Abstract
Spatial distribution of poly(A) RNA, hypophosphorylated Pol IIA, and hyperphosphorylated Pol IIO form of polymerase RNA II was characterized using immunofluorescence, immunogold and fluorescence in situ hybridization (FISH) techniques in relationship to transcriptional activity in the microspore and developing pollen of H. orientalis. During the course of pollen development our results reflected much higher transcriptional activity in the vegetative cell than in the generative cell. The highest levels of transcription in pollen cells were observed in young pollen grains, successively decreasing during pollen maturation, reaching a minimum just before anthesis. Levels of poly(A) RNA were higher in the vegetative cell than in the generative cell during all observed stages of pollen development. Accompanying physiological inhibition of the RNA synthesis in mature pollen cells was a strong accumulation of poly(A) RNA in the cytoplasm, especially in the vegetative cell. Alterations in transcriptional activity of differentiating pollen cells were accompanied by changes in the level and localization pattern of both forms of Pol II. During high transcriptional activity in the pollen nuclei, both forms of RNA Pol II occurred at the periphery of chromatin masses, as well as in the areas between them. A strong decrease in Pol IIO levels was observed in generative and vegetative nuclei as transcriptional activity of pollen cells apparently became inhibited. Finally, just before anthesis, an almost complete lack of the Pol IIO was observed in both pollen nuclei. In contrast, the level of Pol IIA significantly increased during the later stages of pollen development, in spite of apparent transcriptional inhibition in both pollen cells. This rich pool of the hypophosphorylated form of Pol II was located mainly over the central areas of condensed chromatin clumps, which was especially visible in the generative nucleus. Spatial and temporal aspects of RNA synthesis, including poly(A) RNA, as well as organization of transcriptional machinery appear to be closely related in developing pollen cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Eukaryotic protein-encoding gene expression involves transcription, primary transcript (pre-mRNA) processing, and export of a mature mRNA molecule to the cytoplasm, where translation occurs. The processing of most pre-mRNAs includes covalent modifications at both 5′ and 3′ ends as well as splicing (Sims et al. 2004). Almost all mRNA transcripts (with the exception of histone transcripts) contain poly(A) tail at the 3′ end, which protects the mRNA from exonucleases, enables the stability of the mRNA molecule and plays an important role during initiation of translation (Proudfoot and O’Sullivan 2002).
The transcription of protein-encoding genes (pre-mRNA) is catalyzed by a large molecular complex containing a 12-subunit RNA polymerase II core enzyme, general transcription factors (TFIIs), and one or more multisubunit complexes called coactivators or mediators. The largest subunit of RNA polymerase II contains a unique carboxy-terminal repeat domain (CTD) (Lee and Young 2000). The functions of the CTD are closely associated with the phosphorylation state of the domain. So far, it is known that the CTD exists in two phosphorylation states—hypophosphorylated (Pol IIA) and hyperphosphorylated (Pol IIO). RNA polymerase II with hypophosphorylated CTD domain is found in initiation complexes, whereas elongating polymerase molecules contain hyperphosphorylated CTDs. Biochemical and molecular data strongly suggest that the phosphorylated CTD domain plays an important role in recruiting pre-mRNA processing factors, including capping enzymes, polyadenylation factors, as well as splicing machinery molecules (for review see Lee and Young 2000; Reed 2003; Kornblihtt et al. 2004; Bentley 2005).
Coordinated gene expression in each cell is critical for successful organ and tissue development, as well as the whole organism. This rule is especially evident during germ line cell differentiation and during the early stages of zygote and embryo development. Studies concerning these problems have, in the past, been carried out using animal and human cells. There is considerably less known about corresponding events in plant cells. The intracellular localization of both forms of RNA polymerase II has been described for different cell types (Zeng et al. 1997; Grande et al. 1997; Dirks and Snaar 1999; Xie and Pombo 2006). In interphase nuclei, hypophosphorylated forms of Pol II (Pol IIA) were localized in the form of small foci (Bregman et al. 1995). In turn, the hyperphosphorylated form of Pol II (Pol IIO) occured as speckles that also contained diverse pre-mRNA splicing factors. When Pol IIO occurs in small foci throughout the nucleoplasm, it appears to colocalize with sites of active pre-mRNA synthesis (Zeng et al. 1997). Localization of both Pol II forms changes upon transcriptional inhibition. After treatment with transcription inhibitors 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole, α-amanitin or actinomycin D, Pol IIA was relocalized into speckles and Pol IIO into enlarged speckled-like nuclear domains (Bregman et al. 1995). On the basis of these results, the spatial distribution of RNA polymerase II appears to be positively related to the spatial distribution of nascent RNA in the interphase nuclei and depends on the transcriptional state of the cell. In turn, analysis of the spatial distribution of the hypo- and hyperphosphorylated forms of RNA polymerase II during mitosis in different cell lines showed that Pol IIO was present at the surface of chromosomes, as well as in the small foci in the cytoplasm (Dirks and Snaar 1999). The amounts of Pol IIA were, in turn, strongly reduced in mitotic cells suggesting that this form of enzyme undergoes hyperphosphorylation at the onset of mitosis (Dirks and Snaar 1999).
There is little and often fragmentary information about the transcriptional machinery in plant cells, thus the nuclear distribution of Pol II and its regulation in plant cells still remains an open question. The literature concerning the spatial distribution and abundance of mature transcripts in the cells of the germline are more numerous. The cytoplasm of animal female gametes is believed to contain a rich pool of mature transcripts and translation of these transcripts is specifically initiated only after fertilization (Wilt 1977; Colot and Rosbash 1982). This mechanism of gene expression enables the early stages of embryo development to be controlled by the genome of the maternal cell (Juge et al. 2002).
Pollen grains represent an unusually convenient model for investigating transcription and storage of mature transcripts from the microspore to pollen maturity. Pollen differentiation is the result of the distinct gene expression patterns in the two brother cells that occupy the pollen grains. The vegetative cell serves as a growth habitat for the generative cell and germinates, forming a pollen tube—a gametophytic organ that delivers male gametes into the embryo sac during the progamic phase. In contrast, the generative cell performs a reproductive role, since it divides and gives rise to two sperm cells, which will undergo fusions with the target cells of the embryo sac—the egg and central cells.
Transcriptional activity of the microspore and pollen cells was first studied in the 1970s and 1980s (Mascarenhas 1975, 1990), demonstrating that the microspore is highly active in RNA synthesis and, after mitosis, the newly formed pollen cells differ very significantly in their levels of transcription. Both autoradiographic and biochemical studies show that the vegetative cell is much more transcriptionally active than the generative cell (Mascarenhas 1975; Tupy et al. 1983; Bednarska 1984; McCormick 1993). During pollen development, a gradual decrease in transcriptional activity in both pollen cells takes place. Just before anthesis, both in the vegetative and generative cell, the level of RNA synthesis decreases to low or undetectable levels (Mascarenhas 1975; Tupy et al. 1983; Bednarska 1984). The described pattern of RNA synthesis is believed to be common to all three major classes of RNA: rRNA, tRNA and mRNA (Mascarenhas 1990).
