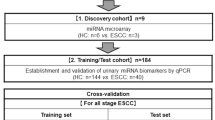
We matched 306 participants by age and sex [153 pairs consisting of patients with GC and healthy controls (HCs)], then randomly divided them across three groups: (1) the discovery cohort (4 pairs); (2) the training cohort (95 pairs); and (3) the validation cohort (54 pairs).
Results
There were 22 urinary miRNAs with significantly aberrant expressions between the two groups in the discovery cohort. Upon multivariate analysis of the training cohort, urinary expression levels of miR-6807-5p and miR-6856-5p were significantly independent biomarkers for diagnosis of GC, in addition to Helicobacter pylori (H. pylori) status. A diagnostic panel that combined these 2 miRNAs and H. pylori status distinguished between HC and GC samples with an area under the curve (AUC) = 0.736. In the validation cohort, urinary miR-6807-5p and miR-6856-5p showed significantly higher expression levels in the GC group, and the combination biomarker panel of miR-6807-5p, miR-6856-5p, and H. pylori status also showed excellent performance (AUC = 0.885). In addition, this biomarker panel could distinguish between HC and stage I GC patients with an AUC = 0.748. Urinary expression levels of miR-6807-5p and miR-6856-5p significantly decreased to undetectable level after curative resection of GC.
Conclusions
This novel biomarker panel enables early and non-invasive detection of GC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer (GC) is the third most frequent cause of cancer death in the world [1]. One reason for this is that it is difficult to diagnose GC at an early stage where symptoms may be mild or absent. Early GC detection by mass screening is important for reduction of mortality. With this purpose, upper gastrointestinal imaging (UGI) and upper gastrointestinal endoscopy (GIE) are often used during medical check-ups in East Asian countries with high GC incidence. Although neither examination was associated with reduced GC mortality in a prospective trial, GIE is generally superior to UGI for detection of GC [2, 3]. Pathological diagnosis using endoscopic biopsy samples is considered the gold standard for GC diagnosis. However, GIE requires trained endoscopists and is highly invasive with great potential for patient discomfort. Consequently, non-invasive screening methods for GC are needed; however, serum tumor markers, including carcinoembryonic antigen and carbohydrate antigen 19-9, have not been used in clinical practice due to their low sensitivity, especially for early-stage disease [4].
MicroRNAs (miRNAs) are short non-coding RNAs that consist of 20–25 nucleotides, which are believed to be involved in carcinogenesis and other immune-related diseases [5,6,7]. The pathway of circulating miRNAs is so complicated that they are transported as complex forms that bind with Argonaute proteins and high-density lipoproteins or with microvesicles such as exosomes [8], which are protected from degeneration. While many reports demonstrated that serum and plasma levels of miRNAs may be diagnostic biomarkers for GC [9,10,11,12,13,14,15,16,17], few reports have examined urinary miRNAs. Since office visits and special tools are needed for blood sampling, urine is an ideal health check screening medium because of its non-invasiveness and low cost. There have been only two reports examining urinary miRNA biomarkers for GC. One study found that urinary levels of miR-21-5p were elevated in patients with GC and decreased after surgery [18]. Another study found that urinary miR-376c showed 60% sensitivity and 64% specificity with an area under the curve (AUC) of 0.70 for diagnosis of GC [19]. These results suggest that urinary miRNA may be a useful diagnostic biomarker for GC. However, this use remains hypothetical because the two previous reports were pilot studies with very small sample sizes (around 30 individuals) and both studies lacked the validation of an independent cohort. Hence, the current study sought to establish a reliable urinary miRNA biomarker for diagnosis of GC. We herein report the efficacy of a novel GC diagnostic biomarker panel that used urinary miRNA.
Methods
Patients and study design
This study included 372 urine samples from 197 patients with GC and 175 healthy controls (HCs). All samples were collected from September 2012 to May 2017 at three Japanese institutions. We included males and females aged between 20 and 90 years old. Patients with GC (GC group) had an existing cancer diagnosis, established by histological and endoscopic findings, and no prior treatment on entry. HC participants had no neoplasms, as confirmed by a medical checkup including UGI or GIE, blood test, fecal occult blood test, chest radiograph, abdominal ultrasound, and medical examination (HC group). Persons with a previous cancer history (within 5 years) or multiple malignancies were excluded from participation. To ensure the accuracy and comprehensiveness of reporting in this case–control biomarker study, the present study complied with both the REMARK guidelines [20] and the STROBE statement [21]. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki (6th revision, 2008) and was approved by the ethics committee at each participating institution (Nagoya City University Hospital Institutional Review Board, Nagoya Daini Red Cross Hospital Institutional Review Board, and Okazaki Public Health Center Ethical Committee). Written informed consent was obtained from all participants. This study was registered with the University Hospital Medical Information Network Clinical Trials Registry (UMIN000021350).
