Abstract
Gametophytic apomixis is an asexual mode of reproduction by seeds. This trait is present in several plant families and is strongly associated with polyploidy. Paspalum rufum is a forage grass with sexual self-incompatible diploids (2n = 2x = 20) and aposporous-apomictic pseudogamous tetraploids (2n = 4x = 40). In previous work embryological observations of the diploid genotype Q3754 showed 8.8–26.8% of the ovaries having one meiotic plus an aposporous-like embryo sac, suggesting some capability for apomictic reproduction. The objective of this work was to characterize progenies derived from Q3754 to determine if aposporous sacs were functional and generated progenies via apomixis at the diploid level. Re-examination of Q3754 ovaries showed that 12.5% of them contained one sexual plus an aposporous sac confirming previous results. Progeny tests were carried out on two experimental families (H1 and S1) employing heterozygous RAPD marker loci. Family H1 was obtained crossing Q3754 with a natural diploid genotype (Q3861) and S1 derived from the induced self-pollination of Q3754. Genetic analysis of H1 showed that all individuals derived from sexual reproduction. However, 5 out of 95 plants from S1 showed the same heterozygous state as the mother plant for 14 RAPD loci suggesting a clonal origin. Further experiments, designed to test the functionality of aposporous sacs by flow cytometric analyses, were carried out on a third family (M1) obtained by crossing Q3754 with the tetraploid plant Q3785. Histograms of 20 M1 plants showed 15 diploids (75%), 4 triploids (20%) and 1 tetraploid (5%). Triploids and the tetraploid may have originated from functional aposporous embryo sacs. Likewise, the reconstruction of the developmental route of 40 individual seeds demonstrated that 11 of them (27.5%) derived from fertilized aposporic sacs. The results presented in this work indicate that gametophytic apomixis is effectively expressed at the diploid level in Paspalum rufum and could be the foundation of a recurrent auto-polyploidization process in the species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Paspalum is a large genus of the Poaceae family that includes about 400 species mainly distributed in tropical and subtropical areas of the Americas (Zuloaga and Morrone 2005). Ploidy levels within the genus range from diploid to 16-ploid, while modes of reproduction range from allogamy to apomixis (reviewed by Quarin 1992). Species are generally organized as agamic complexes, where diploid sexual self-incompatible cytotypes, have pseudogamous, self-compatible apomictic polyploid counterparts (Quarin 1992). Paspalum rufum Nees is a classical example of such a species. Diploid races (2n = 2x = 20) reproduce sexually and are mainly self-sterile, while tetraploids (2n = 4x = 40) are pseudogamous aposporous apomicts (Norrmann et al. 1994). In spite of these differing modes of reproduction diploid and tetraploid forms are morphologically similar, with crosses between them giving rise to hybrids at a very low frequency (Norrmann et al. 1994).
Apomixis, i.e., asexual reproduction through seeds, is a genetically determined character that generates clonal progenies (Nogler 1984). In one of its forms, gametophytic apomixis, an embryo-sac of non-reduced cells (2n) is formed through a mitotic process. It can arise from the megaspore mother cell itself after a failure in meiosis (diplospory) or from a nuclear cell (apospory). The parthenogenetic development of the embryo from the egg cell and the formation of the endosperm autonomously, or after the fertilization of the polar nuclei (pseudogamy), complete seed formation (Savidan 2000).
Gametophytic apomixis tends to occur in polyploids, and then most often at the tetraploid or greater levels (Asker and Jerling 1992). It is accepted that apomictic mutants may have arisen among polyploids of hybrid origin as an escape from sterility (Darlington 1939). According to de Wet and Stalker (1974) the principal adaptative advantage of apomixis is that it can restore fertility in sexually sterile individuals. However, in the Panicoid grasses, apomixis has been also associated with autoploidy (Quarin 1992; Quarin et al. 1998).
While there is a strong link between polyploidy and apomixis, only some unusual cases of gametophytic apomixis have been reported among diploids of Potentilla argentea (Müntzing and Müntzing 1945; Asker 1970; Asker and Jerling 1992) and Boechera holboellii (formerly Arabis holboellii) (Böcher 1951; Kantama et al. 2007). Nevertheless, further re-examinations of Potentilla argentea questioned previous results (Holm and Ghatnekar 1996; Holm et al. 1997). Other cases of alleged apomictic diploids involved species with a basic number of chromosomes high enough to have most probably derived from tetraploids (reviewed in Asker and Jerling 1992). Within grasses only a few exceptional cases of species showing one or more components of apomictic reproduction at the diploid level have been reported. Aposporous embryo sacs and parthenogenetic embryos at an early developmental stage were observed in diploid accessions of Brachiaria decumbens. However, no apomictic seeds were recovered due to the lack of endosperm development (Naumova et al. 1999). Occasional ovules bearing an aposporous embryo sac together with the typical meiotic one have been reported in several diploid Paspalum species, i.e., Paspalum cromyorrhizon, Paspalum equitans, Paspalum intermedium, Paspalum quadrifarium, Paspalum haumanii, Paspalum brunneum, Paspalum rufum and Paspalum notatum (Quarin 1986; Quarin and Norrmann 1987; Norrmann et al. 1989; Quarin et al. 2001). However, no information about the functionality of the apomeiotic embryo sacs is available so far for these species.
