Abstract
The realized host range of herbivores is expected to increase with herbivore population density. Theory also predicts that trait similarity and phylogenetic relatedness between native and exotic plants is expected to increase the susceptibility of introduced plants to feeding by native herbivores. Whether the ability of native herbivores to extend their host range to introduced species is density dependent is still unknown. We addressed this question by monitoring pine processionary moth (PPM, Thaumetopoea pityocampa) attacks during nine consecutive years on 41 pine species (8 native and 33 introduced) planted in an arboretum. The survey encompassed latent and outbreak periods. A total of 28 pine species were attacked by PPM. There was no difference in the probability of attack between native and introduced pine species. Host range increased and was more phylogenetically clustered during outbreak than latent periods. When population density increased, PPM expanded its diet breadth by attacking introduced pine species that were closely related to native hosts. This study demonstrates the density dependence of host range expansion in a common pine herbivore. Importantly, it supports the idea that the degree of phylogenetic proximity between host species can be a better predictor of attacks than the introduction status, which may help to predict the outcomes of new plant–herbivore interactions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Non-native plants are increasingly introduced outside of their natural range. Yet, their establishment may be hindered by damage from herbivores native to the area of introduction (Cappuccino and Carpenter 2005; Parker et al. 2012). For instance, a recent survey showed that 590 European insect herbivore species successfully colonised exotic tree species introduced in Europe (Branco et al. 2015). Predicting the outcomes of novel plant–herbivore interactions is, therefore, of great theoretical and applied interest (Pearse et al. 2013; Branco et al. 2014). However, exactly what determines the likelihood of herbivores to incorporate new hosts in their diet remains controversial (Harvey et al. 2010; Forister and Wilson 2013).
Theory predicts that host range may be density dependent, increasing with consumer abundance (Svanbäck and Bolnick 2007; Araújo et al. 2011; Carrasco et al. 2015). Herbivores may be more likely to exploit new host plants when population densities are high, as a result of a simple sampling effect: intraspecific variability in host use may increase with population density (Bolnick et al. 2011). Alternatively, increased population density may induce changes in the quality of host plants through induced resistance (Underwood 2010) making them less suitable to herbivores, or intra-specific competition may force herbivores to feed on less preferred or less suitable, but ‘competitor free’, hosts (White and Whitham 2000; Plath et al. 2011; Nakládal and Uhlíková 2015). Such density dependent effects on host use may have profound consequences on population dynamics of herbivores by alleviating intra-specific competition, but exposing them to new inter-specific competitors (reviewed in Bolnick et al. 2011).
Phylogenetic relationships among plants can also explain patterns of utilization by a given herbivore (Nakadai and Murakami 2015). Closely related plant species are more likely to share herbivores (Parker et al. 2012; Gilbert et al. 2015). Pearse and Hipp (2009) showed that insect damage on oaks introduced to the US decreased with their phylogenetic distance to native American oaks. This is consistent with phylogenetic niche conservatism for traits involved in plant–herbivore interactions (Srivastava et al. 2012; Nakadai et al. 2014). However, not all traits show a phylogenetic imprint (Whitfeld et al. 2012) and even closely related plant species may diverge in traits relevant to defence against herbivores (Desurmont et al. 2011). Although biologically meaningful and of practical interest, phylogenetic distance among host plants may not be the best predictor of novel plant–insect interactions (Bezemer et al. 2014).
For a new plant to be incorporated in the diet of a given insect, its traits (e.g. phenology, palatability, defenses) have to match those that are involved in the insect’s host plant colonisation and exploitation behavior, without the need for previous coevolutionary processes, that is ecological fitting (Agosta 2006). Only those exotic plants that display such traits, permitting both recognition and sustained feeding by native herbivore may be used as novel hosts (Pearse 2011; Pearse et al. 2013). By contrast, non-native plants may act as ecological traps (Schlaepfer et al. 2002) if they are more or equally attractive to native herbivore adults but are less palatable or more effectively defended, resulting in poorer performance of the offspring (e.g., D’Costa et al. 2014). In particular, the fitness cost experienced by herbivores shifting from one host plant to another was shown to increase with phylogenetic distance between the two hosts (Bertheau et al. 2010).
So far, the consequences of biotic introduction on plant–insect interactions and the effect of increased population density on insect host range expansion have been studied separately (Araújo et al. 2011; Forister and Wilson 2013; Pearse et al. 2013; Bezemer et al. 2014; but see Branco et al. 2014). Bridging these two frameworks can provide important insights into our understanding of the ecological consequences of intentional plant introductions, and, more generally, plant invasions.