Samples and definition
Urine and serum samples were collected from each patient with GC before tumor resection or chemotherapy. Samples were immediately stored at − 80 °C until analyzed, as previously reported [22,23,24]. All patients with GC were classified based on tumor node metastasis (TNM) staging (the Union for International Cancer Control version 7) [25]. Helicobacterpylori (H. pylori) status was analyzed using the serum anti-H.pylori immunoglobulin G antibody, the 13C-urea breath test, and the RAPIRUN test (Otsuka Pharmaceutical Co., Ltd., Tokyo, Japan).
Gastric cancer cell and exosome collection
Human GC cell lines, MKN45 and MKN74 (ATCC, Manassas, VA) were maintained in RPMI 1640 medium containing 10% fetal bovine serum (FBS) in a humidified atmosphere of 5% CO2 at 37 °C. We collected exosomes from cell culture media using ultracentrifugation [26]. Briefly, the cells with 60–80% confluency were washed twice with PBS and maintained in the media without FBS for 24 h. Then, 40 ml of conditioned media was collected and centrifuged at 300×g for 10 min. Next, the supernatant was centrifuged at 2000×g for 10 min followed by collecting and centrifuging the supernatant at 10,000×g for 30 min. The supernatant was ultracentrifuged at 100,000×g for 70 min using CP-100WX (Koki Holdings, Tokyo, Japan), followed by saving 200 µl for miRNA extraction. After discarding the supernatant, we added 40 ml of PBS and ultracentrifuged the tube at 100,000×g for 70 min. We discard the supernatant and added 200 µl of PBS to collect exosomes.
miRNA extraction
Urine was centrifuged at 10,000 rpm after thawing on ice and miRNAs were extracted from 200 µl (600 µl for microarray use) of the supernatant using miRNeasy Serum/Plasma Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol, with the exception that the column was washed three times using 80% ethanol before small RNAs were eluted in 14 µl of RNase-free water. Cellular miRNA and exosomal miRNA extraction was performed using the miRNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol and small RNAs were eluted in 60 µl RNase-free water. miRNAs from formalin-fixed paraffin-embedded (FFPE) tissues were extracted using the miRNeasy FFPE Kit (Qiagen). For extraction of miRNAs from culture media, we also used miRNeasy Serum/Plasma Kit.
Microarray assay
Cyanine-3 (Cy3)-labeled cRNA was prepared from urinary RNA (42 µl of the urine miRNA-containing solution from the GC and HC groups) using the miRNA Complete Labeling and Hyb Kit (Agilent, Santa Clara, CA, USA) according to the manufacturer's instructions. T4 RNA ligase and Cy-3-pCp were used for labeling the miRNA samples. Cy3-labeled miRNA samples were hybridized in a reaction volume of 45 µl containing 1 × Hi-PRM hybridization buffer and 1 × GE blocking agent following the manufacturer's instructions. The mixed samples were hybridized to Agilent Human miRNA Microarrays (G4872A) for 20 h at 55 °C in a rotating Agilent hybridization oven. After hybridization, microarrays were washed 5 min at room temperature with Gene Expression Wash Buffer 1 and 5 min with 37 °C GE Wash buffer 2 and then dried. The slide was scanned immediately after washing on the Agilent DNA Microarray Scanner (G2539A) using one color scan setting for 8 × 60 k array slides (Scan Area 61 × 21.6 mm, Scan resolution 3 µm, Dye channel is set to Green and Green PMT is set to 100%). The scanned images were analyzed with Feature Extraction Software 11.0.1.1 (Agilent) using default parameters (protocol AgilentHD_miRNA and Grid: 070156_D_F_20141006) to obtain background subtracted and spatially detrended processed signal intensities. The microarray raw data have been deposited in the Gene Expression Omnibus (GEO) database under number GSE128720.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Complementary DNA (cDNA) was synthesized using the TaqMan Advanced MicroRNA cDNA Synthesis Kit (Applied Biosystems, Foster, CA, USA) and 2 µl of the miRNA-containing solution, according to the manufacturer’s protocol. After creating a tenfold dilution of the solution, 2 µl of cDNA was used for qPCR. PCRs were performed in duplicate using the TaqMan Advanced MicroRNA Assay (Applied Biosystems) by 7500 Fast Real-Time PCR system (Applied Biosystems), totaling 10 µl of reaction solution with the following thermal cycle: 95 °C for 20 s, 40 cycles at 95 °C for 3 s, and 60 °C for 30 s. Cycle threshold (Ct) values were calculated to quantify miRNA expressions using the 2−∆Ct method. Internal normalization controls for qPCR of miRNAs in urine, serum, and samples from in vitro examinations were determined using a global mean normalization method with the microarray results [27]. Beta-actin was used as a normalizer when evaluating miRNA expression in FFPE tissues. The miRNA assays used in qRT-PCR are listed in Table S1.