As a result of the collection trips carried out by our Institution during the last 20 years, several diploid and tetraploid races of Paspalum rufum became available for investigation. Embryological observation of the diploid genotype Q3745 revealed that between 8.8 and 26.8% of the ovaries included a meiotic plus an aposporous-like embryo sac (Norrmann et al. 1989). Moreover, experimental crosses involving over 13,000 spikelets of diploid Q3754, dusted with pollen of the tetraploid plant Q3785 of the same species, produced three hybrids with 40 chromosomes (Norrmann et al. 1994). This result gave some clues that aposporous sacs might have been functional at the diploid level in Q3754.
Apomixis and sexuality are not mutually exclusive processes, since they can occur simultaneously in the same plant and even in the same ovule (Harlan et al. 1964; Nogler 1984). An apomict that is able to generate at least part of its progeny by sexual means is regarded as facultative. Progenies from facultative apomicts segregate as BII hybrids (n + n) derived from sexual seed development or maternal (2n + 0) derived from the apomictic pathway. Moreover, intermediate forms can also occur. A BIII hybrid may arise following the fertilization of an unreduced egg cell (2n + n) or a plant with decreased ploidy (polyhaploid) may arise from parthenogenetic development of a reduced egg cell (n + 0) (Savidan 2000). It is also possible to differentiate between BII and BIII hybrids derived from cross-fertilization from SII and SIII individuals derived from self-fertilization events (Bicknell et al. 2003).
Progeny tests are valuable methods for estimating the level of agamospermy in facultative apomicts that avoid a number of the deficiencies inherent in the classical cytological methods (Marshall and Brown 1974). In Paspalum notatum, genetic fingerprinting with RAPD and RFLP molecular markers were used for determining the degree of apomictic reproduction of different tetraploid accessions and the effect of the pollination time on the proportion of sexual and apomictic offspring (Ortiz et al. 1997; Espinoza et al. 2002). Recently, a simple method for screening rare maternal progenies in diploid transgenic Arabidopsis based on heterozygous SSLP-markers was reported by Kantama et al. (2006). On the other hand, flow cytometric seed screen (FCSS) methods are powerful tools for distinguishing between different modes of reproduction of plants involving sexual and apomictic development (Matzk et al. 2000). This method is based on estimations of the relative DNA content of the embryo and endosperm of single seeds. Species of Paspalum are particularly suitable for these studies because pseudogamy in aposporous embryo sacs involves the fertilization of two cytologically unreduced polar nuclei, giving a DNA content ratio between embryo and endosperm of 2/5 which differs from the 2/3 ratio from the regular sexual reproduction (Quarin 1999).
The objective of this work was to characterize experimental progenies derived from the diploid genotype Q3754 of Paspalum rufum for determining if aposporous embryo sacs observed in this plant were functional and progenies originated via apomixis.
Materials and methods
Plant material
The plant material used in this work consisted of the natural diploid (2n = 2x = 20) genotype of Paspalum rufum Q3754 and three experimental populations derived from it. Accession Q3754 was a single plant collected from a natural population at Villa Ana, Santa Fe, Argentina (Norrmann et al. 1989). Cytoembryological observations revealed that this plant reproduces sexually. Moreover, it set seed normally when was cross-pollinated with a different diploid genotype of the same species (Norrmann et al. 1989). Notwithstanding, analyses carried out at two different flowering periods showed that between 8.8 and 26.8% of the ovules included one meiotic and one aposporous sac indicating some capacity for apomeiotic development at the diploid level (Norrmann et al. 1989). Family H1 (44 individuals) was generated by crossing Q3754 as the female, with the natural diploid genotype Q3861 of Paspalum rufum (from Paso Lucero, Corrientes, Argentina) as pollen donor. Family S1 (95 individuals) was generated by inducing the self-pollination of Q3754 through dusting spikelets at anthesis with pollen of Paspalum urvillei and then bagging the inflorescences. Pollen from Paspalum urvillei does not fertilize Paspalum rufum, but partially breaks down the natural self-incompatibility system and allows pollen grains of Paspalum rufum to germinate on its own stigma and fertilize the egg cell (C.L. Quarin, unpublished data). This procedure comes from an old attempt to produce interspecific hybrids that involved 2,453 no-emasculated spikelets of Q3754 dusted with pollen of a tetraploid Paspalum urvillei. Although 89 plants were recovered (3.62%), no hybrids were obtained, and all plants were diploids resembling the maternal phenotype (data not published). Though we failed to produce the targeted hybrids, we had found a way to partially break down the self-incompatibility system and generate self-pollinated progenies. This method is in agreement with previous trials in other Paspalum species like Paspalum equitans and Paspalum ionanthum (Quarin and Norrmann 1987) and Paspalum notatum (Burton and Hanna 1992) where the addition of foreign pollen to the stigmata increased the capacity of self-pollination. Moreover, since some hybrids have been reported from crosses involving different ploidy levels (Norrmann et al. 1994) family M1 was obtained by pollinating spikelets of Q3754 at anthesis (without emasculation) with the tetraploid (2n = 4x = 40) genotype Q3785. Seeds were germinated in Petri dishes with sterilized sand after incubation at 40°C for 12 h. Plantlets were transferred to pots and maintained in a greenhouse.
