Abstract
Infertility affects one out of seven couples around the world. Therefore, the best possible management of the in vitro fertilization (IVF) treatment and patient advice is crucial for both patients and medical practitioners. The ultimate concern of the patients is the success of an IVF procedure, which depends on a number of influencing attributes. Without any automated tool, it is hard for the practitioners to assess any influencing trend of the attributes and factors that might lead to a successful IVF pregnancy. This paper proposes a hill climbing feature (attribute) selection algorithm coupled with automated classification using machine learning techniques with the aim to analyze and predict IVF pregnancy in greater accuracy. Using 25 attributes, we assessed the prediction ability of IVF pregnancy success for five different machine learning models, namely multilayer perceptron (MLP), support vector machines (SVM), C4.5, classification and regression trees (CART) and random forest (RF). The prediction ability was measured in terms of widely used performance metrics, namely accuracy rate, F-measure and AUC. Feature selection algorithm reduced the number of most influential attributes to nineteen for MLP, sixteen for RF, seventeen for SVM, twelve for C4.5 and eight for CART. Overall, the most influential attributes identified are: ‘age’, ‘indication’ of fertility factor, ‘Antral Follicle Counts (AFC)’, ‘NbreM2’, ‘method of sperm collection’, ‘Chamotte’, ‘Fertilization rate in vitro’, ‘Follicles on day 14’ and ‘Embryo transfer day.’ The machine learning models trained with the selected set of features significantly improved the prediction accuracy of IVF pregnancy success to a level considerably higher than those reported in the current literature.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Infertility among couples is a significant problem nowadays. It has been reported that one out of seven couples tends to be infertile [11], and a recent survey estimated that around 48.5 million couples were affected by infertility in 2010 [27]. In vitro fertilization (IVF) is a popular method to treat infertile couples. IVF is a complex medically aided reproductive procedure which has been improving since the first pregnancy accomplished through this method in 1979 [16]. The IVF procedure is executed in the following steps [24]:
Collect oocyte and sperm of the couples separately for fertilization;
Fertilize sperm and oocytes in vitro;
Follow assisted fertilization methodologies, e.g., intracytoplasmic sperm injection (ICSI);
Transfer the embryo into the woman uterus;
Wait for 2 weeks and do a pregnancy test.
In the practice of IVF treatment, a rule of thumb is often applied that the collection of more than one oocyte for fertilization can results in increased success rate of fertilization. In order to obtain multiple healthy oocytes, ovulatory stimulants such as follicle stimulating hormone (FSH), clomiphene and GnRHa (gonadotropin releasing hormone analog) are often prescribed [16, 24]. Even though IVF treatment has passed over 30 years since its invention, according to the Centers for Disease Control and Prevention, USA, the overall pregnancy rate varies mostly between from 25 to 35% [16]. There are a number of factors, e.g., age, luteinizing hormone (LH) level and FSH level that influence the pregnancy outcome for the IVF patients [12]. All the influencing factors need to be considered to provide optimum and informed advice in relation to an individual couple’s case in taking up IVF treatment. The lack of efficient methods to prognosticate infertile couple might be the one of the reasons for such a low success rate. Researches have attempted to devise optimum decision-making models using data from the IVF patient records [6, 20]. Such sophisticated models would identify the contributing factors and their impacts, and extract the trend in IVF treatment which would otherwise remain unobserved by the physicians and clinicians. The knowledge gained from such models will help the medical practitioners to design treatment plans in order to increase the success rate for each infertile couple. However, the existing studies in the literature considered only limited (and fixed) number of attributes in predicting pregnancy outcome [16]. For effective analysis and patient advice, an automated method is required that will prognosticate infertile couples through analysis of the factors (attributes), e.g., FSH, sperm quality, fertilization rate, rate of cleavage, embryo quality and number of embryos transferred [24].
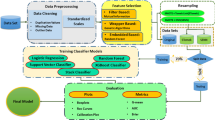
The complex modeling of the IVF treatment for outcome prediction and patient advising can be automated using machine learning models to aid practitioners to make informative decision. The models have the power to investigate and study the pattern in a large dataset’s population [15]. In addition, these models help to extract the implicit knowledge from the datasets. A variety of machine learning techniques, for example, artificial neural networks (ANN), support vector machines (SVM), decision tree (DT) and random forest (RF), and many others classifiers are widely used for prediction. In this research, we investigate both parametric and nonparametric classifiers to analyze IVF datasets. The nonparametric classifiers are accurate and robust in classification [3, 4, 32] and thus offer the advantage to rank the input features. We also employ a feature selection technique which when used in conjunction with a classifier increases its prediction accuracy. Overall, we make the following contributions in this paper: (i) identification of the most influential IVF attributes that affect pregnancy outcome, (ii) devising a machine learning-based system capable of predicting pregnancy outcome form a patient’s data with high accuracy, and (iii) such system will be extremely useful to practitioners to guide patients for IVF treatment.
The rest of this paper is organized as follows. Firstly, Sect. 2 discusses some related works on the predictive models for IVF treatment outcome, while Sect. 3 provides a brief overview of the implemented classification models in this research. Section 4 presents the hill climbing approach and its implementation in our problem, and Sect. 5 elaborates on the dataset and the performance metrics used in our experiments. Section 6 presents results showing the classifiers’ ability to predict IVF outcomes and their analyses, and finally Sect. 7 concludes the paper.
