Abstract
Recombinant protein synthesis in Pichia pastoris is generally controlled by the strong methanol inducible AOX1 promoter which is repressed by glucose and glycerol. In shake flasks, commonly one or two methanol pulses are added per day for induction. Such pulse feeding procedure leads to carbon starvation phases, which may enhance proteolytic activities and, therefore, cause product losses. Starvation between the methanol pulses could be avoided with a continuous enzymatic feed of glucose from a glucose-based polymer. The amount of glucose was low enough to prevent AOX1 repression by glucose. Energy and carbon were continuously supplied for cell maintenance resulting in significantly increased cell densities and product activities, as shown here at the example of a fungal lipase expressed in P. pastoris. A threefold improvement in measured product activity was obtained by applying enzymatic glucose feed and a further improvement was achieved by applying a defined mixture of ammonium compounds. The strategy described here simplifies the general procedure in shaken cultures by allowing the direct continuation of the cultivation from glucose to the methanol-based production phase without a medium change. It is easily applicable to multiwell plates and thus beneficial for high throughput applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Methanol inducible alcohol oxidase 1 (AOX1) promoter is one of the most frequently used promoters for expression of heterologous proteins in Pichia pastoris. This strongly inducible promoter can drive the production of AOX1 enzyme to high levels in relation to the total soluble protein in cells (up to 30 %, [1]) as well as for recombinant proteins. Since P. pastoris has a low maintenance demand on methanol, high cell densities, over 130 g l−1 dry cell weight, can be achieved in methanol fed-batch bioreactor cultivations [2]. The combination of high cell densities and strong transcription may lead to high product concentrations, especially with secreted proteins (For a review see [3]). The AOX1 promoter is repressed by glucose and glycerol. The repression is relieved under glucose/glycerol limitation or starvation, which allows recombinant protein expression to low levels. For the full induction, methanol addition is required [4]. Due to the repression effect, growth medium is usually changed to a glucose/glycerol free medium before induction in shaken cultivations (e.g., [5, 6]).
The use of methanol as a sole carbon source is not straightforward. In shaken cultivations, methanol is usually added as pulses in 12 or 24 h intervals, which however, results in long starvation phases between the pulses [7]. P. pastoris is reported to tolerate as high methanol concentrations as 3 % [8], but the higher concentrations can be growth inhibiting [8, 9]. Katakura et al. [8] concluded that high concentrations affected rather on DNA replication and membrane synthesis than on cell metabolism and protein synthesis. Zhang et al. [9] observed significant reduction in growth when higher than 0.365 % methanol concentrations were applied. Curvers et al. [10] showed that the productivity of P. pastoris cultures collapsed immediately if the cells were even temporarily exposed to toxic levels of methanol. They also observed that the cells reverted to the wild-type growth characteristics. Growth on methanol may also induce cell lysis and reduced product yields due to increased proteolytic activity [11]. Zhu et al. [12] concluded that protein overexpression interferes with carbon utilization when methanol is the sole carbon source. Consequently, improved cell and product yields may not be achieved by simply increasing the methanol concentration.
Increased yields can be achieved by adding other carbon source to the growth medium in a controlled manner to increase the carbon concentrations available for biomass. Mixed feeding strategies utilizing carbon sources like sorbitol, alanine, mannitol, trehalose or yeast extract have been implemented with success [13–19]. Despite the repression, cultivations with mixed feeding strategies where methanol and glucose have been fed together during induction have been also successfully conducted [5, 20–22]. This indicates that the derepression does not necessarily require complete glucose starvation, and low amounts of glucose can be present.
The abovementioned mixed feeding strategies have not been applied so far for well plate formats. Well plates have increased their importance in the last decade, while the interest towards high throughput process development in biotechnology has been increasing. High throughput methods include rapid process development and use of robotics where miniaturization, automation and parallelisation are required (for review see [23]). One possibility to support carbon to the growth medium in small scale can be the recently described slow glucose feeding [24, 25]. The described EnBase® method is especially suitable for small-scale cultivations since external feeding devices are not needed: glucose is enzymatically released to growth medium in a way which mimics the fed-batch process scheme. Fed-batch is the technique used to control the growth rate of the cell in bioreactor cultivations by controlling the feeding of the growth limiting substrate, e.g., glucose or glycerol. In small scale, however, the use of external feeding devices is more complicated due to small feeding amounts required. Pulse feeding techniques could easily lead to a repression of the AOX promoter. The enzymatic glucose release, however, allows a real continuous controlled feeding of glucose in a growth limiting manner, i.e., concentrations in the order of magnitude of the K S value without any specialized instruments. Different glucose feeding rates can be achieved by varying the amount of the glucose-releasing enzyme, and thus the system can be easily optimized.