Cytoembryological analysis
Inflorescences at anthesis were collected and fixed in FAA (70% ethanol, glacial acetic acid, formaldehyde, 90:5:5) for 24–48 h. Pistils were dehydrated in a tertiary butyl alcohol series and embedded in paraffin. Samples were sectioned at 12–15 μm and stained with safranin-fast green. Observations were carried out with a light transmission microscope. Classification of the embryo sac types was performed according to Norrmann et al. (1989).
DNA extraction and molecular markers assays
Genomic DNA was isolated as described in Martínez et al. (2003), from 5 to 6 g of fresh leaf tissue. Sixty arbitrary RAPD oligonucleotides from the University of British Columbia (UBC) series were screened to detect primers giving multiple band-amplification patterns in Q3754 and specific markers of Q3861 genotypes, respectively. PCR reactions were performed in 25 μl total volume containing 25 ng of genomic DNA, 1× Taq reaction buffer (Promega), 200 μM of each dNTP, 1.5 mM MgCl2, 30 ng primer and 1.25 U of GoTaq polymerase (Promega). Amplifications were carried out using a MJ Research, PTC-100 Programmable Thermal Controller including 2 min at 95°C followed by 45 cycles of 1 min at 95°C, 1 min at 36°C and 2 min at 72°C and a final incubation of 5 min at 72°C. PCR products were analysed in 2.5% agarose gels stained with ethidium bromide and visualized under ultraviolet light.
Screening for informative markers was carried out using DNA samples from Q3754, Q3861 and five individuals of their progeny (H1). This scheme was used to detect heterozygous loci in both genotypes, i.e., polymorphic markers segregating in the progeny with a P > 0.90 (Mather 1951). Thereafter, segregation analyses of selected markers were performed including all individuals of each progeny family. Markers were evaluated for 1:1 or 3:1 presence/absence ratios in the H1 and S1 populations, respectively. A Chi-square test was used to determine the goodness of fit between the observed and the expected number of genotypes for each class of segregation ratio. Ratios that differed from the expected value (at P ≤ 0.05) were classified as distorted. Moreover, for avoiding the inclusion in the study of markers clustered in specific genomic regions, a linkage analysis was carried out by using Joinmap 1.4 (Stam 1993). Segregation data were tested as dominant loci derived from a pseudo-test cross or an F2 for families H1 and S1, respectively, at a minimum LOD score of 3.0 according to Ortiz et al. (2001).
Estimation of the frequency of agamospermy by progeny test
An estimation of the frequency of apomictic reproduction of Q3754 was based on segregation data following the method described by Marshall and Brown (1974). It involves progeny testing of known genotypes polymorphic for specific marker loci. The model applied to species reproducing by mixed agamospermy (c), selfing (s) and random outcrossing (t) (c + s + t = 1) was used (Marshall and Brown 1974). This method assumes that autosegregation is absent, and thus, all progenies derived from agamospermy are identical to the mother plant. Moreover, the probability (j) to find in a progeny family i individuals with the same heterozygous state as the mother plant for n alleles after self-pollination was estimated by j = [p n]i, where P = 0.75 (frequency for a dominant allele derived from an heterozygous diploid individual).
Flow cytometry
Flow cytometry analyses were used to determine the ploidy level of experimental plants and to reconstruct the reproductive pathways of mature seeds. In all cases, the fluorescence intensity of DAPI-stained nuclei was determined using a Partec PA II flow cytometer (Partec GmbH, Münster, Germany) with the detector operating at 355 nm. About 3,000 nuclei were measured per sample. Data analysis was performed using PA II`s Partec FloMax software. Ploidy level of progenies was determined using samples of leaf tissue following the recommendations of the Partec P kit CySatin UV Precise P 05-5002 manual. Briefly, 0.5 cm2 of leaf material was placed in a small Petri dish with a comparable amount of tissue from an internal standard (the diploid Q3754). After adding extraction buffer (0.5 ml) the tissue was chopped with a sharp razor blade. Following 2 min incubation, samples were filtered through a 50-μm nylon mesh directly into the sample tube where 1.5 ml of DAPI (0.6-diamidino-2-phenylindole) stain solution (Partec P kit CySatin UV Precise P 05-5002) was added. The mixture was incubated for a further 2 min at room temperature and analysed. Ploidy levels were estimated by comparing the DNA peak of the samples to the internal standard. To reconstruct the reproductive pathways of mature seeds, the FCSS method described by Matzk et al. (2000) was used. Individual seeds derived from crosses between Q3754 (diploid) × Q3785 (tetraploid) were finely chopped in 0.5 ml of extraction buffer (Partec P kit CySatin UV Precise P 05-5002), filtered through 30 μm nylon mesh into a sample tube and incubated for 1 min before adding 1.5 ml of DAPI stain (Partec P kit CySatin UV Precise P 05-5002). Ploidy levels of the endosperm and embryo tissues were estimated by comparing the different peak configurations.