Here, we explored the relationships between the population dynamics of the pine processionary moth (Thaumetopoea pityocampa, hereafter referred to as PPM) and change in its host range. PPM feeds on several species within the genus Pinus, and, occasionally on larch (Larix sp.), cedar (Cedrus sp.) or Douglas fir (Pseudotsuga sp.) (Jactel et al. 2015). Within the genus Pinus, PPM can show preferences for some species (Hódar et al. 2002; Stastny et al. 2006; Pérez-Contreras et al. 2014). However, there is no general agreement on rank order for host use, probably because of local adaptation of PPM populations (Zovi et al. 2008; Jactel et al. 2015) or because host quality may change during outbreaks (Li et al. 2015). Several tree traits have been suggested to drive host selection and use by PPM, including tree height, needle morphology or volatile organic compounds (Paiva et al. 2011; Jactel et al. 2015).
We monitored PPM infestation on 41 pine species planted in an arboretum during nine consecutive years including an outbreak, i.e., covering periods with very high or very low population densities. We tested whether there were significant changes in host use by PPM as a result of population density and whether host range expansion was density dependent. Specifically, we tested the three following hypotheses: (1) host range increases with PPM population density; (2) novel hosts are not chosen at random but preferably among species closely related to natives and/or exhibiting similar traits relevant to host selection or exploitation and (3) native pines are more likely to be attacked than exotic pines.
Materials and methods
Monitoring PPM densities at Arboretum National des Barres
PPM caterpillars feed at night during winter and spend the daytime in silky nests in tree crowns. These so-called winter nests shelter 50–100 larvae and are commonly used to monitor PPM infestation on trees as caterpillars are very conspicuous in tree crowns. The presence of a nest indicates that early instar larvae fed on and survived on the pine tree on which one female moth oviposited.
PPM abundance in the Arboretum National des Barres (N. 47.838, E. 2.7596, Paris Basin, France) was monitored in the arboretum between 1999 and 2007, by counting the number of winter nests on all individual pine trees planted at the arboretum. Hereafter we refer to the total number of nests per year on all sampled trees as ‘PPM population density’.
At the time of the survey, 2556 trees were present in the arboretum, of which 219 belonged to one of 46 Pinus species. Pines from hybrid species, or which died or were newly planted before the end of the survey were discarded from the analyses. Likewise, we did not distinguish between subspecies. The final dataset, therefore, contained 192 individual pines from 41 species of which eight were native to Europe, 11 introduced from Asia and 22 from North America (see Table S1, Supplementary Material). Although arboreta are usually not primarily designed for ecology studies, there were enough tree replicates of European, Asian and American pine species in the Arboretum National des Barres to allow testing our hypotheses.
Tree and needle characteristics
The height of all individual pines was measured in 2002. Tree height data were used to assess the effect of height on PPM attacks (Castagneyrol et al. 2014; Régolini et al. 2014). Although it may have changed over the time of the survey, we assumed that the rank order in tree height among individuals and species was maintained throughout the study, an assumption that should hold over a relatively short period.
Although there is no consensus on needle traits correlated with PPM abundance on different pine species (Jactel et al. 2015), pine needles with a shape that allows female moths clasping their tarsi around and holding on to them are expected to be more suitable for oviposition (Démolin 1969). Mean needle length and width per pine species were retrieved from the literature (Richardson 2000). For the three species for which this information was missing (Pinus cembra, P. arizonica, P. ayacahuite), 30 needles were collected from five trees of the arboretum, and needle length and width values were averaged at the species level.
Pine phylogeny
A phylogenetic tree for the 41 pine species included in this study was obtained from the conifer phylogeny developed by Delzon et al. (personnal communication). This reconstruction used sequences for chloroplast genes rbcL and matK, nuclear gene phyP, and the ITS1-5.8S-ITS2 region of ribosomal DNA, downloaded from the online database GenBank (Benson et al. 2011) and aligned using the pipeline PHLAWD (Smith et al. 2009). The complete dataset included over 300 species of conifers, and used three cycad species as outgroup. Following the so-called supermatrix approach (Dequeiroz and Gatesy 2007), a full likelihood search with 1000 bootstrap replicates was conducted on the whole dataset in RAxML (Stamatakis 2006), with each region treated as a separate partition with a distinct GTR + CAT substitution model. A chronogram was then constructed on the best tree from the likelihood search, using a relaxed-clock model and the chronos function from the ape package in R (Paradis et al. 2015), with a set of fifteen calibration points (derived from the fossil record) from Leslie et al. (2012).