The last several years have marked a period during which rapid progress has been made in understanding male gametophyte gene expression. Genome-wide analyses of the pollen transcriptome of Arabidopsis thaliana based on microarray experiments showed that not only the male gametophyte is unique in its composition but also characteristic changes in gene expression occur during microsporogenesis and microgametogenesis (Honys and Twell 2003, 2004; Becker et al. 2003; Pina et al. 2005; Alves-Ferreira et al. 2007). Transcriptome profiling throughout pollen development has also provided important information about gametophytic transcription of distinct pollen expressed protein-encoding genes.
Investigations assessing quantitative poly(A) RNA levels as marker of mature mRNA during pollen development are few and ambiguous. In maturing tobacco pollen grains a 20-fold increase in poly(A) RNA levels was indicated; moreover, it was shown that in the total pool of pollen RNA, the content of poly(A) RNA increased from 0.3% in young pollen grains to 2.7% in mature pollen (Tupy 1982). Indeed, using fluorescence in situ hybridization (FISH), we showed that, in spite of an apparent lack of current RNA synthesis, the mature pollen grain of Hyacinthus orientalis L. also contains a rich pool of poly(A) RNA (Zienkiewicz et al. 2006). Raghavan (1981, 1989) showed that in Hyoscyamus niger and Oryza sativa the pool of poly(A) RNA decreased during pollen development and was not detectable at anthesis.
The aim of the present study was the analysis of the spatial distribution of poly(A) RNA and different forms of RNA polymerase II. Transcriptional activity of differentiating H. orientalis L pollen cells was analyzed by FISH, immunocytochemical and Western-Blot techniques revealing strong decreases in transcription near and at the time of anthesis.
Materials and methods
Material for investigations
Bulbs of H. orientalis L. cv Pink Pearl (Torseed, Torun, Poland) were planted in plastic pots and held in a refrigerator for 8 weeks at 6°C in the dark. Next, pots were moved into a growth-chamber, and each bulb was covered with a paper cone. Plants were grown at 26°C under 16-h-light/8-h-darkness cycles that provide LD conditions (130 μmol m−2 s−1 cool white fluorescent tubes, Polam, Warsaw, Poland) for 2–4 weeks. Anthers containing differentiating pollen grains were collected every 24 h for 14 days. This time interval allows the collection of all stages of microspore and pollen grain development previously described by Bednarska (1984).
Localization of transcription
Directly after isolation from flower buds or open flowers, anthers were incubated in distilled water for 5 min and rinsed in a 0.01% water solution of Triton X-100 for 10 min. Material was then washed with distilled water for 5 min and incubated in a 20 mM water solution containing 5-bromouracil for 1 h in the dark. Anthers were then washed three times with phosphate-buffered saline buffer (PBS), pH 7.2 for 15 min and fixed in a mixture of 4% paraformaldehyde and 0.25% glutaraldehyde in PBS overnight at 4°C. The material was dehydrated in increasing concentrations of ethanol, with several changes of absolute ethanol, and then infiltrated and embedded in BMM resin (butyl methacrylate, methyl methacrylate, 0.5% benzoin ethyl ether, 10 mM dithiothreitol; Fluka Chemie GmbH, Buchs, Switzerland). Embedded material was cut into semithin (1.5 μm) sections, which were mounted on microscope slides covered with Biobond (British Biocell International, Cardiff, UK). Incorporated 5-bromouracil was detected by incubating with monoclonal primary antibody anti-BrdU (F. Hoffmann-LaRoche Ltd, Rotkrenz, Switzerland) in 1% BSA in PBS (diluted 1:100), pH 7.2, overnight at 4°C, and the goat-anti-mouse secondary antibody conjugated with Alexa Fluor 488 in 1% BSA in PBS (diluted 1:500) for 1 h at 37°C. Control reactions were performed without the primary antibody. DNA was labeled with 4′,6-diamidino-2-phenylindole (DAPI) (Fluka).
Immunogold labeling of RNA polymerase II
For immunocytochemical methods, freshly collected anthers were immediately fixed in a mixture of 4% paraformaldehyde and 0.25% glutaraldehyde in PBS buffer pH 7.2 overnight at 4°C. Samples were then rinsed in PBS, dehydrated in alcohol and embedded in LR Gold (Sigma–Aldrich). Material was cut into ultrathin sections using a Leica Ultramicrotome and placed on formvar-coated nickel grids. For immunolabeling experiments, ultrathin sections were blocked for 15 min in 3% BSA in PBS, followed by incubation in a humid chamber at 4°C overnight (diluted 1:50 in 1% BSA in 0.01 M PBS) with either H5 mAb, recognizing the hyperphosphorylated RNA Pol II, or H14 mAb, which is specific to hypophosphorylated RNA Pol II (Agrisera AB, Vännäs, Sweden). Grids were then washed in PBS and incubated with IgM secondary antibodies conjugated with 15 nm gold (British BioCell International) in PBS containing 1% BSA at 37°C for 1 h. Controls were performed by omitting incubation with the primary antibody. Grids were counterstained to enhance contrast with 1% PTA (phosphotungstic acid) in water and 5% uranyl acetate and then analyzed with a JEOL 1010 electron microscope at 80 kV.
Fluorescent in situ hybridization of poly(A) RNA
Poly(A) RNA was detected using the oligonucleotide probe (Oligo dT), which is complementary to the poly(A) tail of mRNA (Department of Bioorganic Chemistry, Centre of Molecular and Macromolecular Studies, Polish Academy of Sciences). The probe was labelled with digoxygenin 11-dUTP (DIG, F. Hoffmann-LaRoche Ltd; Switzerland), chemically bound at the 5′ end and enzymatically bound at the 3′ end. Hybridization was carried out as described in Zienkiewicz et al. (2006). In order to visualize the probe, mouse anti-DIG primary antibodies (F. Hoffmann-Roche) were added overnight at 4°C and goat-anti-mouse Alexa Fluor 488 secondary antibodies (Molecular Probes, Inc., USA) for 1 h at 37°C. The control reaction was performed in the same way using the hybridization buffer with a sense probe.
