Abstract
Prosopis cineraria, an important multipurpose tree and vital component of the otherwise fragile ecosystem of arid and semiarid regions of India. It is highly drought tolerant and sprouts profusely during the extreme dry summer months when most other trees are leafless. P. cineraria is known to exhibit comparable genetic variations at intra-specific and inter-population levels reflected through morphological and cytogenetical diversity in regions, where this plant grows naturally. In the present study, single primer amplification reaction (SPAR) methods have been used for determination of diversity at DNA level in 30 accessions of P. cineraria collected from different districts of Rajasthan. The analyses include the use of six minisatellite core sequence primers for direct amplification of minisatellite DNA (DAMD), eight inter simple sequence repeats (ISSR) and 20 arbitrary primed decamer sequences for random amplification (RAPD) reactions. Upon analysis of the data generated, all the three SPAR methods, either independently and/or in combination, revealed wide range of genetic variation among accessions. Comparison of matrix of individual SPAR method using MxComp component of NTSYS-pc 2.02 K software proving that analysis of natural genetic variation using combination of SPAR methods particularly ISSR and DAMD, rather than an isolated approach, is very effective. Such an approach also yields better information and reflection of the relatedness and affinities at intra-species and inter-population levels. Therefore, it is opined that in order to reveal the intrinsic intra-specific variation, SPAR approaches involving more than one DNA marker may reveal more authentic genetic variation in tropical tree species like P. cineraria.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Prosopis cineraria (L.) Druce of family Fabaceae is one of the 44 species grown extensively in arid and dry regions of tropics including western region of India (Mann and Shankarnarayan 1980). P. cineraria is a versatile species, providing fodder, fuel for timber, and shade, as well as affecting soil improvement and sand dune stabilization (Bhandari 1978; Burdak 1982). P. cineraria (syn., P. spicigera) occurs naturally in the dry and arid regions of India, Pakistan, Afghanistan, Iran, and Arabia. It is commonly used in dryland agroforestry in India and neighboring Pakistan. The tree is locally known as ‘khejri’ or ‘jandi’ (India), ‘jand’ (Pakistan), and ‘ghaf’ (Saudi Arabia) (Kaul 1967). P. cineraria is a small thorny, irregularly branched, 5–10 feet in height, semi-deciduous tree. It forms an open crown and has thick, rough gray bark with deep fissures (Burkart 1976). It is extremely drought tolerant, growing in areas with as little as 75 mm annual rainfall, with dry seasons of 8 months or more. Slightly frost hardy and tolerant of temperatures up to 50°C, it grows at altitudes from sea level to 600 m. It is one of the principal species on higher and older alluvium in the Indus river valley. The tree is found in alluvial and coarse, sandy, alkaline soils where the pH may reach 9.8.
P. cineraria is also an excellent multipurpose tree that provides excellent firewood (calorific value, ca. 5,000 kcal/kg) and charcoal. Its wood is favored for cooking and domestic heating (Mahoney 1990; Leakey and Last 1980). Hard and reasonably durable, the wood has a variety of uses for house building, posts, tool handles, and boat frames, although poor tree form limits its usefulness as timber. The leaves and pods are excellent as nutritious fodder for many animals (Gates and Brown 1988). Due to multifaceted utility of the species, as mentioned above, the pressure for it and its diverse products is mounting alarmingly. Therefore, there is an urgent need for thorough exploration and exploitation of all the available natural variation in P. cineraria so as to accomplish sustainable utilization. Hence, the present investigation has been undertaken with the main objective of understanding the genetic diversity to define and characterize the natural variation at intra-specific levels in P. cineraria.