Analyses
Test of phylogenetic signal in tree height and needle traits
We tested whether there was a phylogenetic signal in pine trait values expected to drive PPM preference. Among the wide array of indices developed to test for the presence of phylogenetic signals in traits, Pagel’s λ performs well when it is used to compare different traits within a single phylogeny (Münkemüller et al. 2012). Pagel’s λ can theoretically be greater than one (traits of related species are more similar than expected under Brownian motion) but usually varies between zero (no phylogenetic signal) to one (strong phylogenetic signal). Although we did not expect any phylogenetic signal in tree height, given differences in tree age among species, we used Pagel’s λ to test for phylogenetic signals in needle traits (needle length and needle width), but also in tree height for sake of completeness.
Testing factors controlling for pine species use by the PPM
We first tested if patterns of PPM attacks differed between pine trees of different introduction status (native vs. introduced) or from different regions of origin (Europe, North America, Asia) using Generalized Linear Mixed-effect Model (GLMM) with a binomial error family. Tree height, needle length and needle width were included as predictors. We used marginal testing to test the significance of introduction status and region of origin once the effects of traits were accounted for, and conversely the effect of traits while controlling for tree origin. The response variable was presence/absence of attack (winter nest) on a given tree. GLMM allowed accounting for repeated measurements on the same individuals. Random factors were calendar year, pine species identity and individual tree identity nested within species. Because every tree was surveyed every year, tree and year were crossed random factors. Variance explained by fixed effects (introduction status or region of origin) and random factors were estimated by calculating marginal R 2 values (\(R^{2}_{\text{m}}\), for fixed effects) and conditional R 2 values (\(R^{2}_{\text{c}}\), for fixed plus random effects) (Nakagawa and Schielzeth 2013).
Using GLMM with species as a random factor only partially accounts for phylogenetic non-independence among residuals. Therefore, we then used Phylogenetic Generalized Least Square (PGLS) models which are extensions of ordinary least square regressions that account for phylogenetic non-independence among individual observations (Paradis 2012). PGLS uses a theoretical variance–covariance matrix among species that quantifies how much species resemble each other (covariance), and how much they diverged from their common ancestor (variance). The variance–covariance matrix was estimated with Pagel’s λ (Pagel 1999) which improves the performances of phylogenetic regression when there is a phylogenetic signal in the independent variable (Revell 2010). For trait analysis, PGLS were used with data aggregated at the species level, using the mean number of PPM nests per year and per species as response variable and species introduction status, tree height, needle length and needle width as predictors.
Separate models were run for each year of the survey. Predictors were scaled and centred to make coefficients comparable within and between years (Schielzeth 2010).
Modeling diet breadth
Under the null hypothesis (H 0) that female PPM select pines at random for oviposition, the number of pine species with one or more PPM nests should increase with both PPM population and pine densities, simply as a result of a sampling effect: (1) the more females, the more nests and the greater the probability of any tree being attacked and (2) the more pine individuals per species, the greater the probability of a particular pine species being infested. Taking these densities into account, we used null models adapted from Jorge et al. (2014) to test H 0.
We defined two potential host pools (PHP). In the first one (PHP1), all pine species were considered potential hosts for PPM. In the second one (PHP2), only those species that were attacked at least once during the seven-year survey were included. Null models were built by randomly drawing N trees from the PHP, where N was the number of PPM nests observed in a given year. This procedure thus accounted for the unbalanced abundance of the different pine species in the arboretum. Trees were drawn with replacement to make it possible that the same tree holds several nests. Based on this random sample, we calculated the number of host species used by PPM under H 0 (S random). This procedure was repeated 1000 times. We eventually compared observed (S observed) and simulated (S random) numbers of host species using a standardized coefficient k (see below, Eq. 1).
To test the hypothesis that the number of novel hosts increased with PPM population density under a non-random process according to species relatedness (i.e., phylogenetic clustering), we used the same null model but this time added phylogenetic information (Jorge et al. 2014; Parker et al. 2015). We calculated the mean phylogenetic distance between pine species used as hosts in a given year (MPDobserved) and compared this value to the 1000 MPDrandom calculated from random samples generated by the null model. We then calculated the weighted mean of all pairwise distances among species (Parker et al. 2015), using the mpd function in package picante (Kembel et al. 2014), where phylogenetic distances were weighted by the number of attacks on each host species.