Quantitative evaluations
Image analysis was performed on serial semithin sections after immunofluorescence staining (BrU) or FISH [poly(A) RNA], with each reaction step performed using consistent values of temperature, incubation times, concentration of primary antibody or probe, and Alexa Fluor 488 secondary antibody. Quantitative analysis of fluorescence intensity for both BrU and poly(A) RNA was carried out for 35 pollen grains from each pollen development stage. Observations were performed using a Nikon Eclipse 80i fluorescence microscope equipped with CPI Plan Fluor 100 × (numerical aperture, 1.4) DIC H/N2 oil immersion lens and narrow band filters (UV-2EC, B-2EC, G-2EC). Results were photographed using a Nikon DS-5Mc color cooled digital camera and Lucia G image analysis software (Laboratory Imaging, Prague, Czech Republic), which allows measurement of intensity of strictly defined fluorescence signal intensities. All measurements were conducted at the same magnification (500×), field area (controlled with a shutter), and positioning of the fiber optics cable. Camera settings were kept constant for exposition time, gain and offset. Levels of fluorescence intensity were measured by observing the nuclei and cytoplasm of the examined microspores and pollen cells. Lucia G software was used to determine the average μm3 signal intensity of each studied cell compartment and is expressed in a.u. The data obtained in three different experiments were accumulated and averaged in relation to the highest values observed in the microspores. For all antigens and developmental stages, the obtained data were corrected for background autofluorescence as determined from negative control signal intensities. Statistical data and graphs were created using SigmaPlot 8 software (Systat Software GmBH, Germany).
Immunoblotting
Freshly collected pollen (15 mg), pollen tubes growing in vitro (cultivated as described in Zienkiewicz et al. 2006) (25 mg), and roots (250 mg) were frozen and ground into a fine powder in liquid nitrogen. For total protein isolates, the ground material was resuspended in extraction buffer [100 mM Tris–Cl pH 7.5, 1 mM EDTA, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride (protease inhibitor cocktail), Sigma] by gentle shaking and clarified by centrifugation at 16,000×g for 30 min at 4°C. Protein concentration was determined using the Bradford method.
A total of 40 μg of protein per lane were separated using 12% SDS-PAGE and transferred onto a PVDF membrane (Amersham Biosciences, Piscataway, USA). The membrane was blocked (1 h) with TBS buffer (10 mM Tris–HCL, 150 mM NaCl) containing 3% non-fat milk powder. After that, the membrane was incubated with IgM H5 or H14 (Agrisera) mouse monoclonal antibody (1:500 dilution), washed with TBS (four times) and incubated with goat-anti-mouse secondary antibody conjugated to horseradish peroxidase (Sigma). Proteins were detected on the membrane using the ECL System (Amersham Biosciences) and X-ray film (Foton, Bydgoszcz).
Results
Localization of transcription in the microspore and differentiating pollen grain
Immunofluorescence analysis of BrU incorporation showed that in the polarized microspore, the accumulation of green fluorescence which indicates newly synthesized RNA was observed in the nucleus and cytoplasm (Fig. 1). Moreover, both compartments were characterized by a different localization pattern of newly synthesized RNA molecules. In the microspore nucleus, small, abundant fluorescence clusters were present, whereas in the cytoplasm the signal was more uniform. In the central vacuole, which is typical for this developmental stage of the microspore, no fluorescence was observed. After microspore division, signal was present in both pollen cells (Fig. 2); however, its level and localization pattern were significantly different in the vegetative and the generative cell. In the vegetative nucleus a strong, uniform fluorescence was observed, whereas in the cytoplasm, a much lower signal was present, except in vacuoles, where there was no fluorescence. In the generative cell of young pollen grains, newly synthesized RNA was present in both the nucleus and surrounding cytoplasm. Fluorescence in the nucleus was restricted to small spots, and localized throughout the nucleoplasm (Fig. 2b, arrowheads). In the generative cytoplasm, a bright, disperse signal was observed. During the separation of the generative cell from the sporoderm, the localization of incorporated bromouracil in both generative and vegetative cells was similar, as in the previous developmental stage (see supplementary data, Fig. S1). After complete separation of the generative cell from the sporoderm, a decrease in fluorescence intensity was indicated in both pollen cells; however, the localization of signal was similar to that observed in earlier stages of development (Fig. 3). During later stages of pollen development, a strong decrease in the transcription level was observed in both pollen cells (see S2). Low signal was present in the cytoplasm of both pollen cells, where it mainly occurred. In contrast, in the nuclei spots of fluorescence were rarely observed. Just before anthesis, most of the pollen cells showed very little or were devoid of fluorescence (Fig. 4).
Figs. 1–6 Immunofluorescence localization of incorporated bromouracil in the microspore and differentiating pollen grain (bars = 10 μm). Fig. 1 Polarized microspore; numerous, bright fluorescence spots can be seen in the nucleus (N). In the cytoplasm (C) a homogeneous signal is visible, except in vacuoles (V). a DAPI staining. Fig. 2 Young pollen grain; vegetative nucleoplasm (VN) displays strong uniform signal, with nucleoli (no) showing low labeling. In the generative cell, few spots of fluorescence can be seen in the nucleus (GN), whereas in the surrounding cytoplasm stronger homogeneous fluorescence is localized. a DAPI labeling of DNA, b inset of generative nucleus (framework in Fig. 2) indicating fluorescence clusters (arrowheads). Fig. 3 Pollen grain at the spheroidal shape generative cell stage. Strong labeling can be observed in the vegetative nucleus (VN) and weaker in the vegetative cytoplasm (VC). In the generative nucleus (GN), a few spots of signal are evident and in cytoplasm low labeling can be seen. a DAPI labeling of DNA, b greater magnification of generative nucleoplasm (framework in Fig. 4) displaying fluorescence spots. Fig. 4 Mature pollen grain (area marked with dashed line) displays no detectable fluorescence in pollen cells (VN, CN, GN). Figs. 5–8 Localization of poly(A) RNA in the microspore and developing pollen grain (bars 10 μm). Fig. 5 Polarized microspore; intense, homogeneous signal can be seen in the cytoplasm (c), excepting vacuoles (V). Punctate fluorescence is present in the nucleus (N). The nucleoli (no) show no labeling. a DAPI labeling of DNA. Fig. 6 Young pollen grain; signal in the form of numerous, small spots can be seen in both pollen nuclei (VN, GN). In the cytoplasm of both pollen cells (VC, GC), a more intense signal occurs in comparison to pollen nuclei. a DAPI staining. Fig. 7 Pollen grain after detachment of the generative cell from the sporoderm; fluorescence occurs in both pollen cells (VN, VC, GN, GC). In the vegetative nucleus (VN), spots of signal are significantly brighter and more numerous than in the generative one (GN). In the cytoplasm of both pollen cells small clusters of intense signal are present (VC, GC). a DAPI labeling of DNA. Fig. 8 Pre-mature pollen grain; weak signal can be seen in both pollen nuclei (VN, GN). In the pollen cells cytoplasm, especially the vegetative one (VC), strong fluorescence, in the form of numerous, small clusters is present. a DAPI staining
A control reaction performed without the anti-BrdU antibody did not show any fluorescence in polarized microspores (see S3).