Results
Embryo sac analysis
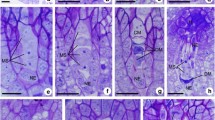
The mode of reproduction of Q3754 was re-examined by cytoembryological observations of embryo sacs at anthesis. Forty-eight ovules were scored for the presence of (1) ovaries carrying one meiotic embryo sac of the Polygonum type, i.e., derived from the reduced functional megaspore and (2) ovaries bearing one meiotic plus an aposporous embryo sac. Observations showed 42 ovules (87.5%) bearing exclusively one sac of the Polygonum type. They were characterized by the presence of the egg cell and two synergids at the micropilar end, a large, widely vacuolate central cell with two polar nuclei, and a group of antipodals at the chalazal end (Fig. 1a). On the other hand, six ovules (12.5%) showed one meiotic plus an aposporous embryo sac. Aposporous sacs were smaller than the meiotic ones and contained four to five nuclei corresponding to the egg cell, one or two synergids and a large cell with two polar nuclei. All aposporous sacs lacked antipodal cells (Fig. 1b–d). The proportion of ovules carrying a meiotic plus an aposporous embryo-sac fell within the range described by Norrmann et al. (1989) and confirmed the potentiality for apomeiosis of genotype Q3754.
Sections of ovules from diploid genotype Q3754 of Paspalum rufum stained with safranin-fast green (×400). a ovule with a mature embryo sac of the Polygonum type (meiotic embryo sac) with the egg cell, two polar nuclei and a group of antipodals. One synergid and several others antipodal cells were present in adjacent sections of the same ovule (not-shown). b–d three consecutive sections of one ovule bearing a mature meiotic embryo sac plus an aposporous embryo sac. a antipodals, e egg cell, p polar nuclei, s synergids, discontinuous lines indicate the boundaries of embryo sacs
Progeny test
Sixty primers from the UBC series were screened for detecting informative markers for both Q3754 and Q3861 genotypes. Twelve decamers (UBC numbers: 301, 308, 313, 319, 322, 329, 349, 359, 361, 364, 368 and 375) were selected because they generated clear multiple amplicons in both genotypes. A total of 46 and 31 RAPD amplification bands were obtained in Q3754 and Q3861, respectively. Segregation analysis using DNA from both genotypes and a sample of five randomly chosen individuals of its progeny (family H1) allowed the identification of 14 and 6 heterozygous markers segregating from Q3754 and Q3861, respectively. Informative markers were used to genotype each progeny plant from both families and estimate genetic parameters of the mating system of genotype Q3754. Segregation data are presented in Tables 1 and 2. Most markers fit with the expected segregation values for dominant alleles of a diploid individual (1:1 or 3:1), but one of them showed a distorted segregation ratio in family S1 (Table 2). Linkage analysis with data derived from both populations (H1 and S1) showed independent segregation for all markers indicating they mapped at random on unlinked genomic regions (data not shown).
Progeny plants from both populations (H1 and S1) were quite uniform and resembled the maternal parent in their phenotypes. No obvious morphological traits were detected for determining the way they originated. The sexual or asexual origin of the progeny plants was determined by comparing the amplification pattern for each individual (RAPD fingerprints) with the maternal profile. All 44 progenies of the H1 family produced at least one of the six markers derived from the paternal plant and segregated a band from the maternal genotype. This outcome indicated that the whole progeny derived from the sexual reproduction pathway. The combination of fingerprints obtained with five RAPD markers was sufficient to determine the hybrid origin of all H1 individuals (Table 1).
Analysis of family S1 (95 individuals) showed that 90 plants segregated for at least one maternal marker, but five individuals exhibited the same amplification pattern as Q3754 for the 14 heterozygous RAPD markers loci tested. These five plants consistently maintained the maternal profile for all markers in at least three independent experiments. This result strongly suggests fixed heterozygosity in these plants, since the probability for finding five progenies, with the same maternal allelic configuration for the 14 alleles tested based on a random Mendelian segregation, would be as low as 1.79 × 10−9. The proportion of maternal plants in the population resulted of 0.052 (5/95). Moreover, an overall estimation of the average frequency of selfing (ŝ) and agamospermy (ĉ) of genotype Q3754 derived from genetic data of the 13 undistorted markers segregating in S1 resulted in 0.926 and 0.073, respectively (Table 2). The five maternal progenies detected in this experiment were grown until maturity in a greenhouse. All of them set seed normally under open pollination conditions in the presence of other diploid genotypes.
