Abstract
Schizophrenia, a serious psychological disorder, causes auditory and visual hallucinations, and delusions in a person. Several studies have shown that schizophrenia imposes structural changes in human brain. Relative changes in the grey matter volume of the schizophrenia patients in comparison to healthy controls have been well explored. However, identification of relevant brain regions that exhibit grey matter atrophy and also aid in the classification of schizophrenic patients and healthy controls has not been extensively investigated. In this study, a novel application of the non-dominated sorting genetic algorithm has been developed to select a set of relevant features (voxels) that show grey matter changes in the brain regions attributable to schizophrenia. This study uses MRI data of 32 healthy controls and 28 schizophrenia patients. The results show notable shrink in the gray matter volume in the brain of the schizophrenia patients, mostly in inferior frontal gyrus, superior temporal gyrus, middle occipital gyrus, and insula. The proposed approach yields a mean classification accuracy close to 90% with a feature set having around 70 voxels. This study may open a means of investigation of underlying neurobiology of schizophrenic brain for effective clinical intervention.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Schizophrenia is a serious mental illness presenting symptoms of hallucinations and delusions related to auditory, visual, social and emotional dullness [5, 6]. Psychological and neurological disorders such as schizophrenia may induce structural changes in the human brain. In this direction, several neuroimaging studies [16, 29, 30, 44] have been reported, which investigate the structural changes with respect to grey matter volume (GMV) [16, 44], white matter (WM) tracts [25], and cerebrospinal fluid (CSF) [9]. In schizophrenia studies, grey matter analysis is an active area of research. Grey matter is a major component of the brain, mainly comprising of neuronal cell bodies. It includes the regions of the brain which control muscles, and sensory functions such as visual, auditory, speech, emotions, memory, and decision making. Grey matter deficits in the range of 2–4% have been found in the patients suffering from schizophrenia as compared to healthy subjects [23, 47]. Volumetric grey matter loss in several regions like insula and superior temporal gyrus regions of the brain has been reported in several cross-sectional studies on schizophrenia patients [16, 41, 44] using magnetic resonance imaging (MRI) [14, 26, 29, 32, 42].
Because of its ability to depict anatomy and soft tissue contrast in a non-invasive manner, MRI is used for studying structural changes. Wright et al. [46] studied the global and regional grey matter changes in the whole brain with the help of structural MRI data of 42 schizophrenia patients and 52 healthy controls. They found significant reductions in regional grey matter in the medial temporal lobe. Job et al. [17] reported grey matter deficiency in schizophrenia patients compared to healthy controls using voxel-based morphometry (VBM) analysis of grey matter volume determined from MRI of 34 first episode schizophrenia patients and 36 controls. Grey matter volumes of subcortical brain regions were studied by Khodaei et al. [18]. With the help of whole brain segmentation, they compared the brain volumes of the MRI data of 12 schizophrenia patients with an equal number of healthy subjects. They identified a reduction in brain volume in the patient group.
Gender influences brain anatomy and age-related structural changes associated with it. Ruigrok et al. [39] found significant differences in size and volume of the brain of males and females in terms of grey matter. Bora et al. [1] conducted a meta-analysis study on schizophrenia patients with bipolar disorder to observe the effect of gender on the grey matter. The authors reported that grey matter abnormalities were more pronounced in data sets having a skewed gender ratio in favour of male patients. In a region-of-interest based study to investigate the influence of gender on first-episode schizophrenia patients, Duggal et al. [11] found greater reduction in insular volume in female patients in comparison to male subjects. Similarly, aging also influences the structural properties of the human brain. Premkumar et al. [38] found that longer duration of illness was associated with GM deficit in several regions of schizophrenic brain.
The above-mentioned studies were based on group analysis of schizophrenic patients. However, individual differential diagnosis of schizophrenic patients and healthy subjects have also been an active area of research. Since MRI captures a detailed 3D image of brain anatomy, MRI images are voluminous (\(\approx {10} ^ 6\) voxels). The number of subjects being low in comparison to number of voxels, discriminative analysis cannot be carried out efficiently. To handle this problem of high dimensionality, several works [34, 40] employed feature selection techniques before proceeding to classification of schizophrenia patients and healthy controls. Efficient feature selection techniques lead to high classification accuracy and also helps in better identification of affected brain regions [8]. Lu et al. [27] proposed a classification of schizophrenia and healthy controls using support vector machine (SVM) with recursive feature elimination to find the changes in GM and WM in the brain. The classification model yielded 88.4% accuracy. Nieuwenhuis et al. [34] used whole grey matter density images for classification of schizophrenia and healthy subjects. Feature selection was performed by selecting the top-ranked features, based on absolute values of the element of weight vectors of the SVM model.