2 Related works
The early prediction of the pregnancy outcome of an IVF treatment is significant not only for patients but also for medical practitioners to assess possible outcomes. There are some studies in the literature that used machine learning techniques for IVF pregnancy prediction. One of the earlier studies was done by Kaufmann et al. [23] which utilized ANN. They first used t test to identify four attributes (patient’s age, number of eggs recovered, number of embryos transferred and whether there was embryo freezing) which they considered as the most influential ones and used those as the input features to train a neural network and attained an accuracy of 59% on the test dataset. Later on, Jurisica et al. [22] proposed a case-based reasoning system that exploited the previous experiences to suggest possible modifications to an IVF treatment plan in order to improve the overall success rates in future. Uyar et al. [36] investigated multiple types of classifiers to predict the outcome of embryo implantation in IVF using 18 features in total; nine of those were related to patient characteristics and another nine were related to embryo. The classifiers studied were SVM, DT, naive Bayes (NB), k-nearest neighbors (KNN), multilayer perceptron (MLP), and radial basis function network (RBF). They utilized receiver operating characteristics (ROC) to evaluate the performance of the classifiers, and better results were achieved using NB and RBF classifiers that attained an area under the ROC curve (AUC) of 0.739 (± 0.036) and 0.712 (± 0.036), respectively. I n a later study, the authors reduced the feature set to 11 by ranking the features as per their relative weight of importance and this produced slightly better performance by NB attaining an accuracy of 80.4% and AUC of 0.756 (± 0.036). In [30], the authors considered ten features and experimented with three different types of base classifiers, namely SVM, ANN and DT as well as their variants. They employed sequential forward floating selection (SFFS) method for feature selection and concluded that an ensemble method based on random subspace of DTs with only three features (patient’s age, subendometrial volume and endometrial vascularization/flow index) attained better prediction accuracy than other classifiers. Their best model attained 0.85 in AUC measure; however, their study was based on a dataset of relatively small number of treatment cycles.
Durairaj and Thamilselvan [13] used ANN to predict the success rate of IVF treatment. They collected a dataset with 14 attributes from which eight attributes were selected for training the model, and an overall accuracy of 73% was achieved. The work was later extended by developing an integrated a data mining approach of rough set theory (RST) and ANN using a bigger dataset with more attributes. RST was used for the data cleaning and reduction which produced nine influential attributes among the 27 attributes from the original dataset. The RST-based nine attributes were then used to train an ANN classifier which could successfully predict pregnancy outcomes in 90% cases. Milewski et al. [28] compared the capability of multivariable logistic regression (MLR) and multilayer perceptron (MLP) models in pregnancy prediction. Their study revealed that though MLR analysis was more suitable for theoretical interest, the MLP classifier was more useful in clinical prediction. An MLP classifier with 26 inputs, three layers of 40-6-2 hidden structure and one output to predict IVF outcome outperformed the MLR in the AUC measure by a margin of 0.061. Güvenir et al. [17] proposed a new technique, called success estimation using ranking algorithm (SERA), for estimating the success of a treatment using a ranking-based algorithm. The particular ranking algorithm used in their study is RIMARC (ranking instances by maximizing the area under ROC curve). They tested three models, namely SERA, NB and RF, where SERA outperformed others with an accuracy of 84.4% and AUC 0.833. Hafiz et al. [18] experimented with five techniques that included SVM, RF, recursive partitioning (RPART), adaptive boosting and one-nearest neighbor to predict the IVF outcome using a dataset containing 29 attributes, and reported that the RF and RPART outperformed other methods attaining AUC of 0.8423 and 0.8205, respectively. In a recent study, Ramasamy and Durairaj [33] proposed a hybrid algorithm which combined the ant colony and relative reduct, called ACRR algorithm, to preprocess the IVF data that reduced the number of attributes from 41 to 18. An application of ACRR in conjunction with NB, MLP, RBF and J48 classifiers demonstrated that NB attained the best performance with an accuracy of 90.35%.
In [9], the authors proposed a Bayesian network model based on an embryo-uterine model (EU) which assumes that both receptive uterus and viable embryo are necessary for a successful pregnancy. In their model, embryo viability and uterine receptivity were represented in probabilistic measure by two variables E (embryo) and U (uterine), respectively, and a Bayesian network was constructed consisting of a directed graph connecting the nodes that represent these variables. The parameters of the networks were estimated by the expectation–maximization (EM) method that maximizes the posterior probability of the parameters (i.e., probability of the parameters given some observed data). They concluded that their method of parameter estimation yielded better AUC (0.834) than the traditional MAP (most probable a posteriori) estimation. However, their conclusion was also based on a small dataset. In [8], the authors proposed a boosted tree-based predictive model for a patient’s first IVF treatment success, personalized to each patient’s clinical data. The model was built and tested using data from multiple clinics without requiring each clinic to satisfy specific mandatory format or a set of clinical protocols. The model could identify 42% of patients to have a personalized predicted live birth success rate > 45%. Although the study compared predictive power of their model with age-based predictive probabilities, no comparison was made with other artificial intelligence-based models proposed in the literature.