Flow cytometry
Flow cytometry experiments were set up to determine the DNA content of experimental progenies and screen for C-values corresponding to seeds derived from sexuality and apomixis. Initially the ploidy level of a sample of ten plants from both H1 and S1 families (in the later case including the five maternal plants detected in the progeny test) was analysed. The histogram of all individuals corresponded to diploids (not-shown) as expected for progenies derived from sexuality in the case of H1 and S1 as well as for apomixis in S1.
Experiments carried out using a sample of 20 progenies from family M1 showed 15 plants (75%) with peaks corresponding to diploids, 4 (20%) displayed values of triploids and 1 (5%) with a peak of a tetraploid (Fig. 2a–c). Diploids must have originated by the union of both female and male reduced gametes (n + n) from Q3754 and should correspond to SII progenies, although the possibility that some of them derived from apomictic reproduction cannot be discarded. Triploids might have originated by the fertilization of a reduced female gamete (n = x) with a reduced male gamete (n = 2x) from the tretraploid parent Q3785 or by the fusion of an unreduced maternal gamete (2n = 2x) derived from apomeiosis, with a normal reduced male gamete (n = x) from Q3754, corresponding to SIII individuals. We were unable to distinguish between these two possibilities in this analysis. The tetraploid plant could only have originated by the union of an unreduced female gamete (2n) from Q3754 with a normal reduced male gamete from the tetraploid genotype (2n) (Fig. 2c).
Flow cytometric analyses of progeny plants of family M1 (derived from a cross between Q3754 (2n = 2x = 20) with the tetraploid (2n = 4x = 40) genotype Q3785). a flow cytometry histogram of a diploid progeny and the control with a high peak at 2C. b flow cytometry histogram of a triploid individual showing a peak at 3C and the control Q3754 (2C). c Flow cytomety histogram of a tetraploid plant with a high peak at 4C and the diploid control Q3754 (2C)
Further experiments were carried out to reconstruct all possible ways of seed development in Q3754 by using the flow cytometric method (Matzk et al. 2000) on a sample of 40 individual seeds from M1. Table 3 shows the theoretical ways of development and the numbers of seeds scored in each class. Most of them (72.5%) showed two main peaks corresponding to 2C (embryo) and 3C (endosperm). They must have arisen by self-pollination of meiotic embryo sacs (Table 3, Class I, Fig. 3a). Nine seeds (22.5%) occurred in the developmental Class V (Table 3, Fig. 3b). They showed peaks corresponding to 3C and 5C and must have originated by the fertilization of an unreduced female gamete (2n) of Q3754 by a normal reduced male gamete of the same genotype (SIII progenies). This kind of seed indicated that aposporous embryo sacs had been effectively fertilized by a normal reduced male gamete of the same plant. The relative high proportion of this type of seed may be explained by the functionality of the aposporous sacs and the endosperm balance 4:1, which is functional in aposporous pathways of seed development in Paspalum (Quarin 1999). Moreover, two seeds (5%) showed histograms displaying peaks corresponding to 4C and 6C (Table 3, Class VI, Fig. 3c). The only possibility for obtaining this kind of seeds is by the fusion of an unreduced female gamete of the Q3754 (2n = 2x) with a reduced male gamete (n = 2x) from 4x Q3785 and should represent (BIII) hybrids between diploid and tetraploid cytotypes. No seeds corresponding to Classes II, III and IV were recovered.
Cytometric histograms of individual seeds from family M1 (derived from a cross between Q3754 (2n = 2x = 20) and the tetraploid (2n = 4x = 40) genotype Q3785). a histogram of a seed corresponding to Class I (Table 3). b histogram of a seed matching to Class V (Table 3). c histogram of a seed corresponding to Class VI (Table 3)
Discussion
In previous embryological studies it was noticed that several sexual outcrossing (self-sterile) diploids of Paspalum occasionally develop an aposporous embryo sac beside the regular meiotic one (Quarin 1986; Quarin and Norrmann 1987; Norrmann et al. 1989). These observations indicated that some potential for apomictic reproduction can be expected in the genus at the diploid level. Because apomixis is closely related with polyploidy, the determination of its expression in diploids could lead to the definition of a new hypothesis about the genetic control of the trait as well as the stability and evolution of agamic complexes. Re-examination of Q3754 ovules confirmed the formation of aposporous embryo-sacs in this plant. Similar results were reported in Brachiaria (Naumova et al. 1999). The presence of aposporous sacs in these plants indicated that the factor/s responsible for apospory is/are occasionally expressed in diploids. Consequently, the development of maternal plants may be possible if the other components of apomictic reproduction (parthenogenesis of the embryo and endosperm development) could be also functional at the diploid level. The flowers analyzed in this work were fixed at the same time that Q3754 was crossed to produce the three families studied. Thus, the results obtained with progeny tests and flow cytometry could be evaluated from the perspective of the embryological data.