Feature selection of MRI data has also been performed using meta-heuristic approaches. Recently, in a study, Tas et al. [42] used Ant Colony Optimization (ACO) as a feature selection tool in discrimination of schizophrenia and schizo-obsessive disorder patients. Classification of 23 schizophrenia and an equal number of schizo-obsessive disorder patients was performed using MRI data. However, to the best of our knowledge, evolutionary approaches have not been explored for grey matter analysis of schizophrenia patients. Since traditional feature selection methods get stuck in local optima and/or incur high computational cost [48], an efficient global search technique may be better suited to solve the feature selection problem for MRI data sets. Evolutionary approaches which possess global search ability have established themselves as an important class of search algorithms that have problem-invariant searching capabilities, i.e., they can efficiently search for optimal or near optimal solutions in any problem domain.
Again, most of the schizophrenic studies cited above either focused on improving the classification accuracy or on the identification of relevant brain regions having grey matter atrophy. However, to the best of our knowledge, no study has modeled the problem as a multi-objective optimization problem in which one can investigate the effect of a minimal set of features on the classification accuracy of schizophrenic patients and healthy subjects. Also, the analysis of the trade-off feature set obtained throws light on the most relevant discriminating features that exhibit grey matter atrophy. Non-dominated Sorting Genetic Algorithm (NSGA-II) [10] has been the most popular evolutionary multi-objective algorithm for obtaining trade-off solutions in a variety of problem domains.
In this study, we explore the suitability of NSGA-II algorithm for generating trade-off solutions in the domain of neuroimaging for studying grey matter atrophy. It yields a high classification accuracy by using a very small set of features. Thus, we are able to obtain features (voxels) that identify the relevant regions of the brain exhibiting grey matter atrophy.
2 Material and methods
2.1 Dataset
The MRI data used in this study was obtained from the Functional Biomedical Informatics Research Network (FBIRN) Phase II dataset.Footnote 1 This was a multi-site neuroimaging dataset acquired across different centers [37]. In this data, all subjects had sufficient eyesight, regular hearing levels, and were able to perform the cognitive tasks. Healthy subjects were excluded if they had a recent or past history of head injury or any major medical illness. All the healthy subjects were free from antipsychotic exposure and they had no recent history of medication effect. Patients with schizophrenia and schizoaffective disorder meeting Diagnostic and Statistical Manual of Mental Disorders, fourth version revised (DSM-IV-TR) criteria were included in this FBIRN study. The research group of FBIRN had determined the symptom scores by using the Schedule for the Assessment of Positive Symptoms (SAPS) and Negative Symptoms assessment measures [19]. In this study, we have employed T1-weighted MRI images of schizophrenia and healthy subjects acquired using machine having 1.5 T with the scanning parameters as follows: \(256 \times 256\) matrix, 24 cm field of view (FOV), 160–170 slices to cover the entire head in sagittal orientation, and slice thickness ranging from 1.2 to 1.5 mm [37]. Demographic details of the utilized dataset is shown in Table 1.
2.2 Proposed approach
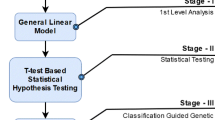
Our approach to identify grey matter atrophy in schizophrenic patients is outlined in Fig. 1. The steps of the proposed approach are described below.
2.2.1 Preprocessing
For preprocessing the raw MRI dataset, we have used the Statistical Parametric Mapping version 8 (SPM8, Wellcome Trust Centre for Neuroimaging, University College London, UK)Footnote 2 [36] toolbox in MATLAB-R2014a (Mathworks Inc., Natick, MA, USA).
Firstly, normalization of the MRI data was performed using the SPM’s default T1-weighted image by preserving the total amount of signal in the images keeping the voxel size as \(2 \times 2 \times 2\) mm\(^3\). Next, we have segmented the normalized images into modulated probability map volumes of grey matter, white matter and cerebrospinal fluid. Finally, as the present study focuses on the analysis of grey matter atrophy, smoothing was done on the modulated normalized grey matter volumes using full-width half maximum (FWHM) Gaussian filter at window size \(10 \times 10 \times 10\). Voxel values in the smoothed segmented volume lie between 0 and 1. Higher the voxel value, higher is the probability of the voxel to be a part of grey matter. Resulting smoothed modulated normalized grey matter volumes were then used for further analysis.
2.2.2 The application of voxel based morphometry
Voxel-based morphometry (VBM) is a technique to identify the structural changes in the brain. It is based on the statistical analysis of brain in voxel-by-voxel manner to trace the structural changes between two groups (population) of data. It is used to localize the changes, and make inferences about the group differences. The resulting output from the VBM analysis is a statistical parametric map which shows brain regions that differ significantly between the two populations. For statistical analysis, the method uses the general linear model (GLM) approach, which further uses the Gaussian random fields [13, 45].