The genetic variability among accessions of Prosopis species was earlier determined using randomly amplified polymorphic DNA (RAPD) by Goswami and Ranade (1999). Ferreyra et al. (2004, 2007) reported that RAPD patterns able to differentiate some Argentinean Prosopis species of section Algarobia. Similarly, variation in isozyme pattern (Saidman and Vilardi 1987), and quantum of seed lipids (Lamarque et al. 1994), mineral, crude protein, and structural carbohydrate (Arya et al. 1995) have also been studied for distinguishing species. Other than the above reports of inter-specific variations, not much is known about intra-specific genetic variability in various Prosopis species, especially at the DNA level (Hunziker et al. 1986). In the recent years, the PCR based single primer amplification reaction (SPAR) methods are gaining prominence as effective tools for genetic diversity studies in plants and they collectively provide a comprehensive description of the nature and extent of the diversity (Bhattacharya et al. 2005; Ranade et al. 2009). Three methods involving single primer for amplification reactions are commonly used in diversity analysis of higher plants which include (a) direct amplification of minisatellite DNA regions (DAMD) (Heath et al. 1993); (b) inter simple sequence repeat (ISSR) (Gupta et al. 1994) and (c) random amplified polymorphic DNA (RAPD) (Williams et al. 1990; Welsh and McClelland 1990). Therefore, in the present investigation, we compare the efficiencies of these three SPAR methods, to determine the natural genetic variation among the thirty accessions of P. cineraria, individually as well as collectively.
Materials and methods
Plant material
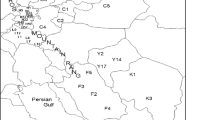
To locate the populations of P. cineraria, several field trips were conducted in five provinces viz. Jodhpur, Barmer, Jaisalmer, Bikaner, and Nagaur of Rajasthan state (Table 1) where maximum diversity is observed. The voucher specimens of all the selected trees were deposited with herbarium at Department of Botany, J. N. V. University, Jodhpur and accession numbers were obtained (Table 1). Leaf samples of one selected tree from each of the thirty accessions of P. cineraria, spreading over five districts of Rajasthan, were used for genomic DNA extraction.
DNA isolation
Frozen leaves were ground and powdered in a pre-chilled mortar using liquid nitrogen, and the DNA was then extracted by the method described by Murray and Thompson (1980). The DNA extracted from the plant material, purified for protein fraction, treated with RNase A, was re-precipitated with pre-chilled absolute ethanol and subsequently dissolved in Tris–EDTA (TE) buffer. The quality of DNA was checked by mupid gel electrophoresis with 0.85% (w/v) agarose in 1× TAE.
RAPD, ISSR, DAMD primers
Two kits (A and C) comprised of 20 decamer random primers per kit were procured form Operon Technologies, Alameda, CA, USA. Fifteen ISSR primers and eight DAMD primers were custom synthesized from Metabion Inc. Ltd., Germany (Table 2).
Amplification reactions
PCR optimization, primer survey and final amplification followed by gel electrophoresis
Varying concentrations of (i) template DNA (20, 30, 40, 50, and 60 ng), (ii) Taq DNA polymerase (0.5–2 U), and (iii) Mg++ salt (1–5 mM) were used to optimize the reaction conditions of the PCR. Of the five different concentrations of template DNA, 50 ng was found to be the most ideal as it yielded maximum number of reproducible bands. In addition, 1.5 mM of MgCl2, and 1 U Taq DNA polymerase have given ideal results among various concentrations tested.
Three randomly selected accessions viz JNVU/RI/2005/1, JNVU/RI/2005/15, and JNVU/RI/2005/30 were chosen for primer survey and screening. Forty RAPD primers from the OPA and OPC series, fifteen ISSR and eight DAMD primers were assayed with these three accessions to identify the primers that were reproducible and generate the most polymorphic patterns. Following the results of initial primer screening experiments, twenty RAPD decamer primers, eight ISSR and six DAMD primers (Table 2) were selected, respectively, for final amplification programme as others produced sub-optimal, indistinct or monomorphic amplification products for the analysis of 30 accessions of P. cineraria. Preliminary experiments for SPAR reproducibility helped to establish the optimal amplification conditions for all the SPAR methods, namely, RAPD, ISSR, and DAMD. Once these conditions were established, amplification reactions for all the genotype primers were thereafter carried out as per these optimized conditions.
On the basis of PCR optimization and primer survey, all further reactions were performed in 25 μl final volume and contained 50 ng template DNA, 20 pmol (RAPD)/40 pmol (ISSR and DAMD), 200 μM of each dNTP, 1.5 mM Mg++, and 1 U Taq DNA polymerase (Bangalore Genei, India). Reactions were thermal cycled for PCR cycle at 94°C for 3 min followed by 44 cycles at 94°C for 1 min, 35°C (in case of ISSR and DAMD amplification, annealing temperature was varied according to primer’s Tm ranging from 45 to 55°C) for 1 min 30 s, and at 72°C for 2 min, with a final extension at 72°C for 7 min in a thermal cycler 2720 (Applied Biosystems, USA). After completion of the amplification, 2.5 μl 10× blue dye was added to the samples, and the amplified DNA was analyzed on 1.5% (w/v) agarose gel in 1× TAE buffer at 65–70 V for 3–4 h.