The enzymatic glucose feeding is an interesting choice also for methylotrophic P. pastoris with AOX1 promoter. Here, we show that the glucose feeding method can be used in well plate cultivations of the yeast and demonstrate a successful recombinant protein production. Increased cell densities and 3–6 times higher recombinant protein activities were obtained in comparison to conventional pulse feeding of methanol in a buffered minimal medium.
Materials and methods
Pichia pastoris strains
The strain X33 pPICZαA-ROL (single copy) [26, 27], kindly provided by Prof. Pau Ferrer from Universitat Autònoma de Barcelona, was used for expression of Rhizopus oryzae lipase (ROL) under the methanol inducible alcohol oxidase 1 (AOX1) promoter. As a negative control for recombinant protein expression, the wild-type X33 strain (methanol utilization plus, Mut+, Invitrogen) was used.
Cultivation media and growth conditions
For culture maintenance, the strains were grown on YPD plates (1 % yeast extract, 2 % peptone, 2 % glucose, 2 % agar and 0.1 mg ml−1 zeocin with the recombinant strain). For long-term storage, glycerol stocks were prepared according to Invitrogen’s instructions [6] and stored at −70 °C.
For the cultivation experiments, precultures were prepared by cultivating the Pichia strains in shake flasks at 30 °C in buffered minimal glycerol medium (BMG) prepared according to Invitrogen’s guidelines [6] (100 mM potassium phosphate, pH 6.0, 1.34 % yeast nitrogen base (Invitrogen), 4 × 10−5 % biotin, 1 % glycerol, 0.1 mg ml−1 zeocin) for 20 h. Preculture was centrifuged with 1,677g, 24 °C for 5 min (Eppendorf centrifuge 5804R), and suspended to the BSEB medium without soluble substrate (described below) to reach an appropriate cell concentration and to avoid traces of BMG to be transferred to other cultivation media.
Cultivation experiments were performed in three different media: in control medium, and in two media which applied the enzymatic glucose release technology. As control cultivations, Invitrogen’s BMG which was changed to buffered minimal methanol medium (BMM: 100 mM potassium phosphate, pH 6.0, 1.34 % yeast nitrogen base, 4 × 10−5 % biotin, 0.1 mg ml−1 zeocin, 0.5 % methanol) for induction, was used. At the start of induction of ROL expression in control cultivations, the medium change was carried out by centrifugation (1,677g, 24 °C for 5 min, Eppendorf centrifuge 5804R). To apply the enzymatic glucose release system, two different media were used. One, which is here called BMEB (Buffered Minimal EnBase), was prepared by substituting glycerol from BMG by 60 g l−1 a starch-based glucose polymer (EnBase Flo®, BioSilta Oy) [25]. BMEB and BMG/BMM had thus the same medium composition except for the carbon source. The other EnBase medium, named BSEB (BioSilta EnBase) was supplied by BioSilta. According to the manufacturer, this prototype medium version contains in addition a balanced mixture of defined organic and inorganic nitrogen sources. The medium contains 30 g l−1 EnBase Flo substrate as carbon source and 0.1 mg ml−1 zeocin. BSEB and EnBase Flo substrate are proprietary products of BioSilta.
pH and glucose were measured from off-line samples at the time of induction and every 24 h. Glucose concentrations were analyzed with YSI 2700 Select Biochemical Analyzer (YSI Inc., Yellow Springs, USA).
Cultivation experiments
Cultivations were performed on 24-deep well plates (Mediscan GmbH & Co. riplate 10 ml, 43001-1066 with Axygen sealing film BF-400-S) in 3 ml culture volume at +30 °C with 200 rpm shaking (Infors HT Ecotron). A total of 14 different cultivation conditions with 36 cultivations were analyzed. Three replicates were done from most of the cultivations, but from BMEB and BSEB cultivations two replicates were done without induction. The EnBase media contained glucose-releasing enzyme in various concentrations (0.5–3 U l−1) to achieve optimal glucose release from the glucose polymer. Each culture was inoculated with the preculture to the starting OD600 of 0.1. The ROL expression was induced by adding methanol to final concentration of 0.5 % (v/v), according to Invitrogen’s instructions [6]. Cultures were induced after 17 h of the start of the cultivation when OD600 was about 3 [26]. To keep the protein expression ongoing, methanol was added to the same end concentration every 24 h. No methanol was added to the cultures where ROL expression due to derepression of AOX1 promoter in glucose-limited conditions was studied. In total, the cultivations were continued for 97 h. Optical density, ROL activity, glucose concentration and pH were monitored daily during the whole experiment.