Genetic studies carried out in the H1 family indicated that the whole progeny derived from sexual reproduction. All individuals showed the presence of at least one paternal marker transmitted by pollen. Moreover, no BIII hybrids were detected because all plants showed the segregation of at least one maternal band as a consequence of the female meiosis. This outcome indicated that in spite of the presence of aposporous embryo sacs, Q3754 functioned mainly as an out-breeding individual when pollen from a different diploid genotype is available.
On the other hand, analysis of S1 progeny showed 5 out of 95 plants carrying the same heterozygous status as the mother plant for 14 segregating loci. This genetic constitution has a very low probability to have occurred by sexuality and thus, a clonal origin of these plants can be assumed. The determination of mating parameters considering 13 undistorted markers indicated that Q3754 reproduced mainly by sexuality, but an average frequency of agamospermy of about 0.073 can be expected. Although this kind of reproduction would be infrequent some plants may originate by this means.
Estimations of DNA content of progeny plants and seeds by flow cytometry confirmed the different modes of reproduction occurring in Q3754, i.e., (1) exclusively diploid plants were recovered in families H1 and S1; (2) diploids, triploids and tetraploids plants were produced by crossing 2x Q3754 with 4x Q3785 (family M1) (Fig. 2) and (3) seeds derived from meiotic as well as from aposporous embryo sacs were identified in the cytometric analysis of single seeds of family M1 (Table 3, Fig. 3). These results indicated that all component of gametophytic apomixis are functional at the diploid level in Q3754. There is experimental evidence in Paspalum notatum that a rise of ploidy level from diploid (sexual) to tetraploid by colchicine treatment induced the expression of apomixis (Quarin et al. 2001). These antecedents support the idea that the genetic factor/s for apomixis exists at the diploid level, but their expression is repressed as postulated by Quarin et al. (2001).
Particularly important is the detection of tripoloid and tetraploid seeds (Table 3, Classes V and VI, Fig. 3b, c) within family M1. Triploids of this type would represent the “triploid bridge” between diploids and polyploids proposed for the genus (Quarin 1992). Occasionally apomictic triploid plants have been found in Paspalum intermedium and Paspalum notatum (Quarin 1992), while apomixis and triploidy are the common condition for Paspalum quadrifarium (Quarin and Lombardo 1986). Moreover, occasional apomictic 3x plants were found for Paspalum simplex (Urbani et al. 2002) and Paspalum wrightii, Paspalum unispicatum and Paspalum cromyorrhizon (C.L. Quarin, unpublished data). All these species have sexual diploid as well as apomictic polyploid (mainly tetraploid) representatives (Quarin 1992). Triploid embryos may have arisen directly from a single 2x plant and could be the starting point for the generation of new autotetraploids within a diploid population. This system would act as one-way gene flow process from sexual outcrossing representatives to apomictic tetraploids of the same species. The rise of ploidy level from the 3x to 4x have been observed in several species of Paspalum (Burton and Hanna 1986; Quarin et al. 1989). Likewise, tetraploid seeds (Table 3 Class VI and Fig. 3c) and plants recovered in our experiments would represent a one-step tetraploidization process within a mixed diploid-tetraploid population through the fertilization of an unreduced gamete from a diploid by a reduced gamete of a neighbouring tetraploid.
Considering that new polyploids can originate from diploids by sequential steps of 2n + n hybridization or directly by inter-ploidy crosses (2n + 2n), it is clear that this peculiar strategy allows the maintenance of a certain degree of variability among polyploids, even though they reproduce by apomixis. A relatively high degree of genetic variability was found in a tetraploid apomictic population of Paspalum notatum situated sympatrically with diploid sexuals, compared to a pure tetraploid population localized in isolation (Daurelio et al. 2004). Even though polyploid species of multiple origin do exist in Paspalum, and examples of 4x, 5x and 6x strains of Paspalum dilatatum and 4x races of Paspalum urvillei obtained through interspecific hybridization experiments are well documented (Burson 1992), our results confirmed that there is also a recurrent autopolyploidization process within species of the genus. Different authors have proposed, based on cytological studies, the autoploid origin for diverse Paspalum species. Autoploidy has been confirmed at least for Paspalum simplex and for Paspalum notatum based on tetrasomic inheritance of molecular markers (Pupilli et al. 1997; Stein et al. 2004).