Localization of poly(A) RNA in the polarized microspore and differentiating pollen grains
Using digoxygenin-labeled complementary probe for poly(A) tails, we localized mature RNA particles during H. orientalis L. pollen development.
In the polarized microspore, poly(A) RNA was detected in both the nucleus and cytoplasm, except for vacuoles (Fig. 5). A clear difference in poly(A) RNA distribution was observed between microspore nucleus and cytoplasm. Cytoplasmic fluorescence was uniform, whereas in the nucleus, signal was present in the form of numerous spots dispersed throughout the nucleoplasm. Nucleoli did not show any label (Fig. 5). Just after microspore division, signal indicating the presence of poly(A) RNA was present in both newly formed pollen cells (Fig. 6). In the vegetative and the generative cell, the level of fluorescence was higher in the cytoplasm than in the nucleus; however, differences in signal localization pattern were observed between the nucleus and cytoplasm in both pollen cells. A speckled pattern of fluorescence was observed in the nucleoplasm of pollen nuclei, but not in the nucleoli. In the vegetative cytoplasm, homogeneous fluorescence was present except in vacuoles. In the thin layer of cytoplasm surrounding the generative nucleus the pattern of the poly(A), RNA distribution seemed to be similar to that observed in the vegetative cell. During separation of the generative cell from the sporoderm (see S4), as well as at the spherical-shape generative cell stage (Fig. 7), poly(A) RNA was localized in both pollen nuclei; however, in the generative nucleus, the level of signal was lower in comparison to the vegetative nucleus. In the pollen nuclei, signal occurred in the form of numerous small fluorescent clusters. In both pollen cells, the level of fluorescence in the form of small and numerous spots was much higher in the cytoplasm than in nuclei. During the next developmental stages, signal indicating the presence of poly(A) RNA was still present in both pollen cells; however, the fluorescence level in the vegetative cell was higher than in the generative cell (Fig. 8). Strong green fluorescence was localized in the vegetative cell cytoplasm, whereas in the nucleus, the level of signal was significantly lower. Additionally, an increased level of fluorescence was observed in vegetative cell cytoplasm in comparison to earlier stages of development. In the generative nucleus, where only a few small fluorescence clusters were observed, the signal level seemed to be much lower than in the vegetative nucleus. No fluorescence was observed in the control section of mature pollen grain (see S5).
Analysis of the transcriptional state of the polarized microspore and differentiating pollen cells
Quantitative analysis of the fluorescence intensity emitted from anti-BrU-Alexa Fluor 488 complexes (Fig. 9) indicated considerable variation in the transcriptional activity of pollen cells during H. orientalis pollen development.
Fig. 9 Graph illustrating the level of incorporated bromouracil fluorescence in the polarized microspore and differentiating pollen cells. Highest fluorescence observed in the early microspores in comparison to pollen cells. During the course of pollen development, the signal was significantly higher in the vegetative cell than in the generative one. The highest levels of fluorescence were observed in young pollen cells, until the detachment of the generative cell from the sporoderm. During later stages a successive decrease in the anti-bromouracil labeling intensity was observed in both pollen cells and reached its minimum in the mature pollen grain. The values represent the average of three replicate experiments; the error bars indicate one standard error of the mean. Fig. 10 Histogram showing the intensities of poly(A) RNA fluorescence in the polarized microspore and developing pollen cells. Microspore shows the highest level of the signal in comparison to the developing pollen grain. The intensities of the poly(A) RNA labeling were significantly higher in the vegetative cell than in the generative one during the whole course of pollen development. Young pollen cells exhibited the strongest hybridization signal, whereas after a complete separation of the generative cell from the sporoderm, a successive increase in the signal levels was observed in the vegetative cell. In the generative cell the levels of poly(A) RNA were at similar, steady and low levels. In the mature pollen grains the highest levels of polyadenylated transcripts were observed in the vegetative cell, in comparison to other stages of pollen development. The values represent the average of three replicate experiments; the error bars indicate one standard error of the mean. Fig. 11 Graph comparing the levels of poly(A) RNA between the nucleus and cytoplasm of the polarized microspore and differentiating vegetative pollen cell. In the polarized microspore the cytoplasmic pool of poly(A) RNA was a little higher than the nuclear one. After the first pollen mitosis, level of poly(A) was higher in the cytoplasm than in the nucleus and varied during pollen development. A successive decrease in the signal level, both in the vegetative nucleus and cytoplasm, was observed until the separation of the generative cell from the sporoderm. In turn at late stages of pollen development a significant decrease in the hybridization signal in the vegetative cytoplasm was indicated. In the vegetative nucleus the level of poly(A) RNA gradually decreased and reached its minimum just before anthesis. Fig. 12 Histogram illustrating the intensities of fluorescence indicating poly(A) RNA in the nucleus and cytoplasm of differentiating generative pollen cell. During almost the whole course of pollen development, the level of polyadenylated transcripts was much higher in the nucleus than in the cytoplasm. As pollen developed, both the nuclear and cytoplasmic pool of poly(A) RNA decreased; however, it was more visible in the nucleus. Just before anthesis the accumulation of poly(A) RNA was higher in the nucleus than in the cytoplasm
As shown in Fig. 9, the highest level of 5-bromouracil incorporation occurred in the polarized microspore. The level of fluorescence intensity in these cells was approximately 2-fold higher than in the vegetative cell of the young pollen grain. At the beginning of pollen development, a significant difference in the level of the signal was observed between both pollen cells. During all stages of pollen development, the level of examined fluorescence was much higher in the vegetative cell than in the generative one. In young pollen grains before the separation of the generative cell from the sporoderm, the level of signal successively increased. After this period, when generative cell was fully separated from the sporoderm, the level of fluorescence labeling, indicating incorporated 5-bromouracil in the vegetative cell, gradually decreased. At anthesis, fluorescence intensity in the vegetative cell was very low. In the generative cell, we showed that from the beginning of pollen development, the level of signal indicating incorporated bromouracil progressively decreased during development and was not detectable in the mature pollen grain.
Analysis of poly(A) RNA levels in polarized microspore and differentiating pollen cells
Quantitative analysis of fluorescence intensity emitted by TOR30-anti-Dig-Alexa 488 complexes showed significant changes in poly(A) RNA levels during pollen development (Figs. 10, 11 and 12). Fig. 10 illustrates the levels of poly(A) RNA in polarized microspore and pollen cells during all stages of their differentiation. In the polarized microspore, the poly(A) RNA level was significantly high. In newly formed pollen cells, the level of poly(A) RNA was lower and varied during the next stages of pollen grain maturation. During pollen development, the level of poly(A) RNA was consistently much higher in the vegetative cell than in the generative one. In the vegetative cell, the lowest level of polyadenylated transcripts occurred just after the generative cell separated from the sporoderm. Beginning from this stage, accumulation of poly(A) RNA was observed in the vegetative cell at each successive stage, reaching its maximum just before anthesis. Unlike in the vegetative cell, the level of polyadenylated transcripts in the generative cell did not change significantly, and appeared the lowest just before anthesis.