Scoring and data analysis
Each amplification product was considered a DNA marker and was scored across all samples. Bands were recorded as present (1) or absent (0). Faint bands with low intensity were not considered for final scoring. Molecular weights of the bands were estimated by using Gene Rular 500 bp DNA ladder (MBI Fermantas, UK) as standards. All amplifications were repeated at least twice and only reproducible bands were considered for analysis.
A data set of profile bands was scored manually from the gel profiles and included only the well separated and distinct bands. The data were scored cumulatively first for all the primers in a SPAR method and subsequently the data sets for all the three methods used, were combined together for the final NJ analysis. A pair-wise matrix of distances between genotypes was determined cumulatively for all the three methods using Jaccard coefficient (Jaccard 1901) by the FreeTree program (ver. 0.9.1.5; Pavlicek et al. 1999). This distance matrix was used to compute a single neighbour-joining (NJ) tree after allowing a 1000 replicate bootstrap test using the same program. The tree was viewed, annotated and printed using TreeView (ver. 1.6.5; Page 2001). Co-phenetic and consensus values were recorded discretely for each SPARs and used to commence the MxComp analyses using the computer software NTSYS-pc, version 2.02k (c), Applied Biosystematics Inc. for matrix comparison to calculate r value, generally which can be correlated with reliability of the SPAR methods, either collectively or independently.
Results
SPAR analysis and profile polymorphism
RAPD-PCR
A total of 123 amplification products were scored, which exhibited an overall 83.73% polymorphism (Table 2). The average number of amplification products formed was 6.15, the maximum was 9 with OPA-9 and OPA-16, whereas the minimum was 4 with OPA-20. The size of the amplification products varied in case of each primer and the range was 0.3–2.5 kb. Figure 1a illustrates the extent of polymorphism observed among the 30 accessions of P. cineraria. The amplification products using 20 primers ranged from 40 to 100% in producing polymorphic bands. OPA-9 and OPA-16 were unique among these 20 primers in yielding the highest number of bands (9) with 100% polymorphism (Fig. 1a; Table 1).
Typical SPAR profiles obtained with a one RAPD primer (OPA-9), b one ISSR primer (P1) and c one DAMD primer (HVR) with all 30 accessions of P. cinereria. All profiles were resolved in 1.5% agarose gels in TAE buffer. The lanes marked as Marker are known DNA fragment size markers. The other lanes are marked with accession numbers as in Table 1
ISSR-PCR
A total of 52 amplification products were scored, which exhibited an overall 92.30% polymorphism (Table 2). The average number of amplification products with various primers formed was 6.5 with a maximum of 10 bands with P1, and a minimum of 4 with P7 and P8 primers. The size of the amplification products ranged from 0.7 to 2.3 kb in case of each primer. Figure 1 illustrates the extent of polymorphism observed among the 30 accessions of P. cineraria. The amplification products using eight ISSR primers ranged between 75 and 100% in producing polymorphic bands (Fig. 1b; Table 1).
DAMD-PCR
A total of 29 amplification products were scored, which exhibited an overall 100% polymorphism (Table 2). The average number of amplification products formed was 5.8; the maximum was 7 bands with HBV and YNZ22 primers, whereas the rest yielded five. The size of the amplification products varied in each primer and the range was 0.3–2.2 kb. Figure 1c illustrates the extent of polymorphism observed among the 30 accessions of P. cineraria. The amplification products using five DAMD primers showed 100% in producing polymorphic bands (Fig. 1c; Table 1).