Recurrent polyploidization involving different parental diploids was suggested as the common way of new polyploid formation for most plant species (Soltis and Soltis 1999). We found that some embryos were formed in a 2x × 4x cross via syngamy of 2n (female) + n (male) gametes from the same diploid parent giving rise to a new autotriploid genotype. These results favoured, at least for Paspalum rufum, the conclusion of Ramsey and Schemske (1998) that the rate of autopolyploid formation may be higher than allopolyploidy. A recurrent auto-polyploidization process within a diploid population could occur in the genus Paspalum with the following characteristics: (1) a diverse array of genotypes exists at diploid level of each species due to a genetic self-incompatible system (allogamy); (2) the fertilization of scarce unreduced (2n) gametes formed in some diploids by a normal male gamete (n) would give rise to apomictic triploids and (3) in a second step of fertilization, triploids would generate new apomictic tetraploids by fusing with normal pollen from diploids. Moreover, our results agreed with the theoretical analysis of Yamauchi et al. (2004), who stated that when both triploids and tetraploids reproduce parthenogenetically, the ploidy level with the highest fitness is likely to dominate the population through direct competition among cytotypes. The lower fitness of 3x in Paspalum may be assigned to the low seed set observed in spite of their apomictic system and would be the reason why tetraploids are the prevalent cytotypes in natural populations.
References
Asker S (1970) Apomixis and sexuality in the Potentilla argentea complex. II Crosses within the complex. Hereditas 66:189–204
Asker SE, Jerling L (1992) Apomixis in plants. CRC, Boca Raton
Bicknell RA, Lambie SC, Butler RC (2003) Quantification of progeny classes in two facultatively apomictic accessions of Hieracium. Hereditas 138:11–20
Böcher TW (1951) Cytological and embryological studies in the amphi-apomictic Arabis holboellii complex. Kongel Danske Vidensk Selskab Biol Skr 6:159
Burson BL (1992) Cytology and reproductive behavior of hybrids between Paspalum urvillei and two hexaploid Paspalum dilatatum biotypes. Genome 35(6):1002–1006
Burton GW, Hanna WW (1986) Bahiagrass tetraploids produced by making (apomictic tetraploid × diploid) × diploid hybrids. Crop Sci 26:1254–1256
Burton GW, Hanna WW (1992) Using apomictic tetraploids to make a self-incompatible diploid Pensacola bahiagrass clone set seed. J Hered 83:305–306
Darlington CD (1939) Evolution of genetic systems. Cambridge University Press, Cambridge
Daurelio DL, Espinoza F, Quarin CL, Pessino SC (2004) Genetic diversity in sexual diploid and apomictic tetraploid populations of Paspalum notatum situated in sympatry or allopatry. Plant Syst Evol 244:189–199
de Wet JMJ, Stalker HT (1974) Gametophytic apomixis and evolution in plants. Taxon 23:689–697
Espinoza F, Pessino SC, Quarin CL, Valle EM (2002) Effect of pollination timing on the rate of apomictic reproduction revealed by RAPD markers. Ann Bot 89:165–170
Harlan JR, Brooks MH, Borgaonkar DS, de Wet JMJ (1964) Nature and inheritance of apomixis in Bothriochloa and Dichanthium. Bot Gaz 125:41–46
Holm S, Ghatnekar L (1996) Sexuality and no apomixis found in crossing experiments with diploid Potentilla argentea. Hereditas 125:77–82
Holm S, Ghatnekar L, Bengtsson BO (1997) Selfing and outcrossing but no apomixis in two natural populations of diploid Potentilla argentea. J Evol Biol 10:343–352
Kantama L, Lambert Y, Hu H, de Jong H, de Vries SC, Russinova E (2006) Use of the SSLP-based method for detection of rare apomictic events in a sexual AtSERK1 transgenic Arabidopsis population. Sex Plant Reprod 19:73–82
Kantama L, Sharbel TF, Schranz ME, Mitchell-Olds T, de Vries S, de Jong H (2007) Diploid apomicts of the Boechera holboellii complex display large-scale chromosome substitutions and aberrant chromosomes. Proc Natl Acad Sci USA 104:14026–14031
Marshall DR, Brown AHD (1974) Estimation of the level of apomixis in plant populations. Heredity 32:321–333
Martínez EJ, Hopp E, Stein J, Ortiz JPA, Quarin CL (2003) Genetic characterization of apospory in tetraploid Paspalum notatum based on the identification of linked molecular markers. Mol Breed 12:319–327
Mather K (1951) The measurement of linkage in heredity, 2nd edn. Methuen and Co., London
Matzk F, Meister A, Schubert I (2000) An efficient screen for reproductive pathways using mature seeds of monocots and dicots. Plant J 21:97–108