In the polarized microspore, the level of fluorescence indicating poly(A) RNA was usually a little higher in the cytoplasm than in the nucleus (Fig. 11). In the vegetative cell, during the whole course of pollen grain differentiation, the hybridization signal was much higher in the cytoplasm than in the nucleus and varied significantly (Fig. 11). From the first pollen mitosis to the separation of the generative cell from the sporoderm, a gradual decrease in the poly(A) RNA pool was observed both in the cytoplasm and the nucleus of vegetative cell,. The final stages of pollen development were, in turn, accompanied by a considerable increase in the poly(A) RNA level in the vegetative cytoplasm. In the vegetative nucleus, the level of polyadenylated transcripts successively decreased and reached its minimum in the mature pollen grain. In the generative cell, during almost all stages of pollen maturation, the level of poly(A) RNA was much higher in the nucleus than in the cytoplasm (Fig. 12). As pollen developed, both in the cytoplasm and in the nucleus, a successive decrease in the hybridization signal intensity was observed, although this decrease was much more noticeable in the nucleus than in the cytoplasm. Just before anthesis, the accumulation of poly(A) RNA was, however, higher in the generative cytoplasm than in the nucleus.
Ultrastructural localization of RNA polymerase II in the polarized microspore and differentiating pollen grains
Different antibodies were used for ultrastructural localization of Pol IIA (H14 mAb) and Pol IIO (H5 mAb) in developing pollen grains of H. orientalis. In the microspore nucleus, the labeling of Pol IIO was localized at the periphery of condensing chromatin masses as well as around the chromatin fibers located between them (Fig. 13). After first pollen mitosis, gold particles indicating Pol IIO were indicated in both pollen nuclei (Figs. 14, 15). In the vegetative nucleus (Fig. 14), strong labeling was localized throughout the nucleoplasm, whereas in the generative nucleus (Fig. 14) numerous colloid gold grains were observed at the periphery of condensed chromatin areas, as well as between them, around the dispersed chromatin fibers. During and after the separation of the generative cell from the sporoderm, localization patterns and the level of labeling did not change significantly in the vegetative nucleus (Figs. 16, S6). In turn, in the generative nucleus gold particles were mainly present at the edges of condensed chromatin masses as well as over their central areas (Figs. 17, S7). Labeling of the interchromatin regions was low. At later stages of pollen development, a significant decrease in labeling levels was indicated in both pollen nuclei (see S8 and S9); however, gold particles were more abundant in the vegetative nucleus than in the generative nucleus. Just before anthesis, both in the vegetative and generative nuclei, an almost complete lack of labeling was observed after H5 mAbs staining (Fig. 18).
Immunogold localization of the hyperphosphorylated form of RNA polymerase II (POL IIO) in the polarized microspore and differentiating pollen cells (bars 0.5 μm) (N nucleus, C cytoplasm, VN vegetative nucleus, VC vegetative cytoplasm, GN generative nucleus, GC generative cytoplasm). Fig. 13 Pre-mitotic microspore; grains of colloidal gold occur mainly in the area of uncondensed chromatin fibers and at the periphery of condensed chromatin masses. Figs. 14–15 Pollen grain just after the first pollen mitosis. Fig. 14 The vegetative nucleus; the labeling can be seen over the chromatin fibers as well as at the periphery of electron dense areas of the nucleus. A few gold particles are also present on the periphery of the nuclear body (arrowhead). Fig. 15 The generative nucleus. Gold grains are localized mainly in the areas between condensed chromatin clumps and at their periphery. Figs. 16–17 Pollen grain after complete detachment of the generative cell from the sporoderm. Fig. 16 The vegetative nucleus; intense labeling can be observed over electron dense areas of the nucleoplasm. The remaining area of the nucleus shows low labeling. Fig. 17 The generative nucleus; colloid gold grains occur at the periphery of condensed chromatin masses. A few single gold particles can be seen over the central areas of chromatin clumps. Interchromatin regions of the nucleus show very low labeling. Fig. 18 Mature pollen grain; very low labeling of both pollen nuclei can be seen
In the microspore nucleus, labeling indicating Pol IIA appeared to be associated with the periphery of condensing chromatin areas (Fig. 19); in the spaces between condensing chromatin areas, signal was usually low. The number of gold particles indicating Pol IIA in the microspore nucleus seemed to be slightly lower than that observed after H5 mAbs staining (compare Figs. 13, 19). After microspore division, both pollen nuclei showed the presence of gold particles (Figs. 20, 21). In the vegetative nucleus, which displays considerable decondensation of the chromatin, distribution of gold particles was dispersed, indicating Pol IIA was observed in the nucleoplasm (Fig. 20). In the nucleus of a parietally situated generative cell, where a high degree of chromatin condensation was indicated, dense chromatin clumps were decorated with the anti-Pol IIA labeling (Fig. 21). Colloidal gold particles were also indicated in the areas of the nucleus not occupied by condensed chromatin. A very similar Pol IIA localization pattern, as well as the level of labeling was observed until the detachment of the generative cell from the sporoderm (see S10 and S11). When the generative cell was fully separated from the wall of the pollen grain, labeling of the interchromatin regions in the generative nucleus was significantly reduced (Fig. 23). In this stage of pollen development, the vegetative nucleus, displayed, dispersed, single gold particles throughout the nucleoplasm (Fig. 22). It should be noted that, at these stages of development, both cells of pollen grain displayed labeling that indicated Pol IIA was generally lower than observed for Pol IIO (compare Figs. 16, 17, 22, 23, respectively). Final stages of pollen development were accompanied by significant changes in the level and labeling pattern indicated (see S12 and Figs. 24, 25). In the vegetative nucleus, abundant colloidal gold particles were usually seen in groups composed of 2–3 grains localized in the various parts of the nucleoplasm (Fig. 24). In the generative nucleus, strong labeling was observed mainly over dense chromatin masses. There were no gold particles over the interchromatin regions of the nucleus (Fig. 25). Control reactions for both H5 and H14 mAbs showed no presence of gold particles in pollen nuclei (see S13 and S14).