For VBM analysis, normalized, segmented, and smoothed grey matter images were used. For the purpose of statistical analysis, we performed two sample t test over the two groups of data. While the first group of data comprised of schizophrenia patients, the second group was of healthy subjects. As we have hypothesized that reduction of grey matter concentration takes place in schizophrenia patients vis-a-vis healthy subjects, we set [\(-1 \& 1\)] as t-contrast that represents a right-tailed (group2 > group1) t test. We have taken the level of significance (\(\alpha\)), as \(\alpha = 0.001\), uncorrected, and 25 as threshold value for cluster size to eliminate less significant voxels.
Age is an important parameter as ageing eventually deteriorates the anatomical structure of the brain and also leads to functional decline due to the gradual death of neurons [31]. Also, the female human brain is slightly smaller in size than the male human brain [39]. Further, the male human brain is around 100 g heavier than a female human brain [28]. Thus, it is important to neutralize the effects of both age and gender in a study on the anatomical changes in the human brain caused by schizophrenia. Therefore, we have introduced two variables namely, age and gender, as covariates during the VBM analysis.
It is evident from Table 1 that the dataset has a mean age of 40.4 and 42.3 years for healthy and schizophrenic subjects respectively. The high standard deviation values suggest that there is a significant variability in age. Similarly, an imbalanced gender ratio can be seen in Table 1.
Finally, VBM analysis generates voxel-wise statistical parametric map that identifies the brain voxels that represent anatomical changes in the grey matter volume. This map has been used for further analysis.
2.2.3 The application of NSGA-II
As the VBM analysis produces a statistical parametric map involving a large number of affected or malfunctioning voxels (features) in different parts of the brain, we have used Non-dominated Sorting Genetic Algorithm (NSGA-II) [10] for identification of relevant voxels that exhibit grey matter atrophy.

NSGA-II is a popular multi-objective genetic algorithm. In addition to the genetic operators—selection, crossover and mutation, NSGA-II uses non-dominated sorting and crowding distance measure to generate trade-off solutions. The algorithm starts with initialization of a population of size N. After initialization, we identify the most potent chromosomes by employing tournament selection and the resulting chromosomes are crossed over and finally, mutated to generate an offspring population of size N. A new population comprising of the parent population and current offsprings is formed and sorted using the non-dominated sorting mechanism to select the best N chromosomes for next generation. The population of chromosomes thus sorted splits the entire combined population into multiple fronts (\(F_1,F_2, F_3,...\)). The first front (\(F_1\)) is the first level non-dominant set, while the second front (\(F_2\)) is dominated only by \(F_1\), and so on. This process is repeated for a given number of generations or till a specific criterion is met.
In our experiment, we have applied the NSGA-II as a feature selection tool in a bi-objective framework [7]. We have identified the following two conflicting objective functions for the study:
-
\(f^1\): Maximization of classification accuracy
-
\(f^2\): Minimization of number of features.
Based on experimental exploration with selection of different numbers of features, finally top 300 features (rank wise) obtained from the VBM analysis were selected. We created a population of binary chromosomes, each having a size of 300 bits. A one (1) at a position in the chromosome indicates the presence of the corresponding feature and zero (0) bit indicates its absence. Initially, for each chromosome, 20% of the randomly selected bits were set as one (1), and the remaining 80% of the bits as zero (0). In these experiments, the population size (N) was kept as 200. The fitness value of a chromosome for the first objective function, \(f^1\), was evaluated with the help of SVM with linear kernel. The fitness value of the second objective function, \(f^2\), for a chromosome was computed by counting the number of ones in the chromosome. Candidate chromosomes for the child population were generated using binary tournament selection. One-point crossover was applied on the selected chromosome set, followed by application of mutation at the rate of 0.01. The fitness values of the resulting child population (\(M_i\)) were computed, and a pooled population (\(T_i\)) comprising of the initial (\(P_i\)) and child population (\(M_i\)) was created. Non-dominated sort was performed on the pooled population (\(T_i\)) to find the set of non-dominated solutions. The best N chromosomes representing the trade-off solutions were passed to the next generation population (\(P_{i+1}\)). Algorithm 1 outlines the steps of standard NSGA-II used in this study. The maximum number of iterations (MaxGen) was set to 500. We have used 80–20 hold-out scheme for learning the decision model, i.e., for every run of the experiment, 80% of the data was randomly selected for training and the rest 20% for testing. The experiment was repeated 20 times to examine the variability in the results.