ROL analysis
The enzymatic activity of ROL as units per ml (U = μmol resorufin methyl ether hydrolyzed per minute) was measured spectrophotometrically at 560 nm using the Lipase colorimetric assay kit from Roche and the method adapted from Resina et al. [28]. In this method, 60 μl of substrate [1,2-O-dilauryl-rac-glycero-3-glutaric acid-(6-methylresorufin) ester] was mixed with 190 μl of 400 mM Tris–HCl, pH 7.25 buffer and 10 μl ROL sample in the total reaction volume of 260 μl. ROL was analyzed from culture media. The enzymatic reactions were set up on microwell plates (Perkin Elmer Spectra Plate™—TC 96, Waltham, USA) and the activity was monitored at 560 nm for 5 min. The volumetric activities were calculated according to Beer–Lambert law. The extinction coefficient ε 405.6 M−1 cm−1 was determined by measuring the absorbance of resorufin methyl ether (SIGMA) for various concentrations at 560 nm and ε was calculated based on the slope achieved according to Beer–Lambert law.
The difference of the amount of expressed ROL in different cultivation conditions was further analyzed by Western blotting using anti-ROL antibodies raised in rabbits (a kind gift from Prof. Pau Ferrer, Universitat Autònoma de Barcelona). Identical volumes of samples were taken for the analysis from negative control, BMM, BMEB and BSEB cultivations at 48 h after start of the induction. The proteins were separated by SDS-PAGE and transferred onto Immobilon P membrane (Millipore) using standard methods. The membrane was blocked with 1.5 % skimmed milk in TBST buffer (10 mM Tris–HCl, pH 8.0, 150 mM NaCl, 0.05 % Tween 20) for overnight at +4 °C followed by incubation in the primary antibody for 2 h. The anti-ROL antibody was recognized by horseradish peroxidase (HRP) conjugated Goat anti-Rabbit IgG (Millipore) followed by detection with HRP color development reagent (Bio-Rad). For a more sensitive detection utilizing enhanced chemiluminescence, ECL™ Advance Western Blotting Detection Kit (GE Healthcare) was used. Same sample volumes were used in both Western blot analyses.
Analysis of organic acids
Succinate, lactate, formate and acetate were analyzed by high-performance liquid chromatography (HPLC). Medium samples were centrifuged with 1,677g, 4 °C for 5 min (Eppendorf centrifuge 5804R) and stored at −20 °C until HPLC analysis. Before analysis, samples were thawed and filtered with 0.2 μm cellulose filters. The HPLC device (Agilent Technologies, 1200 series) was equipped with Transgenomic ICSep ICE-COREGEL 87H3 column. H2SO4 (0.005 M) was used as eluent. Detection of organic acids was performed with a refractive index detector and diode array detector.
Results
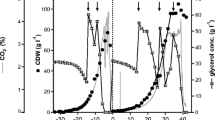
The cultivations in minimal medium with slow enzymatic glucose feed (BMEB) provided 2–3 times higher cell densities in comparison to cultivations in conventional minimal medium (BMM). Optical densities of 19–29 in BMEB, corresponding to cell dry weight of 4.75–7.25 g l−1, and OD600 of 10 in BMM corresponding to CDW of 2.5 g l−1 were obtained (Fig. 1). The final cell densities in BMEB cultivations were higher with methanol induction than without induction (Figs. 1b, 2b), indicating that also the methanol feed increases the biomass.
Product activities and optical densities (a1–c1), and pH and glucose levels (a2–c2) of deep well plate cultivations of P. pastoris X33 pPICZαA-ROL for the production of recombinant fungal lipase. Induction to final concentration of 0.5 % methanol was done after 17 h of cultivation. Methanol addition was repeated in every 24 h of cultivation to keep protein expression ongoing. a Reference cultivation with buffered minimal methanol medium (BMM). b Cultivation with buffered minimal medium including the glucose-releasing polymer (BMEB) with different glucose-releasing enzyme concentrations. c Cultivation with BioSilta’s prototype EnBase medium version (BSEB) with different glucose-releasing enzyme concentrations. pH and glucose were measured from off-line samples once a day. The plots in figures b1 and c1 present biomass densities (OD600), and the bars present the product (ROL) activities, with different glucose-releasing enzyme concentrations. The plots in figure b2 and c2 present pH, and the bars present the glucose concentrations in the medium. The error bars were calculated from three parallel cultivations
Control cultivations of P. pastoris X33 WT (a) and non-induced P. pastoris X33 pPICZαA-ROL (b, c) on deep well plate. The presence of the possible endogenous lipase (a) or recombinant ROL (b, c) was studied. Biomass densities (OD600), pH and glucose levels (g l−1) were followed. BMEB buffered minimal medium including the glucose polymer instead of glycerol. BSEB BioSilta’s prototype EnBase medium version. Methanol was added to the negative control (wild-type strain) (a) after 41 and 64 h of cultivation. The plots in figure a1–c1 present biomass densities and the bars present the measured ROL activities, with different glucose-releasing enzyme concentrations. The plots in figure a2–c2 present pH and the bars present the measured glucose concentrations, with different glucose-releasing enzyme concentrations. The error bars were calculated from two parallel cultivations except the negative control (a) where calculations were done from three parallels
The strict dependence between varying glucose feed and growth rate was observed in non-induced BMEB cultivations (Fig. 2b1). In induced cultivations, this relation was not as clear (Fig. 1b, c) since methanol is also utilized as a carbon source. In BioSilta EnBase medium (BSEB) additionally the nitrogen containing organic carbon sources (see “Materials and methods”) support growth.