Figs. 19–20 Ultrastructural localization of the hypophosphorylated form of RNA polymerase II (POL IIA) in the polarized microspore and differentiating pollen cells (bars 0.5 μm) (N nucleus, C cytoplasm, VN vegetative nucleus, VC vegetative cytoplasm, GN generative nucleus, GC generative cytoplasm). Fig. 19 Pre-mitotic microspore; colloidal gold grains are present over the periphery of condensed chromatin. Low labeling can also be seen over the condensing chromatin masses. Figs. 20–21 Pollen grain just after first pollen mitosis. Fig. 20 The vegetative nucleus; numerous gold grains are present over the chromatin fibers present in the whole area of the nucleus. Fig. 21 The generative nucleus; strong labeling is localized at their periphery and between electron-dense chromatin areas. Figs. 22–23 Pollen grain after complete detachment of the generative cell from the sporoderm. Fig. 22 The vegetative nucleus; the dense set of chromatin fibers is covered by numerous, sparse gold particles. Fig. 23 The generative nucleus; colloid gold grains occur at the boundary of condensed chromatin masses, mainly over the decondensed fibers of chromatin. A few single gold particles can be seen over the central areas of chromatin clumps. Figs. 24–25 Mature pollen grain; a very similar labeling pattern, in comparison to the previous developmental stage, can be seen in the vegetative (Fig. 24) and the generative nucleus (Fig. 25)
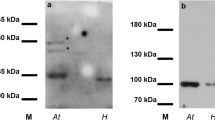
Immunoblot analysis of the antibodies confirmed the presence of single protein bands with a molecular mass of about 210 kDa in H. orientalis roots, mature pollen collected from open flowers and pollen tubes growing in vitro (Fig. 26). In the case of mature pollen, positive results were obtained only for H14 mAb and no cross-reactivity was indicated for the H5 mAb. Note that for pollen tubes growing in vitro for 2 h, a significantly more intense protein band was observed for H5 mAbs. In the case of H14 mAb, the signal was lower in comparison to that observed for protein extracts from mature pollen. As a positive control for H5 antibody, we used protein extract isolated from H. orientalis roots. A single protein band can be observed at 210 kDa.
An immunoblotting analysis of monoclonal H5 and H14 antibodies using H. orientalis pollen, pollen tubes and root extracts. The protein band with a mass of about 210 kDa corresponds to H. orientalis Pol IIA and Pol IIO. In mature pollen extracts only the Pol IIA pool can be seen. In the case of pollen tubes growing in vitro the protein band corresponding to Pol IIA is lower than that corresponding to Pol IIO. As a positive control for H5 antibody a protein extract from H. orientalis roots was used
Discussion
Our results showed significant changes in transcriptional activity of H. orientalis during pollen development. The highest level of RNA synthesis was observed in the polarized microspore. From the first pollen mitosis, throughout H. orientalis pollen development, RNA synthesis was higher in the vegetative cell than in the generative cell. In both pollen cells, the highest level of transcriptional activity was observed in young pollen grains to generative cell separation from the sporoderm.
The obtained results confirm earlier reports about the very high metabolic activity of the microspore. Evidence for high transcriptional activity in microspores, as well as young pollen grains, is in agreement with the recent studies of Honys and Twell (2004). Using microarrays they showed that the majority of protein-encoding genes were actively expressed at the earliest developmental stages of Arabidopsis male gametophyte development. Moreover, genome-wide analyses also showed that the majority of mRNAs present in the microspore encode proteins involved in the general metabolism of the cell, including lipid metabolism, cell wall formation, protein degradation and some steps of gene expression. In turn, the least represented mRNAs genes that encoded transcription factors and receptor proteins (Honys and Twell 2004; Alves-Ferreira et al. 2007).
Early stages of pollen development have crucial importance in the establishment of the microgametogenesis pathway, since the developmental programs of pollen cells are tightly coupled with their biological roles. The vegetative cell acts as an enclosed environment for the development of the generative cell and, after pollination, the vegetative cell germinates into a pollen tube. Such a biological role could easily explain the high metabolic activity of this cell, at least during the early stages of pollen development. In turn, the generative cell gives rise to two sperm cells, which will fuse with the target cells of the embryo sac during double fertilization (Willemse and Van Went 1984). During later stages of pollen development, a successive decrease in RNA synthesis levels was observed in the vegetative and generative cells. Just before anthesis, pollen cells exhibited very low or no transcription.
The decrease of transcriptional activity in both H. orientalis pollen cells, after generative cell separation from the sporoderm, was also shown in other angiosperm species (Mascarenhas 1975; Tupy et al. 1983; Bednarska 1984). Affected RNA classes likely include three major RNA classes: mRNA, rRNA and tRNA (Mascarenhas 1990). Moreover, inhibition of RNA synthesis in both pollen cells was also positively correlated with chromatin condensation in both pollen nuclei of H. orientalis (Bednarska and Górska-Brylass 1987). The vegetative and the generative cells of mature pollen grains of angiosperms are traditionally thought to be metabolically silent and do not synthesize either RNA or proteins (Mascarenhas 1993). More recently, Honys and Twell (2004) using microarray experiments showed also a significant decline in the number of protein encoding mRNAs during final stages of Arabidopsis pollen development in comparison to that observed in young pollen grain. Thus, our results support that H. orientalis pollen grains follow the general trends of transcription stated in several other species examined so far.
Alterations in the levels of RNA synthesis in differentiating H. orientalis pollen cells are in agreement with changes in the level and localization pattern of RNA polymerase II. Monoclonal H14 and H5 antibodies used for an ultrastructural localization of different forms of Pol II are recognized as specific protein bands with a molecular mass of about 210 kDa. Given the fact that molecular weights of eukaryotic RNA polymerase II vary from 210 to 240 kDa (Bregman et al. 1995), we believe that these represent antibodies that are specific for the Pol IIA and Pol IIO isolated from H. orientalis tissues. Both antibodies were also used with success for immunolabeling investigations in several previous studies carried out on animal and human cells (Grande et al. 1997; Zeng et al. 1997). Our results demonstrate the localization of Pol II molecules in the male gametophyte cells of angiosperms.
The ultrastructural localization of Pol IIA and Pol IIO in pre-mitotic H. orientalis microspores showed their presence both over the decondensed chromatin fibers, as well as at the periphery of condensed chromatin masses. Such a localization pattern seems to indicate transcriptionally active sites of the microspore nucleus as, for example, perichromatin fibrils (Fakan 1994). The observation that the Pol IIA labeling level was lower than that observed for Pol IIO seems to confirm the high transcriptional activity of the microspore. Moreover, it should be noted that the hypophosphorylated form of RNA polymerase II is also reflected in the pool of RNA Pol II between cyclic rounds of dephosphorylation (soon after transcription termination) and phosphorylation (in the new initiation complex on the new promotor) (Lee and Young 2000). Thus it seems that in highly transcriptionally active cells, most of the nuclear Pol II pool may be involved in ongoing pre-mRNA synthesis.