By applying NSGA-II as feature selection tool, we obtained a small set of relevant features which yielded high classification accuracy to distinguish the patients from the healthy controls. The features, obtained in each independent run of the experiments, were analysed and accumulated. These accumulated features were then backtracked to the original Cartesian coordinate system. To find the brain voxels corresponding to these coordinates, these Cartesian coordinates were mapped to the MNI coordinate system, and then finally, to Talairach coordinate system to map to the brain space. Thus, we could identify the most affected brain regions in schizophrenia patients.
3 Results
All the experiments conducted in this study have been implemented in MATLAB-R2014a. We have used SPM8 toolbox for preprocessing and VBM analysis of MRI images; libsvm [4] package for the classification task; Talairach Daemon [21] for mapping of brain regions; and Multi-image Analysis GUI (Mango) [22] for visualization of the identified regions in the brain. We have used the C-Support Vector Classification (C-SVC) [2] tool for the classification task. We have experimented with several values of the regularization parameter C in the range [0.01, 1000] in multiple of 10, and obtained the best classification accuracy results for \(C=100\).
In experiments 1(a) and 2(a), the set of voxels obtained from VBM analysis were fed into the SVM classifier. VBM analysis excluding the age and gender as covariates resulted in 16227 voxels. When age and gender were included as covariates, it resulted in 12035 voxels. Experiment 1(a), using 16227 voxels, generated a mean classification accuracy of 52.50%. While experiment 2(a), using 12035 voxels, resulted a mean classification accuracy of 60.83% (see Table 2).
In experiments 1(b) and 2(b), NSGA-II was applied on the 16,227 voxels and 12,035 voxels, respectively. Experiment 1(b) resulted in a mean classification accuracy of 88.33% using a mean feature set of size 72, and experiment 2b resulted in a mean classification accuracy of 90% using a mean feature set of size 65 (see Table 2).
In experiment 1(b), the features over all runs of the experiment were accumulated, and backtracked to the Talairach’s space enabling us to study brain regions at several level of hierarchy starting from hemisphere levels to the cell level. We found 21 brain regions as being affected. We also observed that about 17% of voxels were outside the of grey matter regions (see Table 3). Similarly, experiment 2(b) also resulted in accumulated features which were mapped to the brain regions in Talairach’s space. We identified 26 different brain regions as the most affected (see Table 4). The identified brain regions are visualized in Fig. 2.
Grey matter atrophy was computed as percentage of volume loss in schizophrenia patients as compared to healthy subjects (see Tables 3 and 4). Comparison of the grey matter values for each of the identified brain region showed a drop in grey matter concentration in the patient group. The negative values indicate loss of grey matter. The variation in grey matter loss has been visualized in boxplot diagrams as shown in Figs. 3, 4, and 5. At the hemisphere level, we found that both the left and right hemisphere showed reduced grey matter concentration in schizophrenia patients compared to healthy subjects. We found no changes in grey matter volume (GMV) in the brain stem or cerebellum region. The box plots in the Fig. 3 shows the difference in GMV at hemisphere level of the brain. At the lobular level of the brain, frontal lobe, occipital lobe, sub-lobar region and temporal lobe showed reduced grey matter (see Fig. 3) in the patient group. Gyral regions including inferior frontal gyrus, inferior temporal gyrus, superior temporal gyrus, middle occipital gyrus, middle temporal gyrus, middle frontal gyrus, sub-gyral, insula and extra-nuclear regions were found to have atrophy in grey matter volume (see Fig. 4). At cell level, Brodmann’s area 13, BA 18, BA 19, BA 20, BA 21, BA 37, BA 38, BA 47, BA 6 and BA 9 showed a drop in grey matter concentration of these areas in patient group compared to healthy controls (see Fig. 5). The box plots (Figs. 3, 4 and 5) show the magnitude of the atrophy of GMV in the schizophrenic brain. Tables 3 and 4 show the mean and median values of voxel intensity for schizophrenia patients showing negative (loss) change in grey matter volume compared to healthy controls.
Each of the experiments (experiments 1(b) and 2(b)), as depicted in Fig. 1, was repeated 20 times using 80–20 hold-out scheme to check robustness in classification accuracy and variability of selected features. The variations in mean classification accuracy and the size of feature set for each independent run are shown in Fig. 6a, b. The features obtained from all the runs of the algorithm were aggregated and the brain regions appearing in at least 80% of the runs were identified. For each of the regions so obtained, the Student’s paired t test was applied group wise across healthy subjects and schizophrenia patients, considering 95% confidence level. We found that all the identified brain regions show significantly difference in grey matter volume in schizophrenia patients resulting the P value \(<0.05\). The Table 5 shows the statistically significant difference in grey matter volume when compared between healthy and schizophrenia subjects.