In BSEB cultivations, the growth rate did not significantly decrease due to protein expression. In BSEB with methanol feeding, the growth rate of the non-expressing control strain was approximately the same as with ROL-expressing strain (0.224 and 0.218 OD U h−1, R 2 = 0.958 and 0.953, respectively) when 1 U l−1 of glucose-releasing enzyme was used.
The ROL activity after 64 h of cultivation was about twofold, and after 88 h threefold higher with enzymatic glucose feed (BMEB) compared to the conventional method (Fig. 1a, b, respectively). By providing a balanced mixture of organic and inorganic defined nitrogen compounds, BSEB medium provided even higher ROL activities (3- to 6-fold higher than in BMM medium, Fig. 1a, c, respectively). The higher protein amounts were also clearly seen in Western blot analysis (Supplementary Fig. 1a, b) where the strongest bands were seen in BSEB cultivations with 2 U l−1 of glucose-releasing enzyme. The strongest ROL band in BMEB cultivations, detected with the HRP color reagent, was with 0.5 U l−1 of glucose-releasing enzyme concentrations while the bands with other concentrations and in BMM were below the detection limit (Supplementary Fig. 1a). The more sensitive detection with enhanced chemiluminescence method revealed ROL also in these conditions (Supplementary Fig. 1b). A thinner band seen above the strong ROL bands indicates that the Anti-ROL antibody recognizes two variants which have slightly different sizes. Guillen et al. [29] observed similar thinner band over strong ROL band from an extract of R. oryzae lipase overexpressed in P. pastoris. N-terminal sequencing revealed that the two forms had similar N-terminal sequence and, therefore, the differences in molecular sizes might be caused by post translational modifications [29].
The 88 h sample from BSEB with 2 U l−1 of glucose-releasing enzyme showed decreased activities compared to other enzyme concentrations (Fig. 1c1). Also a pH decrease to the level of 5 was observed (Fig. 1c2). The organic acid concentrations (succinate, lactate, formate, and acetate) remained under 0.4 g l−1, which is unlikely to cause pH drop from 7 to 5. Since a similar pH decrease was not seen in BSEB cultivations without methanol addition (Fig. 2c2) we deduce that the pH drop was related to protein expression together with higher cell densities. In other cultivations, pH levels were maintained between 5 and 7 (Figs. 1, 2). According to Minning et al. [26] the stability of ROL is in its maximum at pH 7 and at pH 5 the loss in stability caused about 45 % loss in activity.
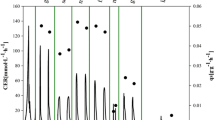
By applying the enzymatic glucose release, ROL activities per OD600 unit could be elevated (Fig. 3). The volumetric product activity was higher even when the cell densities were about the same level after 64 h of cultivation (e.g., BMM OD600 ~17, BMEB 1 U l−1 OD600 ~15). Highest product activities per OD600 unit were achieved in BSEB with 1 U l−1 and BMEB with 0.5 U l−1 of glucose-releasing enzyme after 88 h of cultivation (Fig. 3).
ROL activities in relation to biomass. The activities of expressed ROL were compared to biomass (OD600) in induced BMM cultivation, and cultivations in BMEB or BSEB with different glucose-releasing enzyme concentrations (0.5, 1 or 2 U l−1). The error bars were calculated from three parallel cultivations
With over-optimal glucose-releasing enzyme concentration (5 U l−1) in BMEB, the glucose concentrations were 3.8 g l−1 after 24 h, and 1.4 g l−1 after 48 h (data not shown). In these experiments, methanol was also added every 24 h to a final concentration of 0.5 % to induce ROL expression, but no ROL expression was detected. The presented results suggest that glucose must be maintained in the range of the K S value which is in the order of approx. 100 mg l−1 [2], i.e., in a limiting concentration. This fits well with our data, which show that the repressing glucose concentration is somewhere between 0.2 g l−1 (Fig. 1a2, b2, c2) and 1.4 g l−1.
Discussion
Glucose feed enhances cell densities and product activities
Both the total product activity and the biomass-specific product activity were increased by applying a continuous enzymatic glucose feed. We conclude that the additional carbon has decreased the methanol-caused metabolic burden and relieved starvation phases between the methanol pulses.