Observed and simulated values were compared using a standardized coefficient k (k S for richness and k MPD for phylogenetic clustering) that is equivalent to a z-score (Eq. 1):
where x observed, is either S observed or MPDobserved and m random and σrandom are the mean and standard deviation of the 1000 random simulations of either S random or MPDrandom. As such, k MPD is analogous to the dietary specialization index DSIA developed by Jorge et al. (2014).
For host species richness, negative and positive k S values indicate that the number of pine species on which PPM nests were observed were lower and greater, respectively, than expected by chance.
For MPD, k MPD was multiplied by −1 so that positive k MPD values indicate a greater phylogenetic clustering than expected by chance, while negative values are indicative of phylogenetic overdispersion. If |k| < 2, the observed value is approximately within the range expected by chance. Conversely, if |k| > 2, then the observed value is approximately in the 5 % tail of a normal distribution. We finally tested the effect of PPM population density on k for the two potential host pools using linear regressions. An increase of k MPD with PPM density would indicate that host range tends to be more phylogenetically clustered. We ran a last simulation to confirm this assumption by testing how the phylogenetic distance between the most regularly attacked species and other species varied with PPM density. Pinus nigra, P. sylvestris and P. ponderosa were the most commonly attacked species (see results) and thus constituted the ‘regular hosts pool’. For each year separately, we calculated the mean nearest taxon distance between the regular hosts pool and the pool of newly added host species (‘additional hosts pool’), that is a measure of β-phylogenetic diversity (comdistnt function in package picante). We finally ran the same simulations as above, k coefficient as above for every year, and regressed k against PPM population density.
Although the maximal distance between pines was 952 m (Appendix 1, Supplementary Material), pines were distributed among two areas within the arboretum. We checked for independence between phylogenetic and geographic (Euclidean) distances between trees of the arboretum (Appendix 1). We finally re-ran previous models for each garden separately to confirm the robustness of the main results (Appendix 1).
All analyses were done in R Core Team (2016). We used the functions gls, glmer, r.squaredGLMM from packages nlme, lme4, and MuMIn, respectively (Bates et al. 2013; Pinheiro et al. 2014; Bartoń 2015). For phylogenetic analyses we used the phytools package (Revell 2012).
Results
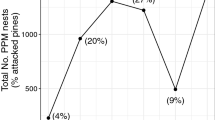
A total of 2309 winter nests were counted on pine trees between 1999 and 2007. Among the 41 pine species planted at the National Arboretum des Barres, 28 were attacked by PPM at least once during the survey period (i.e., 68 %, Fig. 1). PPM population density varied between years but was independent of tree age: it was maximal in 2002, in the middle of the survey, and minimal in 2004 (Fig. 2).
Relationship between mean tree height, needle width, needle length and PPM attack rate with pine species phylogeny and origin. Pink, green and blue colours refer to pines species from Europe, Asia and North America, respectively. Traits values and mean number of nests per year and per species (PPM) were scaled from 0 to 1. The larger the dot, the bigger the value. Crosses indicate pine species that have never been attacked during the survey (color figure online)
PPM attacks were concentrated on P. nigra, P. sylvestris and P. ponderosa. Despite the fact that, together, they represented only 39 % of planted trees, they concentrated between 82 and 85 % of attacks between years of low and high PPM density (% attacks on P. nigra/P. ponderosa/P. sylvestris, low PPM density: 67/15/0; high PPM density: 56/24/5). Although P. ponderosa is non-native to the study area, there was no overall effect of the introduction status (native to Europe vs. introduced, GLMM: χ 2 = 0.05, P = 0.824, \(R^{2}_{\text{m}}\) = 0.18, \(R^{2}_{\text{c}}\) = 0.67) or region of origin (Europe vs. Asia vs. America, GLMM: χ 2 = 0.05, P = 0.974, \(R^{2}_{\text{m}}\) = 0.18, \(R^{2}_{\text{c}}\) = 0.67) of pine species on PPM attack probability (Fig. 1). Only needle width had a significant effect: the wider its needles, the greater the chance was that a pine species was attacked (χ2 = 9.57, P = 0.002 and χ 2 = 10.58, P = 0.001 in GLMMs including introduction status and region of origin as factors, respectively, Fig. 3). Random factors explained more than 66 % of the variance in PPM attack probability.