Cluster/tree analysis
In the present study, three SPAR methods, RAPD, ISSR, and DAMD were used which revealed discrete banding patterns for all the primers, many of which revealed 100% polymorphism across all the accessions studied. The cumulative size ranges for the amplified fragments as well as the numbers of fragments amplified are given in Table 3. The cumulative data of all the three SPAR methods were used to compute pair-wise distances. The NJ tree after a 1000 replicate bootstrap test of robustness is shown in Fig. 2. The NJ tree consists of at least four major clusters marked A, B, C and D with large parenthesis and one distinct out-group marked as OG in Fig. 2. Cluster A may be further resolved into three sub-clusters. A1 includes six accessions of which three are from Jodhpur district (JNVU/RI/2005/10, 21, and 22), two are from Barmer district (JNVU/RI/2005/5 and 14) and only one is from Jaisalmer district (JNVU/RI/2005/12). Sub-cluster A2 comprises two accessions JNVU/RI/2005/13 and JNVU/RI/2005/6 belonging to Jaisalmer and Nagaur districts, respectively. The third sub-cluster A3 consists of three accessions viz JNVU/RI/2005/17, JNVU/RI/2005/18, collected from Nagaur district, and JNVU/RI/2005/16 from Barmer district of Rajasthan. An interesting fact is that cluster A does not comprise a single accession of Bikaner district, whereas in case of cluster B, there is absence of accessions from Nagaur district.
Cluster analysis of SPAR data in case of the 30 accessions of Prosopis cinereria after a 1000 replicate bootstrap analysis. The NJ tree was generated for cumulative band data by all the three SPAR methods, DAMD, ISSR-PCR, and RAPD. The accession numbers are abbreviated as in Table 1, and are indicated to the right of the tree. The branch lengths are based on the distance values computed using NJ method and Jaccard coefficient in the program FreeTree. The numbers at the nodes in each tree are the bootstrap percent values (only values greater or nearby to 50 are shown) for the branches to the right of the node. The large parenthesis to the right labeled with A–D are the four major clusters while the all inner parenthesis showing sub-clusters within the respective cluster
Cluster B can be further subdivided into three sub-clusters. B1 comprises three accessions viz JNVU/RI/2005/1, JNVU/RI/2005/2 and JNVU/RI/2005/11, all of which are collected from Jaisalmer district. Sub-cluster B2 consists of three accessions viz JNVU/RI/2005/3, JNVU/RI/2005/4 and JNVU/RI/2005/15, all belonging to Barmer district, whereas the third sub-cluster B3 comprises of three accessions viz JNVU/RI/2005/23, JNVU/RI/2005/24 (from Jodhpur distritct), and JNVU/RI/2005/26 (from Bikaner district). Similarly cluster C can be further subdivided into sub-cluster C1 comprising four accessions, out of which two are from Nagaur (JNVU/RI/2005/19 and 20) and two are from Jodhpur (JNVU/RI/2005/8 and 9). Sub-cluster C2 had only two accessions JNVU/RI/2005/25 and JNVU/RI/2005/30, both belonging to Bikaner district.
Noticeably, cluster D without any sub-cluster, showed tendency towards out-group with three accessions (JNVU/RI/2005/27, 28 and 29), all collected from Bikaner district’s remote villages. Accessions JNVU/RI/2005/17 collected from Degana area of Nagaur district, conspicuously fall into out-group. The NJ tree, overall, does not show any obvious correlation between geographical or morphological variations.
Comparison of different SPAR methods
To determine the efficiency of the three methods, distance range of each SPAR was calculated by Jaccard’s coefficient and the distance value ranged between 0.17 and 0.94 with an average distance of 0.55 for RAPD, whereas in ISSR it ranged between 0.38 and 0.91 with an average distance of 0.64. For DAMD, distance range lies between 0.16 and 0.95 with an average distance of 0.55. Cumulative data set comprising all the three SPARs suggested that the distance value is between 0.28 and 0.82. Consensus values of SPARs pairs viz RAPD + ISSR, RAPD + DAMD, and ISSR + DAMD are calculated which ranged from 0.33 to 0.67, 0.33 to 0.67, and 0.25 to 0.75, respectively. The co-phenetic values were calculated for the same pairs followed by matrix comparison using MxComp programme of NTSYS-pc 2.02k software to obtain the r values of each pair which are recorded as 0.253, 0.421 and 0.913, respectively.