Cell yields achieved in P. pastoris cultivations often decrease during induction since nutrient flow is largely allocated to recombinant protein production. The production of the heterologous R. oryzae lipase (ROL) has previously shown negative effect on P. pastoris growth [27]. This was not the case with the slow glucose feed cultivations; the result was rather the opposite. This indicates that methanol addition together with glucose feed had promoted the cell growth by providing an extra carbon source.
Simultaneous feed of an alternative carbon source together with methanol has been shown to improve the final cell densities and/or protein yields. Abad et al. [20] cultivated P. pastoris Tv1_mc in a bioreactor with mixed feeding strategies (glucose/methanol or glycerol/methanol feed compared to pure methanol feed). They obtained about 40 % higher biomass yield with both mixed feeding strategies, but observed that the volumetric yield of the studied recombinant protein was higher with pure methanol feed. Several studies with glycerol/methanol feeding have, however, resulted in both higher cell densities and protein yields for Muts and Mut+ strains (for a review see [30]). Katakura et al. [8] used glycerol/methanol mixed feeding strategy in the production phase of human β2-glycoprotein I domain V and obtained a higher specific production with a higher specific growth rate. Similar results were obtained by Mcgrew et al. [31] with trimeric CD40 ligand produced in P. pastoris GS115. In our knowledge, our study is the first case where the mixed feeding strategy has been applied with pulse feeding of methanol in a well plate format. Our results point out that higher cell density together with higher recombinant protein production can be achieved also in well plates using mixed feeding of glucose and pulses of methanol instead of pure methanol feed.
We conclude that the slow glucose feed working on the background in BMEB seems to provide enhanced recombinant protein production due to several reasons. The utilization of glucose provides lower metabolic burden in comparison to pure methanol. Also, the additional glucose provides higher and continuous supply of carbon and eliminates the starvation risks between methanol feeding pulses. The methanol utilization pathway is known to include the formation of several toxic components (formaldehyde, hydrogen peroxide) which are further metabolized inside peroxisomes (for a review see e.g., [3, 32]), and consequently, the amount of peroxisomes increases in the cells (for a review see e.g., [33]). Zhu et al. [12] studied physiology of P. pastoris A3 (multicopy strain) during expression of porcine insulin precursor (PIP) and concluded that pure methanol feeding weakened carbon assimilation during induction. Later, they concluded that this weakened carbon assimilation was the main metabolic burden for the cells during expression of PIP [19]. They also suggested that co-feeding of sorbitol repressed methanol metabolism and, therefore, reduced the stress caused by dissimilation of formaldehyde. Even though they obtained lower PIP mRNA levels rates, the total yield of protein was increased due to increased availability of precursors or energy when compared to pure methanol feed. Although not proven in our research, we assume that the presence of a low amount of glucose may down-regulate the methanol metabolism and thereby substantially decrease the metabolic burden caused by the induction. The amount of the carbon substrate was, however, sufficient to provide higher cell mass and higher volumetric activities of ROL compared to the standard method.
Previously, we had implemented a quasi-continuous feeding of methanol in shake flasks cultures of P. pastoris [7]. On the basis of the dissolved oxygen level measurements, clear starvation phases were observed between the 12 h feed pulses in conventional cultivations. With this quasi-continuous feeding system the production of human collagen II was significantly improved. The big advantage of the enzymatic glucose feeding compared to the earlier feeding system is its simplicity—no extra pumps are needed and the system is applicable to all shaken cultures.
pH levels of cultivations
pH variations during cultivation may impair growth rates and protein yields by preventing the growth, causing extra stress and metabolic burden and by decreasing the stability of the produced protein [25]. Our results point the importance to optimize the cultivation system in relation to glucose feed and pH. The pH of the cultivation decreases when glucose is used as a carbon source and ammonium is consumed from the cultivation medium. In contrast, pH increases when ammonium is released to the medium due to the utilization of organic ammonium sources like amino acids, peptones, or urea as a carbon source [25, 34–36]. The pH of the cultivation is stable when cell mass and product formation, and deamination reactions are in balance. Since the level of deamination increases with increasing cell number, the balance can be achieved by dosing the glucose-releasing enzyme roughly according to biomass. If overproduction of a protein is going on at the same time, it will increase the nitrogen demand. If the product is sensitive to pH, the pH drop may eventually completely inactivate the product. The observed decrease of pH and ROL activity levels in the conditions where the highest protein expression levels were previously measured indicates the complete depletion of organic nitrogen.
Secreted proteins like recombinant ROL are prone to proteolytic activities present in the medium or on the surface of the host organism. Consequently, the stability of the recombinant proteins may be affected by the way how the medium pH affects the activity of proteases [37]. As demonstrated by the current work, the ability of the cultivation system to maintain the pH level can be affected by the medium composition.