Interestingly, some studies focused on the embryo selection process to increase the success rate of IVF treatment. For example, Morales et al. [29] proposed a Bayesian-based classifier utilizing 20 features to help choosing the most promising embryos to be transferred to the woman’s uterus. Moreover, Guh et al. [16] proposed a hybrid method that integrates both genetics algorithm (GA) and decision tree (DT) techniques. Utilizing all the 67 IVF attributes used in the study, the DT achieved sensitivity and specificity of 21.5% and 95.0%, respectively, using fivefold cross-validation. The GA algorithm was used to identify the best set of features and the learning parameters of DT model simultaneously. This integration of GA and DT reduced the number of significant features to 28 (e.g., patient’s age, number of embryo transferred, number of frozen embryos and culture days of embryo). This process generated multiples DT models of which top ten models were selected. The best DT model had a predictive accuracy of 73.2%, and the corresponding sensitivity and specificity were 71.6% and 73.9%, respectively. In [26], the authors employed a neural network-based image analysis technique for quality assessment and scoring of the embryos or oocytes for selection in IVF treatment. The method was tested on 269 oocytes and 269 corresponding embryos from 104 women, and a comparison was made with the authors’ previously proposed algorithm called TRACE (total recognition by adaptive classification experiments). Though preliminary results from their study showed an AUC of 0.83 achieved by an ensemble of neural networks, further study with bigger datasets and comparison with other machine learning methods are necessary. Even though works in the literature show that machine learning techniques are promising in guiding IVF treatment plan, their performance needs to be improved to gain acceptance among medical practitioners.
Table 1 summarizes notable past and recent studies that attempted to apply various machine learning techniques to predict IVF outcome. As it can be seen from the table that the existing works still lack acceptable accuracy of prediction, the developed methods can be used reliably by the practitioners. Therefore, further works are needed to improve prediction accuracy to an acceptable level.
3 Overview of machine learning techniques
In this paper, the performance of different classifiers in predicting IVF pregnancy outcome is computed. In particular, we consider five classification models. Apart from ANN and SVM which are widely used in numerous biomedical problems, three types of decision trees, namely C4.5, CART and random forest, are implemented which have the characteristic of being nonparametric. A distinguishing feature of these trees compared to the other two classifiers is that trees do not need to compute the distance between input vectors to classify them. Below, we briefly describe each of the classification models that are used in this study.
3.1 Artificial neural networks (ANN)
An artificial neural network (ANN) is a computational intelligence method that can realize an input–output mapping even when the exact relationship between the input and output data of observations is not known or hard to formulate mathematically. An ANN usually consists of layers of computational elements called neurons, which are structured as an input layer, one or more hidden layers and an output layer. The layers are interconnected by modifiable weights, represented by links between layers. The size of the input and output layers is determined by the number of variables in input and output data, respectively. The number of neurons in each hidden layer is usually pre-specified by the user.
Multilayer perceptron neural network (MLP) is one of the most popular ANNs used as classifier. Each neuron multiplies its specific input value by the corresponding weight and then sums all the weighted inputs. Usually, a constant (called ‘bias’) is involved in the summation. The final output from the node is calculated by applying an activation function to the sum of the weighted inputs. In this fashion, the input signal is propagated through all layers to the output layer. MLP training algorithm is based on input forward propagation and error backward propagation followed by an update of the weights of the network using gradient descent methods. The error is calculated at the output layer as the difference between the actual and predicted outputs at each node. A detailed description of neural networks and MLP learning algorithm is available in the literature [19].
3.2 Support vector machine (SVM)
Support vector machine is based on the statistical learning theory developed by Vapnik [38]. SVM uses kernel that first maps the input space into a higher-dimensional feature space, and then a hyperplane is constructed in the transformed space to classify the dataset. The input data vectors/instances that appear on edges of the hyperplane are called as support vectors.
The training of a support vector machine demands the solution of a very sophisticated quadratic programming (QP) optimization problem. Sequential minimal optimization (SMO) [31] breaks the QP problem into a series of smallest possible QP problems for SVM training. More importantly, these small QP problems are solved analytically, which means the time-consuming numerical QP optimization as an inner loop is avoided. In addition, SMO requires a linear amount of memory in terms of training set size, making it suitable to handle large amount of training sets. Because the matrix computation is then avoided, computational complexity of SMO lies somewhere between linear and quadratic in the training set size for various problems, while that of the standard chunking of SVM algorithm lies somewhere between linear and cubic in the training set size. The training of SVM is influenced by the choice of regularization parameter (C), slack variables (ξ), and the kernel function used and its parameter. Details on these parameters and types of kernel functions can be found in [38].
3.3 C4.5 decision tree
A decision tree is defined as a data structure that looks like a tree. The tree consists of two components: nodes and leaves. From the leaves, the class label of a data sample can be identified, while each node of the tree selects one attribute that helps to decide its ultimate class label. Thus, the decision tree can be considered as a classifier, i.e., to identify the class label of an input data by analyzing the values of its features or attributes.
Several variations of the decision tree are available in the literature. One of the most widely used variations is C4.5 [32]. In C4.5, nodes are selected using the information gain of the attributes, which can be computed as follows:
Let us consider S to be the set of data samples, and \(C_{i}^{s}\) represents the total number of samples in S that is labeled as class Ci. If the total number of samples in S is |S|, then the entropy of S becomes:
Let us consider that the jth attribute Aj divides the set of data samples S to two subsets S1 and S2 considering a threshold value \(\theta\). Thus, the total entropy for S1 and S2 at \(\theta\) is
The best entropy is identified by varying the threshold \(\theta\) for each attribute whose information gain is computed as:
The attribute that has the maximum gain is selected as a node, and the dataset is split into two subsets which become the two branches of the node. This process is repeatedly executed until the node cannot be divided (i.e., the info gain is zero). These nodes are called as leaf nodes. The class label of a test data sample is identified by the class label of the leaf node for the respective test data sample.