4 Discussion
The study of structural changes in the brain of schizophrenia patients is an important research area. In this cross-sectional study, we have identified the brain regions, typically affected by schizophrenia, showing shrinkage in grey matter volume. With the help of voxel-based morphometry, we obtained a reduced set of features (voxels). When these features were fed as input to the bi-objective NSGA-II, a much reduced set of discriminating features was obtained. Using these features high classification accuracy was achieved. Further, the features obtained using the NSGA-II algorithm were successfully used to identify the affected brain regions.
As mentioned above, the experiments 1(b) and 2(b) were repeated 20 times to observe the changes in different feature sets obtained in independent runs. The most frequently occurring features (brain regions) were taken into account. We found little variation in terms of brain regions at different runs of the experiment. The voxels identified in experiment 1(b) when mapped to Talairach’s space showed 21 brain regions. Similarly, we obtained 26 brain regions from experiment 2(b). It was noted that a significant number of identified regions in experiment 2(b) can be attributed to age and gender as covariates. This suggests that these covariates should be included while conducting VBM analysis. Further, the grey matter loss in the identified regions for experiment 1(b) is less in comparison to experiment 2(b) (Tables 3 and 4).
Table 4 shows that both the left (− 0.20) and right hemisphere (− 0.18) exhibit grey matter loss in the schizophrenia patients. Studies [33, 43] suggest that the hemispheric grey matter loss is noticeable in early-onset schizophrenia patients compared to healthy controls. At the lobe level brain map, frontal lobe (− 0.24), occipital lobe (− 0.20), sub-lobar region (− 0.20) and temporal lobe (− 0.17) show grey matter atrophy in accordance to the previous studies on schizophrenia [3, 15, 20, 33]. Inferior frontal gyrus (− 0.24), inferior temporal gyrus (− 0.18), superior temporal gyrus (− 0.12), middle occipital gyrus (− 0.20), middle temporal gyrus (− 0.15), insula (− 0.21), sub-gyral (− 0.14) and extra-nuclear (− 0.20) regions show a reduction in grey matter concentration. In addition to regions mentioned in literature [3, 33, 46], we find that insula and extra-nuclear region also show a reduction in grey matter volume when compared to healthy controls. Fornito et al. [12] found that insula showed more consistent grey matter reduction. In another study, Lei et al. [24] suggested that lower volume in the extra-nuclear white matter may be a biological sign of schizophrenia. But in our study, we find extra-nuclear region showing prominent grey matter loss. We infer from our study that more research on the extra-nuclear region for identification of a change in grey matter is an open area of investigation. When we see the brain mapping at cell level, we find Brodmann’s areas (BA) showing a change in grey matter concentration. Brodmann’s areas like BA 18 (− 0.23) and BA 47 (− 0.24) show atrophy in grey matter volume. These regions are also mentioned in a previous study [35]. BA 38 (− 0.18) also shows a reduction in grey matter in the patient group. Literature suggests that this region is also affected in schizophrenia [3, 35]. We also found some regions like BA 13 (− 0.19), BA 19 (− 0.19), BA 37 (− 0.14), BA 20 (− 0.19), BA 6 (− 0.12), BA 9 (− 0.21) and BA 21 (− 0.15) showing a prominent loss of grey matter.
This study opens up a new approach for identifying the brain regions that exhibit grey matter atrophy in schizophrenia. Most of the regions identified in this study are in compliance with the existing studies on grey matter changes in schizophrenia patients. The identified regions may serve as cues for further investigation for effective clinical intervention with localized medication. Our study of schizophrenia patients and healthy controls using the MRI data also includes some new findings that might help in devising a more effective treatment strategy for schizophrenia. Although we have considered age and gender as two covariates in VBM, we have used them only to nullify their effect in this study. To understand the specific influences of age and gender on human brain, further studies may be conducted.
5 Conclusion
In this paper, we explored the applicability of evolutionary feature selection approach to study variations in grey matter volume in the brain regions of schizophrenia patients vis-a-vis healthy subjects. We also explored the effect of including age and gender as covariates in VBM analysis. The use of NSGA-II after the VBM analysis phase, led to improvement in classification accuracy from 52.5 to 88.33% when age and gender were not included as covariates during VBM analysis. Whereas, the use of NSGA-II after the VBM analysis, with age and gender as covariates, improved the classification accuracy from 60.83 to 90%. In summary, the NSGA-II based model that included age and gender as covariates during the VBM analysis yielded a better classification accuracy with a smaller set of features. The identified regions exhibiting grey matter atrophy may be further investigated to identify the causes that may play an adverse role in schizophrenia. Our study of schizophrenia patients and healthy controls using the MRI data also includes some new findings that might help in devising a more effective treatment strategy for schizophrenia.