Discussion
P. cineraria is a valuable, multipurpose tree species and highly regarded as ‘State Tree’ by the Government of Rajasthan, India (Sinha 2001; Sahni 1998). Owing to commercial incentives and population pressure, this tree has become threatened in its natural habitat resulting in alarming depletion of its population in recent years. The present study is a preliminary attempt to develop single primer-based DNA amplification methods to distinguish various accessions of P. cineraria exhibiting natural variation at intra-specific level, so as to decide priorities of conservation.
Earlier, Goswami and Ranade (1999) have demonstrated the utility of RAPD analysis in 32 accessions belonging to 11 species of the genus Prosopis and identified genetic variations among them. However, their study deals with inter-specific variation within the genus Prosopis and flings limited information on intra-specific variation. Although 11 species have been included in their study, none of them including P. cineraria is represented by substantial number of accessions collected from various locations to assess variability among them. On contrary, the present study involves 30 accessions from as many locations that spread over five districts of Rajasthan in India where this tree grows extensively in wild habitat. Therefore, this study is expected to offer a cumulative data set using three different single primer-based molecular markers viz RAPD, ISSR, and DAMD with their efficacy to authentically reveal intra-specific variation.
In the present investigation, the three SPAR methods, when tested individually to analyze the natural variation have revealed 83.73, 92.30 and 100% polymorphism in banding pattern, respectively. However, the genetic distance range among 30 accessions calculated through Jaccard coefficient resolved only lower values (Table 3). Therefore, it was decided to pool up the data generated through all the three SPAR methods to calculate a more authentic and relative genetic distance for the 30 accessions presently investigated. The pooled up distance matrixes were used to compute a single neighbour-joining (NJ) tree after allowing a 1000 replicate bootstrap test using the FreeTree program (ver. 0.9.1.5; Pavlicek et al. 1999), which shows clear genetic distance among the accessions of P. cineraria. NJ tree evidently showed that cluster A comprises of 11 accessions and are obtained from all collection-sites except Bikaner. Sub-cluster A1 comprises accessions JNVU/RI/2005/5 and JNVU/RI/2005/14 collected from Barmer district and JNVU/RI/2005/12 collected from Jaisalmer. This is due to possible heterozygosity of genome reflected at DNA level as detected by all SPAR methods. Of the three accessions under this cluster, two are from Barmer region and one from Jaisalmer region. In an earlier effort, attempts have been made to use morphological, biochemical and cytogenetical parameters to characterize them (Rawat et al. 2007; Naranjo et al. 1984). However, both the accessions (JNVU/RI/2005/5, JNVU/RI/2005/14) have shown normal meiosis with no indication of possible genetic variation (Rawat et al. 2007). On the other hand, the third accession (JNVU/RI/2005/12) of this group collected from Jaisalmer showed the presence of occasional quadrivalants in some of the PMCs analyzed. Subsequently, the distribution of the chromosomes of Anaphase I and Anaphase II was also not on the expected lines resulting in abnormal distribution of bivalents/univalents in some PMCs (Rawat et al. 2007). An important point to be noted here is that regardless of the normal/abnormal cytogenetical behavior, the three accessions of sub-cluster A1 have shown the presence of innate variation at DNA level as revealed by SPAR analysis. One of the reasons which can be attributed for the presence of such discrete variations at DNA level is the fact that the regions where these accessions were collected, i.e. Barmer and Jaisalmer, respectively, are the hottest and acute dry regions of Indian Thar desert and also experience harsh and inhospitable climatic conditions (Pant and Hingane 2006). The annual rainfall does not exceed 200 mm per year, while the temperature range from sub-zero to 50°C during summers (Mertia et al. 2006). The presence of discrete variations in certain random sequences of DNA may be considered as an effort by these genomes to adapt to the extreme environmental conditions in which they grow and propagate (Wood et al. 2008; Trivedi 2003; Hickey and Singer 2004). All the remaining accessions have followed more or less the expected pattern of similarity index, and the dendrogram does reflect that all the districts from which samples were collected lies in same geographical distribution area. Our observations are in line with those Strelchenko et al. (1999) who have demonstrated in barley germplasm that genetic diversity can be correlated to geographical distribution and traced through RAPD analysis. However, this is a first attempt to combine and correlate cytogenetical data and SPAR profiles with geographical distribution pattern.