Müntzing A, Müntzing G (1945) The mode of reproduction of hybrids between sexual and apomictic Potentilla argentea. Bot Notiser 98:49–71
Naumova TN, Hayward MD, Wagenvoort M (1999) Apomixis and sexuality in diploid and tetraploid accessions of Brachiaria decumbens. Sex Plant Reprod 12:43–52
Nogler GA (1984) Gametophytic apomixis. In: Johri BM (ed) Embryology of angiosperms. Springer, Berlin, pp 475–518
Norrmann GA, Quarin CL, Burson BL (1989) Cytogenetics and reproductive behavior of different chromosome races in six Paspalum species. J Hered 80:24–28
Norrmann GA, Bovo OA, Quarin CL (1994) Post-zygotic seed abortion in sexual diploid × apomictic tetraploid intraspecific Paspalum crosses. Aust J Bot 42:449–456
Ortiz JPA, Pessino SC, Leblanc O, Hayward MD, Quarin CL (1997) Genetic fingerprinting for determining the mode of reproduction in Paspalum notatum, a subtropical apomictic forage grass. Theor Appl Genet 95:850–856
Ortiz JPA, Pessino SC, Bhat V, Hayward MD, Quarin CL (2001) A genetic linkage map of diploid Paspalum notatum. Crop Sci 41:823–830
Pupilli F, Caceres ME, Quarín CL, Arcioni S (1997) Segregation analysis of RFLP markers reveals a tetrasomic inheritance in apomictic Paspalum simplex. Genome 40:822–828
Quarin CL (1986) Seasonal changes in the incidence of apomixis of diploid and tetraploid plants of Paspalum cromyrrhizon. Euphytica 35:515–522
Quarin CL (1992) The nature of apomixis and its origin in panicoid grasses. Apomixis Newsl 5:8–15
Quarin CL (1999) Effect of pollen source and pollen ploidy on endosperm formation and seed set in pseudogamous apomictic Paspalum notatum. Sex Plant Reprod 11:331–335
Quarin CL, Lombardo EP (1986) Niveles de ploidía y distribución geográfica de Paspalum quadrifarium (Gramineae). Mendeliana 7:101–107
Quarin CL, Norrmann GA (1987) Cytology and reproductive behavior of Paspalum equitans, P. ionanthum and their hybrids with diploid and tetraploid cytotypes of P. cromyorrhizon. Bot Gaz 148:386–391
Quarin CL, Norrmann GA, Urbani MH (1989) Polyploidization in aposporous Paspalum species. Apomixis Newsl 1:28–29
Quarin CL, Norrmann GA, Espinoza F (1998) Evidence for autoploidy in apomictic Paspalum rufum. Hereditas 129:119–124
Quarin CL, Espinoza F, Martínez EJ, Pessino SC, Bovo OA (2001) A rise of ploidy level induces the expression of apomixis in Paspalum notatum. Sex Plant Reprod 13:243–249
Ramsey J, Schemske DW (1998) Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Syst 29:467–501
Savidan Y (2000) Apomixis: genetics and breeding. In: Janick J (ed) Plant breeding reviews, vol 18. Wiley, London, pp 13–86
Soltis DE, Soltis PS (1999) Polyploidy: recurrent formation and genome evolution. Trends Ecol Evol 14:348–352
Stam P (1993) Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. Plant J 3:739–744
Stein J, Quarin CL, Martínez EJ, Pessino SC, Ortiz JPA (2004) Tetraploid races of Paspalum notatum show polysomic inheritance and preferential chromosome pairing around the apospory-controlling locus. Theor Appl Genet 109:186–191
Urbani MH, Quarin CL, Espinoza F, Penteado MIO, Rodríguez IF (2002) Cytogeography and reproduction of the Paspalum simplex polyploid complex. Plant Syst Evol 236:99–105
Yamauchi A, Hosokawa A, Nagata H, Shimoda M (2004) Triploid bridge and role of parthenogenesis in evolution of autopolyploidy. Am Nat 164:101–111
Zuloaga FO, Morrone O (2005) Revisión de las especies de Paspalum para América del Sur Austral (Argentina, Bolivia, Sur de Brasil, Chile, Paraguay y Uruguay). In: Hollowell VC (ed). Missouri Botanical Garden Press, St Luis
Acknowledgments
The authors wish to thank Dr Silvina Pessino and Prof. Michael Hayward for critically reading the manuscript. They also thank Florencia Galdeano for her technical assistance. This study was financed by the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT), Argentina, PICT 2003 No. 13578 and PAV 2003 No. 137/3; Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina, PIP 2004 No. 6805. Centro Argentino Brasilero de Biotecnología (CABBIO 2004 No. 012). L. Siena and M. Sartor received fellowships from CONICET and F. Espinoza, C.L. Quarin and J.P.A. Ortiz are career members of CONICET.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J.S. Heslop-Harrison.
Rights and permissions
About this article
Cite this article
Siena, L.A., Sartor, M.E., Espinoza, F. et al. Genetic and embryological evidences of apomixis at the diploid level in Paspalum rufum support recurrent auto-polyploidization in the species. Sex Plant Reprod 21, 205–215 (2008). https://doi.org/10.1007/s00497-008-0080-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00497-008-0080-1