Statistical analyses
The GC and HC groups were subjected to propensity score (PS) matching, made from a logistic regression model (age and sex). We randomly matched the two groups one by one, using the nearest-neighbor method with a caliper with a width of 0.05 of the standard deviation of the logit of the PS.
Mann–Whitney U test, Student’s t test, and the chi-squared test were used for detecting significant between-group differences, as appropriate. Spearman’s rank method with a coefficient (r) was used for evaluating correlations. Receiver operating characteristic curve analysis was used to calculate AUC for each biomarker, and the representative value was shown as the AUC value with 95% confidence intervals (CIs). Logistic regression modeling was used to estimate the odds ratio (OR) with a 95% CI and construct a composite score. This score was used to calculate the AUC for the combination biomarker. The compensated values (100 × 2−∆Ct) were used to calculate OR. Statistical analyses were carried out using R software (https://www.R-project.org/) or IBM SPSS statistics, version 25 (IBM Corp., Tokyo, Japan). All P values were two sided and P < 0.05 was considered statistically significant.
Results
Participants
Among the 372 participants, there were 197 patients with GC and 175 HCs. This produced 306 age- and sex-matched participants (153 pairs from each group) who were enrolled in this study. After that, the cohort was randomly divided into three groups: 8 participants (4 pairs) in the discovery cohort, 190 participants (95 pairs) in the training cohort and 108 participants (54 pairs) in the validation cohort. Moreover, additional validation was conducted using other samples including 64 participants (32 pairs) with serum samples (Fig. 1). Expectedly, the H. pylori positivity rate was significantly higher in the GC groups than in the HC group. However, there were no significant between-group differences for any other factor. More than 60% of patients with GC were stage I and around 50% could undergo curative endoscopic resection in this study (Table 1).
Urinary miRNA difference between HC and GC groups
First, to comprehensively detect differences in urine miRNAs between the HC and GC groups, we carried out an miRNA microarray analysis. Since one sample from the GC group was determined to be of poor quality given that the resultant signals were quite smaller than the other plates, we finally analyzed the microarray data from seven samples (HC = 4 and GC = 3) after excluding this sample. There were 22 miRNAs in urine with significantly aberrant expressions between the HC and GC groups, including differences of threefold or greater. Patients with GC showed significantly higher expression of 17 miRNAs, compared with the HC group, and lower expression of five miRNAs (Table S2).
Development of urinary miRNA biomarker
Among 22 candidate miRNAs that were identified using microarray analysis, we quantitated 10 miRNAs using qRT-PCR in the training cohort (Figure S1). Based on a global mean normalization method using microarray signals, miR-3610 and miR-4669 were determined as internal controls for qRT-PCR.
On univariate analysis, urinary expression levels of 2 miRNAs including miR-6807-5p were significantly higher in the GC group than in the HC group. On multivariate analysis, including H. pylori status, another significant factor for GC, urinary levels of miR-6807-5p and miR-6856-5p were also independent biomarkers for diagnosis of GC [miR-6807-5p, OR 1.58 (95% CI 1.06–2.34), P = 0.024; miR-6856-5p, OR 1.21 (95% CI 1.01–1.46), P = 0.042], as well as H. pylori status (Table 2). Based on these results, we established a diagnostic biomarker panel of GC consisting of urinary miR-6807-5p, miR-6856-5p, and H. pylori status using a logistic regression model. This urinary miRNA biomarker panel was satisfactorily able to distinguish patients with GC from HCs with an AUC = 0.736 (95% CI 0.663–0.810), which was higher than that observed with miR-6807-5p/6856-5p alone or both without H. pylori status (Fig. 2a). When the cutoff point was determined using Youden’s index, the logistic regression model showed 62.0% sensitivity, 83.2% specificity, and 72.7% accuracy for detection of GC.