References
Bora, E., Fornito, A., Yücel, M., Pantelis, C.: The effects of gender on grey matter abnormalities in major psychoses: a comparative voxelwise meta-analysis of schizophrenia and bipolar disorder. Psychol. Med. 42(2), 295–307 (2012)
Boser, B.E., Guyon, I.M., Vapnik, V.N.: A training algorithm for optimal margin classifiers. In: Proceedings of the fifth annual workshop on computational learning theory. ACM, New York, pp 144–152 (1992)
Cascella, N.G., Fieldstone, S.C., Rao, V.A., Pearlson, G.D., Sawa, A., Schretlen, D.J.: Gray-matter abnormalities in deficit schizophrenia. Schizophr. Res. 120(1), 63–70 (2010)
Chang, C.C., Lin, C.J.: LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol. 2:27:1–27:27 (2011) (software available at http://www.csie.ntu.edu.tw/~cjlin/libsvm)
Chatterjee, I.: Mean deviation based identification of activated voxels from time-series FMRI data of schizophrenia patients. F1000Research 7 (1615) (2018)
Chatterjee, I., Mittal, K.: A concise study of schizophrenia and resting-state FMRI data analysis. Qeios (599711). https://doi.org/10.32388/599711 (2019)
Chatterjee, I., Agarwal, M., Rana, B., Lakhyani, N., Kumar, N.: Bi-objective approach for computer-aided diagnosis of schizophrenia patients using FMRI data. Multimed. Tools Appl. 77(20), 26991–27015 (2018)
Chatterjee, I., Kumar, V., Sharma, S., Dhingra, D., Rana, B., Agarwal, M., Kumar, N.: Identification of brain regions associated with working memory deficit in schizophrenia. F1000Research 8 (124) (2019)
Chua, S.E., Cheung, C., Cheung, V., Tsang, J.T., Chen, E.Y., Wong, J.C., Cheung, J.P., Yip, L., Ks, Tai, Suckling, J., et al.: Cerebral grey, white matter and csf in never-medicated, first-episode schizophrenia. Schizophr. Res. 89(1), 12–21 (2007)
Deb, K., Pratap, A., Agarwal, S., Meyarivan, T.: A fast and elitist multiobjective genetic algorithm: NSGA-II. IEEE Trans. Evol. Comput. 6(2), 182–197 (2002)
Duggal, H.S., Muddasani, S., Keshavan, M.S.: Insular volumes in first-episode schizophrenia: gender effect. Schizophr. Res. 73(1), 113–120 (2005)
Fornito, A., Yücel, M., Patti, J., Wood, S., Pantelis, C.: Mapping grey matter reductions in schizophrenia: an anatomical likelihood estimation analysis of voxel-based morphometry studies. Schizophr. Res. 108(1), 104–113 (2009)
Friston, K.J., Holmes, A.P., Poline, J., Grasby, P., Williams, S., Frackowiak, R.S., Turner, R.: Analysis of FMRI time-series revisited. Neuroimage 2(1), 45–53 (1995)
Galderisi, S., Quarantelli, M., Volpe, U., Mucci, A., Cassano, G.B., Invernizzi, G., Rossi, A., Vita, A., Pini, S., Cassano, P., et al.: Patterns of structural mri abnormalities in deficit and nondeficit schizophrenia. Schizophr. Bull. 34(2), 393–401 (2007)
Giuliani, N.R., Calhoun, V.D., Pearlson, G.D., Francis, A., Buchanan, R.W.: Voxel-based morphometry versus region of interest: a comparison of two methods for analyzing gray matter differences in schizophrenia. Schizophr. Res. 74(2), 135–147 (2005)
Glahn, D.C., Laird, A.R., Ellison-Wright, I., Thelen, S.M., Robinson, J.L., Lancaster, J.L., Bullmore, E., Fox, P.T.: Meta-analysis of gray matter anomalies in schizophrenia: application of anatomic likelihood estimation and network analysis. Biol. Psychiatry 64(9), 774–781 (2008)
Job, D.E., Whalley, H.C., McConnell, S., Glabus, M., Johnstone, E.C., Lawrie, S.M.: Structural gray matter differences between first-episode schizophrenics and normal controls using voxel-based morphometry. Neuroimage 17(2), 880–889 (2002)
Khodaei, A., Hossein-Zadeh, G.A., Ananloo, E.S.: Comparison of volumes of subcortical regions in schizophrenia patients and healthy controls using MRI. In: 2015 2nd International Conference on Pattern Recognition and Image Analysis (IPRIA). IEEE, New York, pp 1–5 (2015)