After the first pollen mitosis, both forms of Pol II were indicated in the vegetative and the generative pollen nucleus; however, the level of the hyperphosphorylated form of Pol II seemed to be higher than the hypophosphorylated form in both pollen nuclei. These results confirm high rates of RNA synthesis in young pollen cells. As pollen developed we observed a successive decrease in the Pol IIO level and a simultaneous increase in the Pol IIA pool in both pollen nuclei. Such data, compared with the results of bromouracil incorporation, suggest that during H. orientalis pollen development not only a successive dephosphorylation of Pol II might occur, but also that dephosphorylated transcriptional machinery is stably stored in pollen nuclei and does not undergo degradation. We cannot exclude that dephosphorylation might be underlying a mechanism of a gradual transcription repression in differentiating pollen cells; however, further studies are expected to confirm this hypothesis.
Changes in the levels of labeling, indicating the presence of Pol IIA and Pol IIO, were accompanied by a successive elimination of both forms of Pol II from the interchromatin spaces to the periphery of condensing chromatin in both pollen nuclei. In the generative nucleus of young pollen grains, which exhibits the highest transcriptional activity at this time of pollen development, both forms of Pol II were localized at the perichromatin regions as well as between them. Most probably such a localization pattern indicates transcriptionally active areas of the generative nucleus. Indeed, in recent years a number of studies have showed that in the transcriptionally active animal and human interphase nuclei, the hyperphosphorylated form of Pol II is localized in speckled domains, which also contain splicing factors and in small foci throughout the nucleoplasm which colocalize with sites of active gene transcription, including perichromatin regions of the nucleoplasm (Bregman et al. 1995; Zeng et al. 1997; Xie and Pombo 2006). During later stages of pollen development, accompanied with a strong decrease in transcriptional activity in both pollen cells, depletion of the Pol IIO pool was observed in the pollen nuclei, as well as redistribution of Pol IIA from the interchromatin areas into chromatin masses. Finally, in the mature pollen nuclei, which are thought to be transcriptionally silent, the hypophosphorylated form of Pol II was observed in central areas of the chromatin clumps and not at their periphery.
This pattern was particularly well-defined in the generative nucleus. Animal and human cells undergoing drug- or heat shock-induced inhibition of RNA synthesis display a strong redistribution of Pol II molecules into enlarged nuclear speckles, which also contain a rich pool of pre-mRNA processing factors (Zeng et al. 1997, Xie and Pombo 2006). Thus, the localization pattern of Pol IIA in the mature pollen nuclei was unexpected. However, such localization of Pol IIO on the chromatin masses can be compared with the results of Dirks and Snaar (1999), which showed similar Pol IIO localization during mitosis of human fibroblasts. The authors speculate that hyperphosphorylated Pol II, unlike Pol IIA, cannot assemble in pre-initiation complexes and that localization of Pol IIO might be a mechanism of transcriptional repression in mitotic cells (Dirks and Snaar 1999). It should also be taken into consideration that the same or similar model of transcriptional repression is also true for pollen cells. Down regulation of RNA synthesis in both mature pollen cells of angiosperms is a part of their developmental program and we cannot exclude that the distribution of gene expression machinery might be specific for the gametophyte cells.
Because only a hypophosphorylated form of Pol II is able to assemble into the transcriptional pre-initiation complexes and the early steps of pollen tube growth appear to be linked with up-regulation of transcription (Zienkiewicz et al. 2008), we propose that the pool of Pol IIA in the mature pollen grain may represent transcriptional machinery ready for activation (phosphorylation) soon after pollen germination (Zienkiewicz et al. 2008). This hypothesis, as well as transcriptional upregulation on activation, can be supported also by the results of immunoblotting experiments. In pollen tubes growing in vitro for 2 h, the pool of Pol IIA was significantly reduced in comparison to that observed in the ungerminated pollen. Moreover, the pool of hyperphosphorylated Pol IIO appears only in growing pollen tubes at a relatively high level.
In H. orientalis polarized microspores and differentiating pollen cells, the presence of poly(A) RNA was indicated in the nucleus and cytoplasm of the examined cells. It is well known that the poly(A) sequence is present in mature mRNA molecules (Gray and Wickens 1998), thus we assumed that the signal observed in the nucleus represents newly synthesized transcripts, just after their processing, whereas the cytoplasmic one indicates mRNA particles during their translation and/or storage in the cytoplasm. In the polarized microspore of H. orientalis, high levels of poly(A) RNA observed in the nucleus as well in the cytoplasm positively correlate with the high transcriptional activity of these cells. Similar levels of poly(A) RNA in the microspore nucleus and cytoplasm suggest also that mRNAs intensively synthesized in the nucleus are actively exported to the cytoplasm. During the whole time of H. orientalis pollen development, the general pool of poly(A) RNA was more abundant in the vegetative cell, where high rates of RNA synthesis were indicated. In comparison, the generative cell exhibits much lower transcriptional activity. Just after the first pollen mitosis, when the level of RNA synthesis in both H. orientalis pollen cells was high, poly(A) RNA was present in both the vegetative and the generative cells, although its highest level was observed in the vegetative cell cytoplasm.
During separation of the generative cell from the sporoderm, the level of poly(A) RNA in the vegetative cell decreased, in spite of its transcriptional activity reaching a peak at this time of development. This suggests that most of the synthesized transcripts are used for current metabolism of the vegetative cell. Further decrease in the poly(A) RNA level in the vegetative cell was observed also after complete detachment of the generative cell from the sporoderm, which may correspond to a temporal cue, as transcriptional repression starts to occur at this stage. At final steps of H. orientalis pollen grain maturation, the level of poly(A) RNA observed in the vegetative cell is significantly increased and negatively correlates with transcriptional activity. Analysis of poly(A) RNA levels in the maturing H. orientalis vegetative cell, compared with its transcriptional activity, clearly indicates an accumulation of mature mRNAs in the cytoplasm. This accumulation was especially evident during the second half of pollen development. During all stages of pollen maturation, the cytoplasmic pool of poly(A) RNA was higher than the nuclear one; however, until the moment of complete separation of the generative cell from the sporoderm, differences in poly(A) RNA levels between the two compartments were not significant. Thus, it seems that most of the newly synthesized mRNA was currently translated. Indeed, the vegetative cell of young H. orientalis pollen grain was shown to be very active in protein synthesis (Bednarska 1984).
The biggest differences in poly(A) RNA levels between the nucleus and the cytoplasm of the vegetative cell were observed just before anthesis, when both pollen cells were shown to be transcriptionally and translationally silent (Bednarska 1984). The highest levels of polyadenylated transcripts in the vegetative cytoplasm were also indicated in the mature pollen grains. The negative correlation between the level of RNA synthesis and the poly(A) RNA pool observed at the end of the pollen maturation course, strongly suggests that the poly(A) RNA pool present in the vegetative cytoplasm of the mature pollen grain, was synthesized earlier and is the result of a successive storage of mature mRNAs. On the basis of our previous results we conclude that the mature transcripts stored in vegetative cytoplasm are delivered to the germinated pollen grain and growing pollen tube (Zienkiewicz et al. 2006). The protein products of these mRNAs most probably are involved in, at least, the early stages of pollen tube growth. Indeed, microarray analysis of the Arabidopsis mature pollen transcriptome revealed a strongly unrepresentative suite of mRNAs encoding proteins involved in cell wall metabolism, cytoskeleton, signal transduction and cell cycle (Becker et al. 2003; Honys and Twell 2003, 2004; Pina et al. 2005).