Validation of urinary miRNA biomarker
Next, we validated this diagnostic panel using an independent validation cohort for general extrapolation. As shown in Table 3, both urinary miR-6807-5p and miR-6856-5p showed significantly higher expression levels in the GC group than in the HC group (P < 0.001), consistent with the training cohort. For the prediction of GC, the combination biomarker panel of urinary miR-6807-5p, miR-6856-5p, and H. pylori status also showed an excellent AUC = 0.885 (95% CI 0.823–0.948) with 76.9% sensitivity, 88.9% specificity and 81.5% accuracy in the validation cohort (Fig. 2b).
This combination biomarker panel showed an AUC = 0.774 (95% CI 0.721–0.827) in the whole cohort (Figure S2). We investigated whether H. pylori infection affects urinary level of miR-6807-5p or miR-6856-5p; however, no significant differences were found for urinary levels of these miRNAs between H. pylori-positive and -negative HCs (Figure S3).
Since early detection is essential for diagnostic biomarker on medical checkup, we next analyzed diagnostic potential for early stage. On comparing HCs with early-stage GC, urinary expression levels of both miR-6807-5p and miR-6856-5p were significantly higher in the stage I GC group than in the HC group (Fig. 3a). This biomarker panel was distinguished between HC and stage I GC with an AUC = 0.748 (95% CI 0.683–0.812), indicating that this biomarker panel can predict the presence of even early-stage GC (Fig. 3b). In addition, urinary levels of both miR-6807-5p and miR-6856-5p were significantly correlated to clinical stage (Figure S4A, B).
Moreover, we compared expression levels of urinary miR-6807-5p and miR-6856-5p before and after curative resection with endoscopy or surgery using urine samples obtained from 14 GC patients (Table S3). Remarkably, urinary levels of these 2 miRNAs decreased to undetectable range in all cases (Fig. 4a–c).
Analysis using serum, tissue sample, cell line and in silico
Since both miR-6807-5p and miR-6856-5p are unfamiliar miRNAs, we also measured expression levels of these miRNAs in other samples including serum and tissue samples. Interestingly, serum levels of miR-6807-5p and miR-6856-5p were also significantly higher in the GC groups compared with the HC group, as well as urinary levels (miR-6807-5p, P = 0.003; miR-6856-5p, P = 0.002) (Table S4). The combination panel of serum miR-6807-5p, miR-6856-5p, and H. pylori status showed excellent power with an AUC = 0.798 (95% CI 0.679–0.917) as well as the panel with urinary miRNAs (Figure S5). In addition, expression level of miR-6807-5p was significantly higher in primary tumor tissues than in adjacent normal tissues, and miR-6856-5p revealed the same tendency without significant level (Figure S6). Characteristics of GC patients for tissue miRNA analysis are shown in Table S5. To investigate whether these miRNAs originated from GC, we evaluated the expression of these miRNAs in exosomes, culture media, and cells bodies using MKN45 and MKN74. As expected, these miRNAs were detectable in all exosomes, culture media, and cell bodies (Figure S7).
To explore function of miR-6807-5p and miR-6856-5p, we conducted gene ontology (GO) analysis using the data of miRNA-target interactions from miRTarBase (https://miRTarBase.mbc.nctu.edu.tw/). In miRTarBase, 577 genes and 253 genes are listed as the target of miR-6807-5p and miR-6856-5p, respectively. The targets of 2 miRNAs showed the same tendency in classification of GO terms, suggesting that both miRNAs may control similar gene expression (Figure S8).
Discussion
Following our large study of three independent cohorts, we identified urinary miR-6807-5p and miR-6856-5p as non-invasive biomarkers for GC. Moreover, a diagnostic biomarker panel combined with H. pylori status demonstrated excellent diagnostic power.
Urine is one of the ideal samples for mass disease screenings due to its non-invasiveness and low cost. miRNAs originating from specific tissues are secreted into the extracellular environment and biological fluids, including urine [28]. Extracellular circulating miRNAs can be carried to the kidneys and discharged into urine by connecting with RNA-binding proteins or setting in microvesicles [8, 29]. Urinary miRNAs are considered relatively stable under various storage conditions [30].
Both miR-6807-5p and miR-6856-5p are novel miRNAs with a limited history of use. There have been no specific reports of the functions and targets related to both miR-6807-5p and miR-6856-5p. Whether detected miR-6807-5p and miR-6856-5p are secreted from the kidneys or other urothelial organs exactly remains unclear. In the current study, urinary levels of both miRNAs showed a positive association with disease stage of GC and remarkably decreased to undetectable range after curative resection even in the patients with early-stage GC, suggesting that these miRNAs would be derived from GC.