Kim, D.I., Mathalon, D., Ford, J., Mannell, M., Turner, J., Brown, G., Belger, A., Gollub, R., Lauriello, J., Wible, C., et al.: Auditory oddball deficits in schizophrenia: an independent component analysis of the FMRI multisite function BIRN study. Schizophr. Bull. 35(1), 67–81 (2009)
Kong, L., Bachmann, S., Thomann, P.A., Essig, M., Schröder, J.: Neurological soft signs and gray matter changes: a longitudinal analysis in first-episode schizophrenia. Schizophr. Res. 134(1), 27–32 (2012)
Lancaster, J.L., Woldorff, M.G., Parsons, L.M., Liotti, M., Freitas, C.S., Rainey, L., Kochunov, P.V., Nickerson, D., Mikiten, S.A., Fox, P.T.: Automated Talairach atlas labels for functional brain mapping. Hum. Brain Mapp. 10(3), 120–131 (2000)
Lancaster, J.L., Laird, A.R., Eickhoff, S.B., Martinez, M.J., Fox, P.M., Fox, P.T.: Automated regional behavioral analysis for human brain images. Front. Neuroinform. 6, 23 (2012)
Lawrie, S.M., Abukmeil, S.S.: Brain abnormality in schizophrenia. a systematic and quantitative review of volumetric magnetic resonance imaging studies. Br. J. Psychiatry 172(2), 110–120 (1998)
Lei, W., Li, N., Deng, W., Li, M., Huang, C., Ma, X., Wang, Q., Guo, W., Li, Y., Jiang, L., et al.: White matter alterations in first episode treatment-naïve patients with deficit schizophrenia: a combined VBM and DTI study. Sci. Rep. 5, 12994 (2015)
Levitt, J.J., Nestor, P.G., Levin, L., Pelavin, P., Lin, P., Kubicki, M., McCarley, R.W., Shenton, M.E., Rathi, Y.: Reduced structural connectivity in frontostriatal white matter tracts in the associative loop in schizophrenia. Am. J. Psychiatry 1102–1111 (2017)
Liu, J., Li, M., Pan, Y., Wu, F.X., Chen, X., Wang, J.: Classification of schizophrenia based on individual hierarchical brain networks constructed from structural MRI images. IEEE Trans. NanoBiosci. 16(7), 600–608 (2017)
Lu, X., Yang, Y., Wu, F., Gao, M., Xu, Y., Zhang, Y., Yao, Y., Du, X., Li, C., Wu, L., et al.: Discriminative analysis of schizophrenia using support vector machine and recursive feature elimination on structural MRI images. Medicine 95(30) (2016)
Magoun, H.W., Marshall, L.: American Neuroscience in the Twentieth Century. CRC Press, London (2005)
Mitelman, S.A., Shihabuddin, L., Brickman, A.M., Hazlett, E.A., Buchsbaum, M.S.: MRI assessment of gray and white matter distribution in brodmanns areas of the cortex in patients with schizophrenia with good and poor outcomes. Am. J. Psychiatry 160(12), 2154–2168 (2003)
Mitelman, S.A., Brickman, A.M., Shihabuddin, L., Newmark, R.E., Hazlett, E.A., Haznedar, M.M., Buchsbaum, M.S.: A comprehensive assessment of gray and white matter volumes and their relationship to outcome and severity in schizophrenia. Neuroimage 37(2), 449–462 (2007)
Morrison, J.H., Hof, P.R.: Life and death of neurons in the aging brain. Science 278(5337), 412–419 (1997)
Nakamura, K., Takahashi, T., Nemoto, K., Furuichi, A., Nishiyama, S., Nakamura, Y., Ikeda, E., Kido, M., Noguchi, K., Seto, H., et al.: Gray matter changes in subjects at high risk for developing psychosis and first-episode schizophrenia: a voxel-based structural MRI study. Front. Psychiatry 4 (2013)
Narr, K.L., Bilder, R.M., Toga, A.W., Woods, R.P., Rex, D.E., Szeszko, P.R., Robinson, D., Sevy, S., Gunduz-Bruce, H., Wang, Y.P., et al.: Mapping cortical thickness and gray matter concentration in first episode schizophrenia. Cerebral Cortex 15(6), 708–719 (2004)
Nieuwenhuis, M., van Haren, N.E., Pol, H.E.H., Cahn, W., Kahn, R.S., Schnack, H.G.: Classification of schizophrenia patients and healthy controls from structural MRI scans in two large independent samples. Neuroimage 61(3), 606–612 (2012)
Pantelis, C., Velakoulis, D., McGorry, P.D., Wood, S.J., Suckling, J., Phillips, L.J., Yung, A.R., Bullmore, E.T., Brewer, W., Soulsby, B., et al.: Neuroanatomical abnormalities before and after onset of psychosis: a cross-sectional and longitudinal MRI comparison. Lancet 361(9354), 281–288 (2003)