Slow enzymatic glucose release with suitable enzyme concentration does not prevent expression
Boettner et al. [5] have shown that simultaneous feed of glucose together with methanol in AOX1 induction is possible, if the glucose concentration is kept low enough. In their study, the Pichia cells were initially grown for 3 days in a medium having glucose as the sole carbon source at the beginning. The medium was then exchanged into a non-glucose containing medium and the feeding of methanol/glucose mixture was initiated to induce recombinant protein expression. Glucose concentration higher than 0.1 % (1 g l−1) resulted in repression of the AOX1 promoter. The non-repressing amount of glucose during methanol induction was in the same range as observed in our study.
Our study, like Boettner et al. [5], shows that the mixed glucose/methanol feed results in higher cell mass and product yield than cultivation in a medium with methanol as the sole carbon source. In our cultivation system, glucose was constantly enzymatically released to the medium from a soluble glucose polymer during the whole cultivation. Thus, there is no need for medium exchange before the start of the induction, which greatly simplifies the cultivation protocol.
Our results point out that the AOX1 promoter is not fully repressed by low glucose concentrations (~0.2 g l−1) and we believe this is related to glucose uptake and signal transduction system in AOX1 repression. Zhang et al. [38] studied the effect of hexose transporters PpHXT1 and PpHXT2 on catabolite repression of AOX. They proposed that hexose transporter PpHXT1 is expressed on higher glucose concentrations and PpHXT2 on lower glucose concentrations. In their study, the levels of PpHXT2 mRNA were fully induced in cells with less than 1 g l−1 of glucose in growth medium, but PpHXT1 mRNA was about one-third of the levels measured with glucose concentration of 5 g l−1. They suggested that PpHXT1 is directly involved in AOX1 repression. We believe that the low glucose concentration obtained in our experiments does not induce PpHXT1 expression and, therefore, one factor in signal transduction system of AOX1 repression is missing. This also could explain why measurable ROL activities were detected even in control cultivations without methanol induction: PpHXT1 is not expressed due to low glucose levels and, therefore, AOX1 is not fully repressed. Consequently, a small amount of recombinant product is observed even without methanol induction.
Based on these findings, we conclude that with adequately adjusted slow enzymatic glucose release the methanol-induction repressing effect of glucose can be partly or fully avoided. In addition, simultaneous glucose feeding provides extra carbon source and energy for both, growth and recombinant protein production.
Application of other promoter systems
The presented cultivation method is clearly applicable for AOX1 promoter strains. Another commonly used promoter derives from the glyceraldehydes-3-phosphate dehydrogenase (GAP). This promoter is constitutively expressed in a glucose containing medium [39] and the presented cultivation system could be well suitable for this promoter. The fed-batch cultivation mode is one of the methods used in larger scales to control growth and recombinant protein expression of the GAP promoter strains (e.g., [40, 41]). To our opinion, it is reasonable to expect positive impacts to growth and protein activities with controlled and continuous glucose feed also in small scale. However, the use of GAP promoter in the expression of toxic proteins is restricted due to the constitutive expression (for a review see [42]).
Conclusions
The presented results show that enzymatic glucose release system is applicable with AOX1 promoter and can be used in simple shaken cultivations. Since the small amount of glucose fed into the cultivations does not repress the AOX1 promoter, the medium change to glucose/glycerol free medium is not required before induction. Therefore, our method simplifies the high throughput cultivations and allows even higher automation level during the whole P. pastoris cultivation. The continuous enzymatic glucose feed also prevents the long starvation phases between methanol pulses during induction and, therefore, improves the cultivation conditions for recombinant protein expression.
The presented method enables increased volumetric protein expression levels compared to the conventional method. Using the slow glucose feed system, a threefold increase in the measured recombinant enzyme activity was observed. Furthermore, by further applying a medium composition containing a mixture of inorganic and organic ammonium compounds, up to sixfold improvement was achieved.
The expression of different proteins may require further optimization of the medium, enzyme dosing and methanol dosing for each application. In applications where a higher growth rate is preferred, a growth medium with additional inorganic and organic nitrogen sources could be advantageous. In contrast to mechanical glucose feeding, the enzymatic glucose feeding minimizes the risk of formation of glucose gradients and provides glucose for the cells all the time even during induction thereby relieving starvation after the cells have consumed the entire added methanol. We thus assume that slow glucose feeding can partly reduce the inhibitive effects of the methanol feeding. The presented method has great potential to speed up the screening of new recombinant proteins and other high throughput applications for P. pastoris, where simple cultivation protocols and high cell and product yields are required.