Moreover, the fact that both miR-6807-5p and miR-6856-5p are elevated in serum samples of patients with GC also might imply that these miRNAs originated from GC. In fact, the expression level of miR-6807-5p was higher in GC tissues significantly, compared with adjacent normal tissues and that of miR-6856-5p had the same tendency. In addition, we confirmed that both miRNAs were detectable in GC cell lines (MKN45 and MKN74) and secreted to culture media as exosomes and other forms. These results suggested that miR-6807-5p and miR-6856-5p might be secreted from GC tissues and excreted into urine. However, whether these miRNAs are specific to GC remains unclear. H. pylori infections play a well-known and crucial role in the carcinogenesis of GC [31]; however, H. pylori status alone is not sufficient for diagnosing GC due to the very high frequency of false positives. Since miR-6807-5p and miR-6856-5p were independent from H. pylori infection in the current study, combining these 2 miRNAs with H. pylori may overcome these issues.
Several reports have indicated that urinary miRNAs might be useful biomarkers for urological cancers and diseases [32,33,34]; however, a few studies showed urinary miRNAs as indicators of non-urological cancers including breast and pancreatic cancers [35, 36]. Only two studies have identified urinary miR-21-5p and miR-376c as diagnostic biomarkers of GC [18, 19]. Although these two studies raised the possibility of urinary miRNA as a GC biomarker, the utility of miR-21-5p and miR-376c remained unrealized due to the relatively small sample sizes and lack of independent validation of these preliminary results. Interestingly, neither urinary miR-21-5p nor miR-376c was detected in the urine of both patients with GC and HCs, for unknown reasons, in the current microarray analysis. This discrepancy may be due to differences in study scale, disease stage, and/or participant ethnicity.
Our current study included around 300 urine samples and consisted of a straightforward method comprising three independent phases: miR-6807-5p and miR-6856-5p were selected in the discovery cohort; a novel significant biomarker panel, including miR-6807-5p, miR-6856-5p, and H. pylori status, was established following multivariate analysis of the training cohort; the diagnostic potential of this biomarker panel was proven in the independent validation cohort. We, therefore, believe that the current study provides high-quality evidence.
The current study has a limitation. Future studies should elucidate the exact mechanisms between miRNAs and GC. However, consistent significant results of both miRNAs through all independent phases may produce strong evidence to support the use of these biomarkers for GC. Moreover, the GC cohort in this study featured many patients with early-stage GC: GCs more than 60% were stage I and most could be treated with endoscopic resection. The fact that a novel biomarker panel could detect very early-stage GC is a notable result of this investigation.
In conclusion, a novel biomarker panel consisting of urinary miR-6807-5p, miR-6856-5p, and H. pylori status enabled early and non-invasive detection of GC.
References
Fitzmaurice C, Akinyemiju TF, Al Lami FH, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2016: a systematic analysis for the global burden of disease study. JAMA Oncol. 2018;4:1553–688.
Tashiro A, Sano M, Kinameri K, et al. Comparing mass screening techniques for gastric cancer in Japan. World J Gastroenterol. 2006;12:4873–4.
Jun JK, Choi KS, Lee HY, et al. Effectiveness of the Korean National cancer screening program in reducing gastric cancer mortality. Gastroenterology. 2017;152:1319–28 e7.
Feng F, Tian Y, Xu G, et al. Diagnostic and prognostic value of CEA, CA19-9, AFP and CA125 for early gastric cancer. BMC Cancer. 2017;17:737.
Calin GA, Croce CM. Chromosomal rearrangements and microRNAs: a new cancer link with clinical implications. J Clin Invest. 2007;117:2059–66.
Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36.
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66.
Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–8.
Liu R, Zhang C, Hu Z, et al. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer. 2011;47:784–91.
Liu H, Zhu L, Liu B, et al. Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer Lett. 2012;316:196–203.
Li C, Li JF, Cai Q, et al. miRNA-199a-3p in plasma as a potential diagnostic biomarker for gastric cancer. Ann Surg Oncol. 2013;20(Suppl 3):S397–405.
Zeng Z, Wang J, Zhao L, et al. Potential role of microRNA-21 in the diagnosis of gastric cancer: a meta-analysis. PLoS One. 2013;8:e73278.
Zhu C, Ren C, Han J, et al. A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br J Cancer. 2014;110:2291–9.