3.4 Classification and regression trees (CART)
CART decision tree [3] produces binary trees where each node consists exactly of two branches, unlike C4.5. First of all, CART recursively partition the records in the training set into smaller subsets, e.g., training and validation. An optimal model can be selected through utilizing all the subsets and pruning the saturated tree. The optimal tree has the appropriate fit to the unknown test dataset (i.e., the data that have not been used in constructing the tree) [4]. Thus, the CART procedure guarantees a simple and robust tree.
3.5 Random forest (RF)
Random forest [5, 10] fits many classification trees to a dataset, and then these trees are combined to predict an outcome in response to an input. The RF algorithm starts off by choosing many bootstrap samples from the data. Approximately 63% of typical bootstrap of the original observations occurs at least once [2]. However, those observations in the original data that do not occur in the bootstrap sample are called out-of-bag observations. A classification tree is mapped to each bootstrap sample. However, at each node there will be only small number of randomly chosen variables that are available for the binary partitioning. Lastly, the trees will be fully grown and each will be utilized to predict the out-of-bag observations. The calculation of the predicted class of an observation is based on the majority vote of the out-of-bag predictions for that observation, with the ties split randomly.
The classification techniques outlined above have their own advantages and disadvantages and their performances vary in various applications and with complexity of data. In this work, our aim is to improve the overall accuracy of IVF pregnancy prediction and our approach is to rank each IVF attribute and gain insight into the most significant ones that might provide more information to improve the fertility rate.
4 Feature selection using hill climbing approach
The performance of a classifier can be enhanced through selection of a combination of important features that represent maximal separation between the classes [1]. In this paper, we applied a hill climbing wrapper algorithm to select the best subset of features for which the respective classifier can achieve an optimal classification performance. This algorithm initially selects the most influential feature from the dataset and then repetitively adds one feature at a time to the selected feature set that positively improves the classification results or provides the least reduction in classification accuracies. The steps of the algorithm are as follows:
- 1.
Initialize a feature set \(\varvec{F}^{{\prime }}\) as an empty set.
- 2.
Compute the classification accuracy for each of the individual features fi paired with \(\varvec{F}^{{\prime }}\).
- 3.
Select the feature \(f_{\text{best}}\) for which the best classification accuracy is achieved and add the feature to selected feature set \(\varvec{F}^{{\prime }}\).
- 4.
Repeat steps 2 and 3 until there is no remaining feature to be added to \(\varvec{F}^{{\prime }}\).
- 5.
Select the smallest subset of features \(\varvec{F} \subseteq \varvec{F}^{{\prime }}\) for which classification accuracy is maximum.
To avoid any biasness in the selection of the optimal feature subset, the whole dataset is first divided into ten folds (the dataset is described in detail in the following section). The data in the nine folds are then used for the training purpose, while the data in the remaining one fold are considered as the test data, which are kept unseen to the approach of feature selection in the current round. For classifier building, eight out of the nine folds are chosen randomly for training and one is used for model validation to prevent overtraining and overfitting the data. For each combination of nine folds–one fold, the steps described in the above algorithm, implemented through the procedure described below, are applied to extract a subset of features that are most influential.
For each round of training–testing, the hill climbing approach in the above algorithm is used where individual feature’s impact on the classification ability of a classifier is first determined. The importance of each feature is measured by considering how much classification accuracy is achieved by training the classifier with that feature alone. Since the process is repeated in ten rounds, the average classification accuracy over all rounds with respect to each feature is determined to assess its importance. Once the most influential feature has been identified, it is anchored in \(\varvec{F}^{{\prime }}\) and then paired with each other individual feature and the classification performance is again analyzed to identify which second feature, paired with the first selected one, provides the best classification performance. This process is iteratively executed to identify the best subset F of features that together yields the best classification accuracy.
It should be noted that, by adding a new feature to the previously selected best feature set might not enhance the overall performance of the classifier due to inherent differences among the features, and thereby, for some combination of the features, along with the underlying technique of the applied classifier, might not enhance the performance of the classifier further. We call the point from where the classification performance does not improve or shows a downward trend as the saturation point. The feature subset at the saturation point is selected as the best feature set for the considered classifier, and the classification performance is noted as the performance of the classifier.
Example
Let us consider the dataset \({\cal D}\) is divided into k folds as: \({\cal D_1}, {\cal D_2}, \ldots, {\cal D_k}\). First, the training dataset \({\cal T_{1}}\) is constructed combining the data in the folds from \({\cal D_1}\) to \({\cal D_9}\) and the remaining fold \({\cal D_{10}}\) is used as the test dataset. Using the individual feature \(f_{i} \left( {i = 1,2, \ldots , n} \right)\) in \({\cal T_{1}}\) to train a classifier \({\cal C}\), the importance of each feature in this round, measured in terms of accuracy attained using that feature alone, is recorded. This process is repeated ten times, each time using a different fold as the test dataset, and the average accuracy attained by each feature is determined. Let us assume this feature as \(f_{3 } \left( {f_{\text{best}} } \right)\). Now, each other feature paired with f3 is used to train the classifier and test its classification accuracy. The feature paired with the first selected feature (f3) that provides the highest classification performance is selected as the second selected feature. Assuming (f1) to be the second feature, the set {f3, f1} becomes the best combination of two features. This process is repeated till there is no influential feature to pair with. Thus, the resultant set for which the highest accuracy is attained is the selected feature subset for the classifier.