References
Couderc R, Baratti J (1980) Oxidation of methanol by the yeast Pichia pastoris. Purification and properties of alcohol oxidase. Agric Biol Chem 44:2279–2289
Jahic M et al (2002) Modeling of growth and energy metabolism of Pichia pastoris producing a fusion protein. Bioprocess Biosyst Eng 24:385–393
Cereghino JL, Cregg JM (2000) Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev 24:45–66
Tschopp JF et al (1987) Expression of the lacZ gene from two methanol-regulated promoters in Pichia pastoris. Nucleic Acids Res 15:3859–3876
Boettner M et al (2002) High-throughput screening for expression of heterologous proteins in the yeast Pichia pastoris. J Biotechnol 99:51–62
Invitrogen (2010) Pichia expression kit for expression of recombinant proteins in Pichia pastoris. K1710-01
Ruottinen M et al (2008) Improved production of human type II procollagen in the yeast Pichia pastoris in shake flasks by a wireless-controlled fed-batch system. BMC Biotechnol 8:33
Katakura Y et al (1998) Effect of methanol concentration on the production of human [beta]2-glycoprotein I domain V by a recombinant Pichia pastoris: a simple system for the control of methanol concentration using a semiconductor gas sensor. J Ferment Bioeng 86:482–487
Zhang W et al (2000) Modeling Pichia pastoris growth on methanol and optimizing the production of a recombinant protein, the heavy-chain fragment C of botulinum neurotoxin, serotype A. Biotechnol Bioeng 70:1–8
Curvers S et al (2001) Human chymotrypsinogen B production with Pichia pastoris by integrated development of fermentation and downstream processing. Part 1. Fermentation. Biotechnol Prog 17:495–502
Jahic M et al (2003) Temperature limited fed-batch technique for control of proteolysis in Pichia pastoris bioreactor cultures. Microb Cell Fact 2:6
Zhu TC et al (2011) Understanding the effect of foreign gene dosage on the physiology of Pichia pastoris by transcriptional analysis of key genes. Appl Microbiol Biotechnol 89:1127–1135
Celik E, Calik P, Oliver SG (2010) Metabolic flux analysis for recombinant protein production by Pichia pastoris using dual carbon sources: effects of methanol feeding rate. Biotechnol Bioeng 105:317–329
Gao MJ et al (2012) Methanol/sorbitol co-feeding induction enhanced porcine interferon-alpha production by Pichia pastoris associated with energy metabolism shift. Bioprocess Biosyst Eng 35:1125–1136
Guerrero-Olazaran M et al (2009) Recombinant shrimp (Litopenaeus vannamei) trypsinogen production in Pichia pastoris. Biotechnol Prog 25:1310–1316
Inan M, Meagher MM (2001) Non-repressing carbon sources for alcohol oxidase (AOX1) promoter of Pichia pastoris. J Biosci Bioeng 92:585–589
Niu H et al (2013) A quantitative study of methanol/sorbitol co-feeding process of a Pichia pastoris Mut +/pAOX1-lacZ strain. Microb Cell Fact 12:33
Thorpe ED, d’Anjou MC, Daugulis AJ (1999) Sorbitol as a non-repressing carbon source for fed-batch fermentation of recombinant Pichia pastoris. Biotechnol Lett 21:669–672
Zhu T et al (2013) Transcriptional investigation of the effect of mixed feeding to identify the main cellular stresses on recombinant Pichia pastoris. J Ind Microbiol Biotechnol 40:183–189
Abad S et al (2010) Stepwise engineering of a Pichia pastoris D-amino acid oxidase whole cell catalyst. Microb Cell Fact 9:24
Holmes WJ et al (2009) Developing a scalable model of recombinant protein yield from Pichia pastoris: the influence of culture conditions, biomass and induction regime. Microb Cell Fact 8:35
Jungo C, Marison I, von Stockar U (2007) Mixed feeds of glycerol and methanol can improve the performance of Pichia pastoris cultures: a quantitative study based on concentration gradients in transient continuous cultures. J Biotechnol 128:824–837
Bhambure R, Kumar K, Rathore AS (2011) High-throughput process development for biopharmaceutical drug substances. Trends Biotechnol 29:127–135
Panula-Perala J et al (2008) Enzyme controlled glucose auto-delivery for high cell density cultivations in microplates and shake flasks. Microb Cell Fact 7:31
Krause M et al (2010) A novel fed-batch based cultivation method provides high cell-density and improves yield of soluble recombinant proteins in shaken cultures. Microb Cell Fact 9:11
Minning S, Schmidt-Dannert C, Schmid RD (1998) Functional expression of Rhizopus oryzae lipase in Pichia pastoris: high-level production and some properties. J Biotechnol 66:147–156
Minning S et al (2001) Optimization of the high-level production of Rhizopus oryzae lipase in Pichia pastoris. J Biotechnol 86:59–70
Resina D, Serrano A, Valero F, Ferrer P (2004) Expression of a Rhizopus oryzae lipase in Pichia pastoris under control of the nitrogen source-regulated formaldehyde dehydrogenase promoter. J Biotechnol 109:103–113