Perceptibly, the accession JNVU/RI/2005/17 which has been resolved as an out-group in the NJ tree may be ascertained from the fact that the trees with this accession number inhabitant a moderate temperature and soil conditions and not subjected to harsh environment/climatic factors. On the other hand, the trees bearing accession numbers JNVU/RI/2005/28/29/30 falling into cluster D distinctly, grow in an extreme environment/climate in terms of temperature, drought, and soil conditions, showing high divergence with other accessions studied presently and thereby giving a proposal to smudge as an out-group by forming a different cluster.
An interesting and significant aspect of the present investigation has been that RAPDs, ISSR, and DAMD, which are all single primer-based amplification strategies, were analyzed and compared not only individually but also pair-wise with each other. It has been clearly observed from our studies that the r value, which reveals the efficacy of these method (Bhattacharya and Ranade 2001; Ranade and Farooqui 2002; Ranade et al. 2009) has been the highest for ISSR and DAMD pair when compared to RAPD either individually or in combination with either ISSR or DAMD. Such observations are also reported earlier in case of other trees, for e.g. pomegranate (Ranade et al. 2009), mango (Srivastava et al. 2005, 2007), and papaya (Saxena et al. 2005) to describe variation. Therefore, it can be authentically concluded that in order to reveal the intrinsic intra-specific variation, SPAR approach reveals more authentic genetic variation in tropical tree species like P. cineraria.
References
Arya S, Bisht RP, Tomar R, Toky OP, Harris PJC (1995) Genetic variation in minerals, crude protein and structural carbohydrates of foliage in provenances of young plants of Prosopis cineraria (L.) Druce in India. Agrofor Syst 29:1–7
Bhandari MM (1978) Flora of the Indian desert. Scientific Publishers, Jodhpur
Bhattacharya E, Dandin SB, Ranade SA (2005) Single primer amplification methods reveal exotic and indigenous mulberry varieties are similarly diverse. J Biosci 30:669–677
Bhattacharya E, Ranade SA (2001) RAPD and DAMD profile differences amongst mulberry varieties. BMC Plant Biol 1:3 (http://www.biomedcentral.com/content/pdf/1471-2229-1-3.pdf)
Burdak LR (1982) Recent advances in desert afforestation. Dissertation submitted to Shri R.N. Kaul, Director, Forestry Research, F.R.I., Dehra Dun
Burkart A (1976) A monograph of the genus Prosopis (Leguminosae, Subfamily Mimosoideae). J Arnold Arbor 57:219–249
Ferreyra LI, Bessega C, Vilardi JC, Saidman BO (2004) First report on RAPDs patterns able to differentiate some Argentinean species of section Algarobia (Prosopis, Leguminosae). Genetica 121:33–42
Ferreyra LI, Bessega C, Vilardi JC, Saidman BO (2007) Consistency of population genetics parameters estimated from isozyme and RAPDs dataset in species of genus Prosopis (Leguminosae, Mimosoideae). Genetica 131:217–230
Gates PJ, Brown K (1988) Acacia tortilis and Prosopis cineraria: leguminous trees for and areas. Outlook Agric 17:61–64
Goswami M, Ranade SA (1999) Analysis of variations in RAPD profiles among accessions of Prosopis. J Genet 78:141–147
Gupta M, Chyi YS, Romero-Severson J, Owen JL (1994) Amplification of DNA markers from evolutionarily diverse genomes using single primers of SSRs. Theor Appl Genet 89:998–1006
Heath DD, Iwana GK, Delvin RH (1993) PCR primed with VNTR core sequences yield species specific patterns and hypervariable probes. Nucleic Acids Res 21:5782–5785
Hickey A, Singer GAC (2004) Genomic and proteomic adaptations to growth at high temperature. Genome Biol 5:117
Hunziker JH, Saidman BO, Naranjo CA, Palacois RA, Poggio L, Burghardt AD (1986) Hybridization and genetic variation of Argentine species of Prosopis. For Ecol Manage 16:301–315
Jaccard P (1901) Etude comparative de la distribution orale dans une portion des Alpes et des Jura. Bull Soc Vaudoise Sci Nat 37:547–579
Kaul RN (1967) Trees or grass lands in the Rajasthan—old problems and new approaches. Indian For 93:434–435