5 Experiments
5.1 Infertility data
The data used in this study were collected from an infertility clinic at Istanbul, Turkey. Data were collected for about 3 years in the period from March 2005 to January 2008 and consist of infertility treatment of 1729 patients, out of which embryos could not be transferred for 178 patients. Table 2 lists the IVF attributes used in this study along with their respective description explaining physical interpretation, type of the values they assume and their range in the collected dataset. Eighteen out of the total 25 attributes attain numerical values, while the rest seven attributes are of categorical in nature. Each numerical attribute was normalized by its maximum value in the dataset as given in the table. The categorical attributes only assume one of the possible two (‘Yes/No’ or ‘Primary or Secondary’) or multiple values as explained in the table. Each record in the dataset is labeled with the outcome of the IVF trial (successful/unsuccessful) and therefore is used as the class label during training and testing of the machine learning-based models. Out of 1551 patients whose embryos were transferred, 503 patients’ data were not useable after preprocessing (described below) leaving 1048 patients’ record to work with.
5.2 Missing values
Usually data collected from the real world might have missing values and/or noises. Data preprocessing is required to clean the data and replace missing values by meaningful values. The following two steps are adopted to clean (i.e., remove noisy data from the dataset) and identify meaningful value to replace any missing value.
- (i)
Data cleaning: The dataset used in this study contained many records where a number of attributes do not have any value/information. We refer to these as missing values. Records with many attributes as missing values were first removed from the dataset as these records did not provide valuable knowledge. In this study, records having more than 50% of attribute values missing were considered as noisy data and removed from the dataset (503 in total).
- (ii)
Data scrubbing: Data records which had missing values in less than 50% of attributes were kept in the dataset. To replace these missing values with some meaningful value, first a record with missing value was identified. Let us consider a data record di had missing value for the attribute \(f_{j}\). The distances between di and other individual data records were then computed. Since the value of \(f_{j}\) for \(d_{i}\) was not known, in computing the distance the respective attribute (i.e., \(f_{j}\)) was not considered. The average of the corresponding values of feature \(f_{j}\) of top k(= 5) data records that were at close distance to \(d_{i}\) was used to replace the value of \(f_{j}\) for data record \(d_{i}\). To compute the distance between two data records, Euclidean distance was used. After cleaning the data and replacing missing attribute, the resultant dataset contained 358 successful and 690 unsuccessful pregnancy cases.
5.3 Evaluation metrics
The clinical research often investigates the statistical relationship between symptoms (or test results) and the presence of a disease. When significant associations are found, it is useful to express the data in ways which are clinically relevant. Therefore, to evaluate the performances of the different classification models, the following performance metrics are adopted in this study. These metrics are widely adopted in many studies involving patient outcome [1].
Let,
TP = the number of true positives, i.e., the classifier predicted a positive pregnancy outcome when the actual outcome was also positive.
FP = the number of false positives, i.e., the classifier predicted a positive pregnancy outcome when the actual outcome was negative.
TN = the number of true negatives, i.e., the classifier predicted a negative pregnancy outcome when the actual outcome was also negative;
FN = the number of false negatives, i.e., the classifier predicted a negative pregnancy outcome when the actual outcome was positive;
N = TP + FP + TN + FN is the total number of samples in the test data.
A confusion matrix given in Table 3 summarizes the number of samples correctly or incorrectly classified by a binary classifier.
- (1)
Classification Accuracy (ACC) is one of the widely used performance metrics to evaluate a classifier. ACC is defined as the ratio of all samples that are classified correctly to the total number of test samples.
$${\text{ACC}} = \frac{{{\text{TP}} + {\text{TN}}}}{N}.$$(1)
-
(2)
Sensitivity (true positive rate) is defined as the ability of the classifier to accurately predict a successful pregnancy outcome from the attributes of the patient.
$${\text{Sensitivity}} = \frac{\text{TP}}{{{\text{TP}} + {\text{FN}}}}.$$(2)
-
(3)
Precision is proportion of the predicated successful pregnancy cases that are actually successful.
$${\text{Precision}} = \frac{\text{TP}}{{{\text{TP}} + {\text{FP}}}}.$$(3)
-
(4)
F-Measure (F) is the harmonic mean of precision and sensitivity, which is defined as:
$$F = \frac{{2*{\text{Sensitivity}}*{\text{Precision}}}}{{{\text{Sensitivity}} + {\text{Precision}}}}.$$(4)
-
(5)
Area under ROC (AUC) plots: The ROC curve is plotted as true positive rates or sensitivity at the y-axis, and the false positive rates (=FP/(TN + FP)) at the x-axis, while the threshold values are changed from 0 to 1. The AUC is the accumulated area covered by the ROC curve. An AUC less than 0.5 refers to a random performance, while the value close to 1 indicates very good performance.
6 Results and discussion
6.1 Model validation and testing
It has been shown in the literature that, for selecting a good classifier from a set of classifiers using a real-world dataset, tenfold cross-validation is the most suitable and widely used method [25], particularly for limited datasets [1]. By employing this technique for model building, validation and testing, the total dataset of 1048 IVF patients was divided into 10 nearly equal-sized mutually exclusive folds: eight folds containing 105 patients each and two folds containing 104 patients. The ratio of the ‘success’ and ‘failure’ in each fold was roughly the same as in the total dataset. As alluded in Sect. 4, each time eight folds were used as the training set to build models, one fold was used as the validation set to estimate the classification ability of the model, and the remaining one fold was used as the test set. The model with the smallest misclassification error rate on the validation set was then selected, and the test set was used to obtain an unbiased estimate of the classification ability of the model [34]. For the tenfold cross-validation scheme, the process was repeated ten times, each time with a different test set (test sets are disjoint). The average of the ten classification accuracy rates from all folds was then taken as an unbiased estimate of the model on the complete dataset and is reported in this section along with the standard deviation over the folds. The results of the ten folds were combined into a single ROC plot and AUC calculation for each classifier.