Shin VY, Ng EK, Chan VW, et al. A three-miRNA signature as promising non-invasive diagnostic marker for gastric cancer. Mol Cancer. 2015;14:202.
Zhou X, Zhu W, Li H, et al. Diagnostic value of a plasma microRNA signature in gastric cancer: a microRNA expression analysis. Sci Rep. 2015;5:11251.
Qiu X, Zhang J, Shi W, et al. Circulating MicroRNA-26a in plasma and its potential diagnostic value in gastric cancer. PLoS One. 2016;11:e0151345.
Azarbarzin S, Feizi MAH, Safaralizadeh R, et al. The value of MiR-383, an intronic MiRNA, as a diagnostic and prognostic biomarker in intestinal-type gastric cancer. Biochem Genet. 2017;55:244–52.
Kao HW, Pan CY, Lai CH, et al. Urine miR-21-5p as a potential non-invasive biomarker for gastric cancer. Oncotarget. 2017;8:56389–97.
Hung PS, Chen CY, Chen WT, et al. miR-376c promotes carcinogenesis and serves as a plasma marker for gastric carcinoma. PLoS One. 2017;12:e0177346.
McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol. 2005;23:9067–72.
Vandenbroucke JP, von Elm E, Altman DG, et al. Strengthening the reporting of observational studies in epidemiology (STROBE): explanation and elaboration. Epidemiology. 2007;18:805–35.
Shimura T, Dagher A, Sachdev M, et al. Urinary ADAM12 and MMP-9/NGAL complex detect the presence of gastric cancer. Cancer Prev Res (Phila). 2015;8:240–8.
Shimura T, Ebi M, Yamada T, et al. Urinary kallikrein 10 predicts the incurability of gastric cancer. Oncotarget. 2017;8:29247–57.
Shimura T, Iwasaki H, Kitagawa M, et al. Urinary cysteine-rich protein 61 and trefoil factor 3 as diagnostic biomarkers for colorectal cancer. Transl Oncol. 2019;12:539–44.
Sobin LH, Gospodarowicz MK, Wittekind C. TNM classification of malignant tumours. 7th ed. Hoboken: Wiley; 2009.
Thery C, Amigorena S, Raposo G, et al. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006; Chapter 3:Unit 3 22.
Mestdagh P, Van Vlierberghe P, De Weer A, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10:R64.
Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–41.
Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9.
Mall C, Rocke DM, Durbin-Johnson B, et al. Stability of miRNA in human urine supports its biomarker potential. Biomark Med. 2013;7:623–31.
Uemura N, Okamoto S, Yamamoto S, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784–9.
Miah S, Dudziec E, Drayton RM, et al. An evaluation of urinary microRNA reveals a high sensitivity for bladder cancer. Br J Cancer. 2012;107:123–8.
Mengual L, Lozano JJ, Ingelmo-Torres M, et al. Using microRNA profiling in urine samples to develop a non-invasive test for bladder cancer. Int J Cancer. 2013;133:2631–41.
Cardenas-Gonzalez M, Srivastava A, Pavkovic M, et al. Identification, confirmation, and replication of novel urinary MicroRNA biomarkers in lupus nephritis and diabetic nephropathy. Clin Chem. 2017;63:1515–26.
Erbes T, Hirschfeld M, Rucker G, et al. Feasibility of urinary microRNA detection in breast cancer patients and its potential as an innovative non-invasive biomarker. BMC Cancer. 2015;15:193.
Debernardi S, Massat NJ, Radon TP, et al. Noninvasive urinary miRNA biomarkers for early detection of pancreatic adenocarcinoma. Am J Cancer Res. 2015;5:3455–66.
Acknowledgements
We thank Yukimi Hashidume-Itoh for handling of urine sample and Takako Onodera for data management of enrolled patients in this study (Department of Gastroenterology and Metabolism, Nagoya City University Graduate School of Medical Sciences). We also thank Masahide Ebi at Aichi Medical University, Tomonori Yamada at Nagoya Daini Red Cross Hospital and other clinical colleagues who assisted for the sample collection. This study was supported, in part, by Japan Agency for Medical Research and Development under Grant Number JP19lm0203005j0003 (to T. S.) and the Takeda Science Foundation (to T. S.)
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Iwasaki, H., Shimura, T., Yamada, T. et al. A novel urinary microRNA biomarker panel for detecting gastric cancer. J Gastroenterol 54, 1061–1069 (2019). https://doi.org/10.1007/s00535-019-01601-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00535-019-01601-w