Lamarque AL, Maestri DM, Grosso NR, Zygadlo JA, Guzman CA (1994) Proximate composition and seed lipid components of some Prosopis (Leguminosae) from Argentina. J Sci Food Agric 66:323–326
Leakey RB, Last FT (1980) Biology and potential of Prosopis species in arid environment with particular reference to P. cineraria. J Arid Environ 3:9–24
Mahoney D (1990) Trees of Somalia—a field guide for development workers. Oxfam/HDRA, Oxford, pp 133–136
Mann HS, Shankarnarayan KA (1980) The role of Prosopis cineraria in an agropastoral system in Western Rajasthan. In: LeHouerou HN (ed) Browse in Africa. International Livestock Centre for Africa, Addis Ababa, pp 437–442
Mertia RS, Prasad R, Kandpal BK, Narain P (2006) Regeneration of Lasiurus sindicus in relation to grazing pressure Rajasthan (India). Trop Grassl 40:40–44
Murray HG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Naranjo CA, Poggio L, Zeiger SE (1984) Phenol chromatography, morphology and cytogenetics in three species and natural hybrids of Prosopis (Leguminosae, Mimisoideae). Plant Syst Evol 144:257–276
Pant GB, Hingane LS (2006) Climatic changes in and around the Rajasthan desert during the 20th century. Int J Climatol 4:391–401
Pavlicek A, Hrda S, Flegr J (1999) FreeTree—freeware program for construction of phylogenetic trees on the basis of distance data and bootstrapping/jackknife analysis of the tree robustness. Application in the RAPD analysis of the genus Frenkelia. Folia Biol (Praha.) 4:597–599
Ranade SA, Farooqui N (2002) Assessment of profile variations amongst provenances of neem using single-primer amplification reaction (SPAR) methods. Mol Biol Today 3:1–10
Ranade SA, Rana TS, Narzary D (2009) SPAR profile and genetic diversity amongst pomegranate (Punica granatum L.) genotypes. Physiol Mol Biol Plants 15:61–70
Rawat D, Kumar A, Rao SR (2007) Studies on cytogenetical variation in Prosopis cineraria (Linn.) Druce—a key stone tree species of Indian desert. Silvae Genet 56:184–189
Sahni KC (1998) The book of Indian trees. Bombay natural history society, Mumbai
Saidman BO, Vilardi JC (1987) Analysis of genetic similarities among seven species of Prosopis Leguminosae: Mimosoideae. Theor Appl Genet 75:109–116
Saxena S, Chandra R, Srivastava AP, Mishra M, Pathak RK, Ranade SA (2005) Analysis of genetic diversity among papaya cultivars using Single Primer Amplification Reaction (SPAR) methods. J Hortic Sci Biotechnol 80:291–296
Sinha BC (2001) Book of state animal, bird, tree and flower of India. Wildlife Institute of India, Dehra Dun, p 24
Srivastava AP, Chandra R, Ranade SA (2005) Applicability of PCR based molecular markers for parentage analysis of three commercial mango hybrids. Indian J Plant Breed Genet 64:275–280
Srivastava AP, Chandra R, Saxena S, Rajan S, Ranade SA, Prasad V (2007) A PCR-based assessment of genetic diversity, and parentage analysis among commercial mango cultivars and hybrids. J Hortic Sci Biotech 82:951–959
Strelchenko P, Kovalyova O, Okuno K (1999) Genetic differentiation and geographical distribution of Barley Germplasm based on RAPD markers. Genet Resour Crop Evol 46:193–205
Trivedi S (2003) Do microsatellites have biased associations. Nucleus 46:61–76
Welsh J, McClelland M (1990) Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res 18:7213–7218
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucl Acids Res 18:6531–6535
Wood HM, Grahame JW, Humphray S, Rogers J, Butlin RK (2008) Sequence differentiation in regions identified by a genome scan for local adaptation. Mol Ecol 17:3123–3135
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by F. Canovas.
Rights and permissions
About this article
Cite this article
Sharma, S.K., Rawat, D., Kumar, S. et al. Single primer amplification reaction (SPAR) reveals intra-specific natural variation in Prosopis cineraria (L.) Druce. Trees 24, 855–864 (2010). https://doi.org/10.1007/s00468-010-0455-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00468-010-0455-4