6.2 Model parameters
In neural networks, the performance can vary with the number of hidden layers, initial weights and learning parameters. We experimented with a three-layer MLP with the number of input units the same as the number of features (after feature selection): one hidden layer with a varying number of units (between 5 and 20) and a single output layer unit representing the class label (successful/unsuccessful). We found that the hidden of units roughly equal to (feature number + class labels)/2 produced the best results. Sigmoid activation function was used at the hidden and output layers, and the conjugate gradient backpropagation with Fletcher–Reeves algorithm [7] was used to train the MLP classifier. Learning rate and momentum factor were chosen as 0.3 and 0.2, respectively, and training was continued until accuracy on the validation set stated decreasing or the maximum epoch of 500 reached. Similarly, in SVM the parameters that influence the performance are the regularization parameter (C), slack variables (ξ) and degree (d) of the polynomial kernel function that we used in this study. The values for C and ξ were optimized using grid search [35], and d = 2 was found suitable. All simulations were carried out in a MATLAB environment using toolboxes (The MathWorks, Natick, MA) and Weka, 2017.
6.3 Influential features
Table 4 lists the important selected IVF attributes for each classifier by applying the hill climbing feature selection algorithm described in Sect. 4. It shows that different classifiers selected different sets of attributes because each attribute has varied influence on each classifier; however, some important attributes were chosen by all classifiers. Out of the 25 features, the number of features selected by MLP, SVM, C45, CART and RF is 19, 17, 12, 8 and 16, respectively. Analysis of the features selected by the different classifiers reveals the following interesting facts:
Two features: ‘Indication’ (of infertility factor) and ‘NbreM2’ (number of mature eggs M2) were selected by all classifiers.
‘Age’ (of the woman) was selected as the highest ranked feature by MLP, SVM and RF. This feature was also ranked as the most influential feature in the study by [37].
Seven features: ‘diagnosis’ (of infertility factor), ‘AFC’, ‘method of sperm collection’, ‘ovulatory stimulants unit’, ‘freeze’ (frozen sperm or oocyte), ‘Embryo transfer day’, ‘embryo transfer quantity’ (total number of embryo) were selected by four out of the five classifiers.
The top five ranked features selected by MLP and SVM classifiers were the same. Four of them were also selected by BF classifier within its top ten ranked features.
The number of same features selected by SVM and MLP was 14, that by SVM and RF was 12 and that by SVM, MLP and RF was 9. These nine features are: ‘age’, ‘indication’, ‘AFC’, ‘NbreM2’, ‘method of sperm collection’, ‘Chamotte’, Fertilization rate in vitro’, ‘Follicles on day 14’ and ‘Embryo transfer day.’
Finding of the above features as important is consistent with other studies in the literature, for example, Uyar et al. [37] found ‘age’ as the most important attribute, while Güvenir et al. found ‘age’ and ‘AFC’ as significant predictors of IVF success. The features we identified above will help the practitioners to advise the patients accordingly to increase the likelihood of pregnancy.
6.4 Prediction performance
Table 5 shows the performance evaluation in predicting pregnancy outcome by five classifiers when all the 25 features were used in building the models. The results presented in the table are the average over the ten folds in cross-validation. Though C4.5 and CART were the least performers, the accuracy of RF was around 90%, while that of MLP and SVM was above 97%. F-measure and AUC metrics also showed similar trend. Both MLP and SVM attained above 0.97 AUC.
Table 6 shows the performance of all classifiers when only the features selected by the respective classifiers were used. With the selected features, performance of all classifiers improved compared to using all features. C.45, CART and RF achieved around 6–7.64% improvement, while SVM and MLP achieved relatively lower improvement. Similar improvement was also achieved in F-measure and AUC. All these show the efficacy of the feature selection technique in building classifiers for IVF pregnancy prediction. Overall, SVM attained the highest accuracy (98.38%), F-measure (0.984) and AUC (0.995) which indicates the strong capability of SVM in predicting IVF treatment outcomes. RF and MLP achieved performance close to the SVM in all the three metrics. As can be seen from the table that the standard deviation of accuracy among folds is within 1.4% in these three classifiers while that in F-measure and AUC is 0.01, which indicates that, with the selected features, the classifiers perform consistently in all folds.