Guillen M, Benaiges MD, Valero F (2011) Comparison of the biochemical properties of a recombinant lipase extract from Rhizopus oryzae expressed in Pichia pastoris with a native extract. Biochem Eng J 54:117–123
Cos O, Ramon R, Montesinos JL, Valero F (2006) Operational strategies, monitoring and control of heterologous protein production in the methylotrophic yeast Pichia pastoris under different promoters: a review. Microb Cell Fact 5:17
Mcgrew JT et al (1997) Expression of trimeric CD40 ligand in Pichia pastoris: use of a rapid method to detect high-level expressing transformants. Gene 187:193–200
Gellissen G (2000) Heterologous protein production in methylotrophic yeasts. Appl Microbiol Biotechnol 54:741–750
van der Klei IJ, Yurimoto H, Sakai Y, Veenhuis M (2006) The significance of peroxisomes in methanol metabolism in methylotrophic yeast. Biochim Biophys Acta-Mol Cell Res 1763:1453–1462
Loureiro-Dias MC (1988) Movements of protons coupled to glucose transport in yeasts. A comparative study among 248 yeast strains. Antonie Van Leeuwenhoek 54:331–343
Sibirny AA et al (1988) Genetic control of methanol utilization in yeasts. J Basic Microbiol 28:293–319
van Urk H, Postma E, Scheffers WA, van Dijken JP (1989) Glucose transport in crabtree-positive and crabtree-negative yeasts. J Gen Microbiol 135:2399–2406
Sinha J, Plantz BA, Inan M, Meagher MM (2005) Causes of proteolytic degradation of secreted recombinant proteins produced in methylotrophic yeast Pichia pastoris: case study with recombinant ovine interferon-tau. Biotechnol Bioeng 89:102–112
Zhang P et al (2010) Catabolite repression of Aox in Pichia pastoris is dependent on hexose transporter PpHxt1 and pexophagy. Appl Environ Microbiol 76:6108–6118
Waterham HR et al (1997) Isolation of the Pichia pastoris glyceraldehyde-3-phosphate dehydrogenase gene and regulation and use of its promoter. Gene 186:37–44
Garcia-Ortega X, Ferrer P, Montesinos JL, Valero F (2013) Fed-batch operational strategies for recombinant Fab production with Pichia pastoris using the constitutive GAP promoter. Biochem Eng J 79:172–181
Heyland J, Fu JA, Blank LM, Schmid A (2010) Quantitative physiology of Pichia pastoris during glucose-limited high-cell density fed-batch cultivation for recombinant protein production. Biotechnol Bioeng 107:357–368
Cregg JM, Cereghino JL, Shi J, Higgins DR (2000) Recombinant protein expression in Pichia pastoris. Mol Biotechnol 16:23–52
Acknowledgments
The authors wish to thank Mrs. Lilja Tuohimaa, Mrs. Tuula Karppinen, B.Sc Henna Karppinen, and M.Sc Ville-Hermanni Sotaniemi for their skillful technical assistance. Prof. Pau Ferrer is gratefully acknowledged for providing the Pichia pastoris strains and the anti-ROL antibody. Dr. David Resina, Dr. Monika Bollok and the other members of the ERA-IB EngBioCat consortium are thanked for many helpful discussions. This study was funded by the Finnish Funding Agency for Technology and Innovation (Tekes), decision number 40345. The project was part of ERA-IB-project implementing an enzyme engineering technology platform for the provision of tailor-made enzymes (EngBioCat). Also Finnish Foundation for Technology Promotion and Tauno Tönning Research Foundation are thanked for the financial support.
Conflict of interest
JPP, AV and PN are co-founders and minor shareholders of BioSilta Oy. AV is R&D director, and PN is a scientific advisor of the company. AM, HO and JK declare no competing interests.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
449_2013_1098_MOESM1_ESM.tif
Supplementary Fig. 1 Western blot analysis of expressed ROL after 64 h of cultivation. Samples were taken from induced cultivations (BMEB or BSEB) made with different concentrations of the glucose-releasing enzyme (0.5, 1 or 2 U l−1), from negative control cultivation (WT) and reference cultivation with BMM medium. Equal volumes of culture supernatant were loaded to the gel. BMEB buffered minimal medium including the glucose polymer but no glycerol. BSEB BioSilta’s prototype EnBase medium version. BMM buffered minimal methanol medium. ROL was detected by using suitable antibodies and a horseradish peroxidase-coupled color reagent (a) and by ECL (b). In b, the higher concentrations of ROL were detected as white bands because of over exposure of the detection film. (TIFF 117 kb)
Rights and permissions
About this article
Cite this article
Panula-Perälä, J., Vasala, A., Karhunen, J. et al. Small-scale slow glucose feed cultivation of Pichia pastoris without repression of AOX1 promoter: towards high throughput cultivations. Bioprocess Biosyst Eng 37, 1261–1269 (2014). https://doi.org/10.1007/s00449-013-1098-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-013-1098-9