In the generative cell of H. orientalis pollen grains, the level of poly(A) RNA was much lower and did not change as significantly during maturation as did the vegetative cell. Prior to the moment of separation of the generative cell from the sporoderm, the majority of the signal in young pollen was observed in the nucleus rather than in the cytoplasm. During the course of pollen development, a successive decrease in the nuclear poly(A) RNA pool was observed. Just before anthesis, the level of polyadenylated transcripts in the generative cell was higher in the cytoplasm than in the nucleus. These results seem to reflect different metabolic profiles of differentiating H. orientalis pollen cells. It seems that in the generative cell, the majority of synthesized transcripts undergo translation during the course of pollen development; however, some of the pool of poly(A) RNA is stored in the generative cell of mature pollen grain. The role of these transcripts remains a poorly understood topic, although in recent years an increasing number of generative cell-specific genes have been identified. We are led to believe that mRNAs indicated in the generative cell of H. orientalis may encode some specific genes and these might be involved in the control of further steps of the male gametophyte development.
References
Alves-Ferreira M, Wellmer F, Banhara A, Kumar V, Riechmann JL, Meyerowitz M (2007) Global expression profiling applied to the analysis of Arabidopsis stamen development. Plant Physiol 145:747–762
Becker JD, Boavida LC, Carnerio J, Haury M, Feijó JA (2003) Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol 133:713–725
Bednarska E (1984) The differentiation of pollen cells in Hyacinthus orientalis L.—ultrastructural and metabolic studies. Post Biol Kom 11:585–587
Bednarska E, Górska-Brylass A (1987) Ultrastructural transformations of nuclei in differentiating Hyacinthus orientalis L. pollen grain cells. Acta Soc Bot Pol 4:667–685
Bentley DL (2005) Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 17:251–256
Bregman DB, Du L, Van der Zee S, Warren SL (1995) Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol 129:287–298
Colot HV, Rosbash M (1982) Behavior of individual maternal pA + RNA during embryogenesis of Xenopus laevis. Dev Biol 94:79–86
Dirks RW, Snaar S (1999) Dynamics of RNA polymerase II localization during the cell cycle. Histochem Cell Biol 111:405–410
Fakan S (1994) Perichromatin fibrils are in situ forms of nascent transcripts. Trends Cell Biol 4:86–90
Grande MA, van der Kraan I, de Jong L, van Driel R (1997) Nuclear distribution of transcription factors in relation to sites of transcription and RNA polymerase II. J Cell Sci 110:1781–1791
Gray NK, Wickens MP (1998) Control of translation initiation in animals. Ann Rev Cell and Dev Biol 14:399–457
Honys D, Twell D (2003) Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol 132:640–652
Honys D, Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5:R85
Juge F, Zaessinger S, Temme C, Wahle E, Simonelig M (2002) Control of poly(A) polymerase level is essential to cytoplasmic polyadenylation and early development in Drosophila. EMBO J 21:6603–6613
Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G (2004) Multiple links between transcription and splicing. RNA 10:1489–1498
Lee TI, Young RA (2000) Transcription of eukaryotic protein-coding genes. Annu Rev Genet 34:77–137
Mascarenhas JP (1975) The biochemistry of angiosperm pollen development. Bot Rev 41:259–314
Mascarenhas JP (1990) Gene activity during pollen development. Annu Rev Plant Physiol 41:317–338
Mascarenhas JP (1993) Molecular mechanisms of pollen tube growth and differentiation. Plant Cell 5:1303–1314
McCormick S (1993) Male gametophyte development. Plant Cell 5:1265–1275
Pina C, Pinto F, Feijó JA, Becker JD (2005) Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol 138:744–756
Proudfoot N, O’Sullivan J (2002) Polyadenylation: a tail of two complexes. Curr Biol 12:R855–R857
Raghavan V (1981) Distribution of poly(A)-containing RNA during normal pollen development and during induced pollen embryogenesis in Hyoscyamus niger. J Cell Biol 89:593–606
Raghavan V (1989) mRNAs and a cloned histone gene are differentially expressed during anther and pollen development in rice (Oryza sativa L.). J Cell Sci 92:217–229
Reed R (2003) Coupling transcription, splicing and mRNA export. Curr Opin Cell Biol 15:326–331
Sims RJ, Mandal SS, Reinberg D (2004) Recent highlights of RNA-polymerase-II-mediated transcription. Curr Opin Cell Biol 16:263–271
Tupy J (1982) Alternation in polyadenylated RNA during pollen maturation and germination. Biol Plantarum 24:331–340
Tupy J, Süss J, Hrabetova E, Rihova L (1983) Developmental changes in gene expression during pollen differentiation and maturation in Nicotiana tabacum. Biol Plantarum 25:231–237
Willemse MTM, Van Went JL (1984) The female gametophyte. In: Johri BM (ed) Embryology of angiosperms. Springer, Berlin, pp 159–196
Wilt FH (1977) The dynamics of maternal poly(A) containing mRNA in fertilized sea urchin eggs. Cell 11:673–681
Xie SQ, Pombo A (2006) Distribution of different phosphorylated forms of RNA polymerase II in relation to Cajal and PML bodies in human cells: an ultrastructural study. Histochem Cell Biol 125:21–31
Zeng C, Kim E, Warren S, Berget S (1997) Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J 16:1401–1412
Zienkiewicz K, Smolinski DJ, Bednarska E (2006) Distribution of poly(A) RNA and splicing machinery elements in mature Hyacinthus orientalis L. pollen grains and pollen tubes growing in vitro. Protoplasma 227:95–103
Zienkiewicz K, Zienkiewicz A, Rodriguez-Garcia MI, Smolinski DJ, Swidzinski M, Bednarska E (2008) Transcriptional activity and distribution of splicing machinery elements during Hyacinthus orientalis pollen tube growth. Protoplasma 233:129–139
Acknowledgment
This work was supported by the grant of Polish Ministry of Science and Higher Education no. No.N303 290434.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Scott Russell.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zienkiewicz, K., Zienkiewicz, A., Smoliński, D.J. et al. Transcriptional state and distribution of poly(A) RNA and RNA polymerase II in differentiating Hyacinthus orientalis L. pollen grains. Sex Plant Reprod 21, 233–245 (2008). https://doi.org/10.1007/s00497-008-0085-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00497-008-0085-9