Figures 1, 2, 3, 4 and 5 plot performance metrics in terms of classification accuracy, F-measure and AUC as features were gradually added in each classifier as per their significance level. The feature (attribute) set for which the highest accuracy was achieved for a particular classifier is listed in Table 4. As can be seen from the figures, after the saturation point as discussed in Sect. 4 was reached, the accuracy either did not increase (SVM in Fig. 2) or started to decrease (other classifiers). In many cases, the accuracy decreased slightly (e.g., in Fig. 1 when feature no. 13 or 17 was added) before it increased again till the saturation point. It has been demonstrated in many studies in the literature that increasing features does not necessarily always improves performance, rather irrelevant and redundant features may degrade classifier’s performance. Feature selection technique preserves the importance of single features, and excessive features can lead to lower classification accuracy [21]. Janecek et al. showed in their experiment with drug discovery dataset that the classification accuracy of all five classifiers decreased when all features were used as opposed to when wrapper-based feature selection was employed which attained highest accuracy using only a subset of features. Similarly, Begg and Kamruzzaman [1] showed that, while using human gait data to differentiate between healthy and abnormal gait, the classification accuracy increased with adding features till the number of selected features reached three, remained unchanged till the number of features added reached 17 and then started decreasing as more features were added. The trend of not improving performance or slight degradation of performance with higher feature number is consistent with the findings in the literature. Though SVM and MLP selected 17 and 19 features to yield best accuracy, they can achieve above 95% accuracy with only the first 10 best features. The other observation is that, though the hill climbing feature selection method was built on the accuracy metric, F-measure and AUC also followed somewhat similar trend as accuracy with increasing features. Note that in our experiment though each feature was selected based on its contribution toward accuracy any other metric such as AUC can also be used in the same way.
It may be noted that an alternative to tenfold cross-validation method in the literature is the holdout method [25] where the whole dataset is partitioned into mutually exclusive subsets—training set (e.g., 70–80% of the dataset) and the test set (e.g., the remaining part 30–20%), and the result on the test dataset is presented as the model’s classification accuracy. We conducted another experiment employing the holdout method where 3/4 of the whole dataset was used as the training set and the remaining 1/4 as the test set using the features selected in Table 4. The results are presented in Table 7. The performance of classifiers using the handout method is similar to the performances achieved using tenfold cross-validation method without much difference, and SVM and random forest perform better than other classifiers attaining above 98% accuracy. This shows the stability of our proposed machine learning approach to produce consistent results in predicting the pregnancy outcome for IVF treatment.
6.5 Comparison with existing works
Since access to the dataset used in this type of study is quite restricted due to confidentiality and other ethical reasons, there exists no benchmark dataset that the research community can use to compare their works. In the absence of such dataset, it is only possible to compare with the reported results of significant works. In our approach, using the dataset described earlier, we have achieved a prediction accuracy of 98.38% and an AUC of 0.995 by the SVM classifier with a reduced feature set of 19 IVF attributes, obtained using feature selection technique. This accuracy is substantially higher than that reported in other studies, such as 90% accuracy using 27 attributes [14] and 90.35% accuracy using 18 attributes [33]. Similarly, the AUC value achieved by our approach outperforms that achieved in similar studies, which has reported AUC mostly close to 0.85 [9, 18, 26, 30, 36]. Overall, our work presents a promising technique to assist health practitioners to predict IVF treatment outcome.
7 Conclusion
In this paper, we propose an automated tool for predicting IVF pregnancy success based on emerging machine learning classifiers, namely MLP, SVM, C4.5, CART and random forest. Furthermore, the hill climbing feature selection technique is employed to evaluate the influence of each attribute of IVF treatment procedure and select the most influential attributes for each classifier to enhance the predictability of pregnancy success. Our experiments revealed several important attributes, e.g., ‘age’, ‘indication’, ‘AFC’, ‘NbreM2’, which were identified by majority of the classifiers. Prediction accuracy of each classifier improved when it is modeled using only the set of selected attributes. Results show that the proposed approach combining hill climbing feature selection method with classifiers like SVM, MLP or RF yielded prediction performance better than those reported in the literature. The proposed approach can be effective to guide medical practitioners to assess pregnancy outcome and advice patients accordingly for better management of IVF treatment. Future study will consider multicollinearity among features to further improve feature selection strategy and thereby pregnancy prediction outcome.
References
Begg R, Kamruzzaman J (2005) A machine learning approach for automated recognition of movement patterns using basic, kinetic and kinematic gait data. J Biomech 38(3):401–408
Binder H, Schumacher M (2008) Adapting prediction error estimates for biased complexity selection in high-dimensional bootstrap samples. Stat Appl Genet Mol Biol 7(1):1–26 (Article 12)
Breiman L (1984) Classification and regression trees. Routledge, New York