Penny, W.D., Friston, K.J., Ashburner, J.T., Kiebel, S.J., Nichols, T.E.: Statistical Parametric Mapping: The Analysis of Functional Brain Images. Elsevier, New York (2011)
Potkin, S., Turner, J., Brown, G., McCarthy, G., Greve, D., Glover, G., Manoach, D., Belger, A., Diaz, M., Wible, C., et al.: Working memory and DLPFC inefficiency in schizophrenia: the FBIRN study. Schizophr. Bull. 35(1), 19–31 (2009)
Premkumar, P., Fannon, D., Kuipers, E., Cooke, M.A., Simmons, A., Kumari, V.: Association between a longer duration of illness, age and lower frontal lobe grey matter volume in schizophrenia. Behav. Brain Res. 193(1), 132–139 (2008)
Ruigrok, A.N., Salimi-Khorshidi, G., Lai, M.C., Baron-Cohen, S., Lombardo, M.V., Tait, R.J., Suckling, J.: A meta-analysis of sex differences in human brain structure. Neurosci. Biobehav. Rev. 39, 34–50 (2014)
Sharma, A., Ramkiran, S.: MRi based schizophrenia patient classification: a meta-cognitive approach. In: 2015 International Conference on Cognitive Computing and Information Processing (CCIP). IEEE, New York, pp 1–6 (2015)
Takahashi, M., Matsui, M., Nakashima, M., Takahashi, T., Suzuki, M.: Callosal size in first-episode schizophrenia patients with illness duration of less than one year: a cross-sectional mri study. Asian J. Psychiatry 25, 197–202 (2017)
Tas, C., Mogulkoc, H., Eryilmaz, G., Gogcegoz-Gul, I., Erguzel, T.T., Metin, B., Tarhan, N.K.: Discriminating schizophrenia and schizo-obsessive disorder: a structural MRI study combining VBM and machine learning methods. Neural Comput. Appl. 1–11 (2016)
Thompson, P.M., Vidal, C., Giedd, J.N., Gochman, P., Blumenthal, J., Nicolson, R., Toga, A.W., Rapoport, J.L.: Mapping adolescent brain change reveals dynamic wave of accelerated gray matter loss in very early-onset schizophrenia. Proc. Natl. Acad. Sci. 98(20), 11650–11655 (2001)
Torres, U.S., Duran, F.L., Schaufelberger, M.S., Crippa, J.A., Louzã, M.R., Sallet, P.C., Kanegusuku, C.Y., Elkis, H., Gattaz, W.F., Bassitt, D.P., et al.: Patterns of regional gray matter loss at different stages of schizophrenia: a multisite, cross-sectional VBM study in first-episode and chronic illness. NeuroImage Clin. 12, 1–15 (2016)
Worsley, K.J., Marrett, S., Neelin, P., Vandal, A.C., Friston, K.J., Evans, A.C., et al.: A unified statistical approach for determining significant signals in images of cerebral activation. Hum. Brain Mapp. 4(1), 58–73 (1996)
Wright, I., Ellison, Z., Sharma, T., Friston, K., Murray, R., McGuire, P.: Mapping of grey matter changes in schizophrenia1. Schizophr. Res. 35(1), 1–14 (1999)
Wright, I.C., Rabe-Hesketh, S., Woodruff, P.W., David, A.S., Murray, R.M., Bullmore, E.T.: Meta-analysis of regional brain volumes in schizophrenia. Am. J. Psychiatry 157(1), 16–25 (2000)
Zhao, Z.A., Liu, H.: Spectral Feature Selection for Data Mining. CRC Press, London (2011)
Acknowledgements
This work was supported by the research fellowship of Indranath Chatterjee from Council of Science and Industrial Research (CSIR), India having Grant number 09/045(1323)/2014-EMR-I. Data used in this work are taken from the Functional Biomedical Informatics Research Networks (FBIRN) data repository, under the following support: for function data, U24-RR021992, Function BIRN and U24 GM104203, Bio-Informatics Research Network Coordinating Centre (BIRN-CC). The data were obtained from the Functional BIRN Data Repository, Project accession number 2007-BDR-6UHZ1.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by B. Prabhakara.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chatterjee, I., Kumar, V., Rana, B. et al. Identification of changes in grey matter volume using an evolutionary approach: an MRI study of schizophrenia. Multimedia Systems 26, 383–396 (2020). https://doi.org/10.1007/s00530-020-00649-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00530-020-00649-6