Breiman L (1993) Classification and regression trees. CRC Press, Boca Raton
Breiman L (2001) Random forests. Mach Learn 45(1):5–32
Bustillo M, Stern JJ, King D, Coulam CB (1993) Serum progesterone and estradiol concentrations in the early diagnosis of ectopic pregnancy after in vitro fertilization-embryo transfer. Fertil Steril 59(3):668–670
Charalambous C (1992) Conjugate gradient algorithm for efficient training of artificial neural networks. IEE Proc G-Circuits Dev Syst 13(3):301–310
Choi B, Bosch E, Lannon BM, Leveille MC, Wong WH, Leader A, Pellicer A, Penzias AS, Yao MW (2013) Personalized prediction of first-cycle in vitro fertilization success. Fertil Steril 99(7):1905–1911
Corani G, Magli C, Giusti A, Gianaroli L, Gambardella LM (2013) A Bayesian network model for predicting pregnancy after in vitro fertilization. Comput Biol Med 43(11):1783–1792
Cutler DR, Edwards TC Jr, Beard KH, Cutler A, Hess KT, Gibson J, Lawler JJ (2007) Random forests for classification in ecology. Ecology 88(11):2783–2792
Demyttenaere K, Bonte L, Gheldof M, Vervaeke M, Meuleman C, Vanderschuerem D, D’Hooghe T (1998) Coping style and depression level influence outcome in in vitro fertilization. Fertil Steril 69(6):1026–1033
Ding CH, Dubchak I (2001) Multi-class protein fold recognition using support vector machines and neural networks. Bioinformatics 17(4):349–358
Durairaj M, Thamilselvan P (2013) Applications of artificial neural network for IVF data analysis and prediction. J Eng Comput Appl Sci 2(9):11–15
Durairaj M, Nandhakumar R (2014) An integrated methodology of artificial neural network and rough set theory for analyzing IVF data. In: 2014 International conference on intelligent computing applications (ICICA), pp 126–129
Fayyad UM, Piatetsky-Shapiro G G, Smyth P (1996) The KDD process for extracting useful knowledge from volumes of data. Commun ACM 39(11):27–34
Guh R, Wu TCJ, Weng SP (2011) Integrating genetic algorithm and decision tree learning for assistance in predicting in vitro fertilization outcomes. Expert Syst Appl 38(4):4437–4449
Güvenir HA, Misirli G, Dilbaz S, Ozdegirmenci O, Demir B, Dilbaz B (2015) Estimating the chance of success in IVF treatment using a ranking algorithm. Med Biol Eng Comput 53(9):911–920
Hafiz P, Nematollahi M, Boostani R, Bahia NJ (2017) Predicting implantation outcome of In Vitro fertilization and intracytoplasmic sperm injection using data mining techniques. Fertil Steril 11(3):184–190
Haykin S (2004) Neural network—a comprehensive foundation. Prentice Hall, New Jersey
Hoover L, Baker A, Check JH, Lurie D, O’Shaughnessy A (1995) Evaluation of a new embryo-grading system to predict pregnancy rates following in vitro fertilization. Gynecol Obstet Invest 40(3):151–157
Janecek A, Gansterer W, Demel M, Ecker G (2008) On the relationship between feature selection and classification accuracy. In: New challenges for feature selection in data mining and knowledge discovery, pp 90–105
Jurisica I, Mylopoulos J, Glasgow J, Shapiro H, Casper RF (1998) Case-based reasoning in IVF: prediction and knowledge mining. Artif Intell Med 12(1):1–24
Kaufmann SJ, Eastaugh JL, Snowden S, Smye SW, Sharma V (1997) The application of neural networks in predicting the outcome of in vitro fertilization. Hum Reprod 12(7):1454–1457
Kaur H, Krishna D, Shetty N, Krishnan S, Srinivas MS, Rao KA (2012) Effect of pre-ovulatory single dose GnRH agonist therapy on IVF outcome in GnRH antagonist cycles; a prospective study. J Reprod Infertil 13(4):225–231
Kohavi R (1995) A study of cross-validation and bootstrap for accuracy estimation and model selection. In: 14th International joint conference on artificial intelligence (IJCAI), vol 14(2), pp 1137–1143
Manna C, Nanni L, Lumini A, Pappalardo S (2013) Artificial intelligence techniques for embryo and oocyte classification. Reprod Biomed Online 26(1):42–49
Mascarenhas MN, Flaxman SR, Boerma T, Vanderpoel S, Stevens GA (2012) National, regional, and global trends in infertility prevalence since 1990: a systematic analysis of 277 health surveys. PLoS Med 9(12):e1001356
Milewski R, Milewska AJ, Wiesak T, Morgan A (2013) Comparison of artificial neural networks and logistic regression analysis in pregnancy prediction using the in vitro fertilization treatment. Stud Log Gramm Rhetor 35(1):39–48
Morales DA, Bengoetxea E, Larrañaga P, García M, Franco Y, Fresnada M, Merino M (2008) Bayesian classification for the selection of in vitro human embryos using morphological and clinical data. Comput Methods Programs Biomed 90(2):104–116
Nanni L, Lumini A, Manna C (2010) A data mining approach for predicting the pregnancy rate in human assisted reproduction. Adv Comput Intell Paradig Healthc 5:97–111
Platt J (1998) Sequential minimal optimization: a fast algorithm for training support vector machines, Microsoft Research Technical Report MSR-TR-98-14
Quinlan JR (1993) C4.5: programs for machine learning, vol 1. Morgan Kaufmann, San Francisco
Ramasamy N, Durairaj M (2017) Feature reduction by improvised hybrid algorithm for predicting the IVF success rate. J Adv Res Comput Sci 8(1):37–40
Schwarzer G, Vach W, Schumacher M (2000) On the misuses of artificial neural networks for prognostic and diagnostic classification in oncology. Stat Med 19(4):541–561
SMO Weka (2017) Optimizing parameters. https://weka.wikispaces.com/Optimizing+parameters. Accessed 20 Dec 2017
Uyar A, Bener A, Ciray HN, Bahceci M (2010) ROC based evaluation and comparison of classifiers for IVF implantation prediction. In: Electronic healthcare, pp 108–111
Uyar A, Ayse B, Ciray HN (2015) Predictive modeling of implantation outcome in an in vitro fertilization setting: an application of machine learning methods. Med Decis Mak 35(6):714–725
Vapnik VN (1998) Statistical learning theory. Wiley, New York
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Hassan, M.R., Al-Insaif, S., Hossain, M.I. et al. A machine learning approach for prediction of pregnancy outcome following IVF treatment. Neural Comput & Applic 32, 2283–2297 (2020). https://doi.org/10.1007/s00521-018-3693-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-018-3693-9