Effects of needles width, needles length and tree height on PPM attack. a Effects of needle width on the probability of PPM attack. Lines represent predictions from GLMM for years as random intercepts. Colour palette refers to PPM population density (and corresponding year). b Standardized model parameter estimates of year-specific PGLS models showing the effects of tree height, needles length and needles width on the mean number of PPM nests per tree. Vertical bars represent standard errors. Stars indicate coefficients that are significantly different from zero
Closely related species tended to be more similar in terms of needle traits as shown by the significant, though rather weak, phylogenetic signal in needle length (λ = 0.26, P = 0.013) and needle width (λ = 0.29, P = 0.015). As expected, pine height was independent of pine phylogeny (λ = 0.00, P = 1.000, Fig. 1).
The effect of needle width remained after phylogenetic non-independence was accounted for (PGLS, Table S2). The strength of the needle width effect increased with PPM population density (Fig. 3). Needle length had no effect on PPM attacks (except in 2007, Table S2). There was a tendency for taller trees being attacked more than smaller ones, but this effect was significant only when PPM population density was low (Fig. 3). Needle length had no effect on PPM attacks (but in 2007, Table S2).
The number of pine trees and pine species attacked by the PPM increased with PPM population density (Pearson’s correlation: r individuals = 0.96, r species = 0.93, Fig. 2). Regardless of the potential host pool (i.e., all pine species, PHP1, or only species attacked at least once, PHP2), PPM attacked significantly fewer species than expected by chance (all k S < 2, Fig. 4a). The realized host range departed more from randomness as PPM population density increased (PHP1: F 1,7 = 632, P < 0.0001, R 2 = 0.99, PHP2: F 1,7 = 179, P < 0.0001, R 2 = 0.96, Fig. 4a).
Effects of PPM nest density on PPM host range. a Results of null models comparing the observed (S obs) vs. simulated (S sim) number of species attacked by the PPM under the null hypothesis; k is proportional to the difference between observed and simulated values. Negative values indicate that the number of attacked species was lower than expected by chance. b Results of null models comparing the observed (MPDobs) vs. simulated (MPDsim) mean phylogenetic distance between species attacked by the PPM under the null hypothesis. To make it consistent with (a), k was multiplied by −1 so that positive k values indicate a greater phylogenetic clustering than expected by chance. Simulations were based on two potential host pools (PHP, see “Materials and methods”). Shaded areas and horizontal dashed lines represent the [−2; 2] interval corresponding to expectations under the null hypothesis
The observed phylogenetic clustering of host trees increased with PPM population density (Pearson’s correlation: r = 0.66). PPM’s host range was more clustered than under the null hypothesis (all k MPD > 2, Fig. 4b) and phylogenetic clustering increased with PPM population density (PHP1: F 1,7 = 11, P < 0.013, R 2 = 0.55, PHP2: F 1,7 = 21, P < 0.003, R 2 = 0.71, Fig. 4b). When PPM population density increased, z-scores corresponding to β-phylogenetic distance between the ‘regular hosts pool’ and the ‘additional hosts pool’ were always positive and increased with PPM density (intercept: 2.05 ± 0.21, t = 9.80, P < 0.0001, slope: 22.83 × 10−4 ± 6.36 × 10−4, t = 3.59, P = 0.009), which confirms that the species newly added to the regular host range were more closely related to regular hosts than expected by chance.
Discussion
Our results clearly show that PPM’s host range expansion depended more on PPM population density and phylogenetic relationships among pine species than on introduction status of pines.
We hypothesized that PPM’s host range should increase during outbreaks, by incorporating pine species closely related to hosts normally used during latent periods. Our results are consistent with this hypothesis and with the literature. Closely related host species are more likely to share common traits and defenses against herbivores (Agrawal and Fishbein 2006; Rasmann and Agrawal 2011; Nakadai and Murakami 2015). It is, therefore, not surprising that the new hosts were chosen from among the close relatives of regular hosts. This result suggests that in years of low PPM abundance, PPM nests were found on a small set of highly suitable hosts compared to years of high PPM abundance. Increased intraspecific competition during outbreaks may have increased divergence in host use and forced PPM to exploit less preferred, but still suitable hosts (Araújo et al. 2011). Why these optional hosts are only used during outbreaks may be explained by the fitness loss of herbivores feeding on suboptimal plants (Bertheau et al. 2010; Rasmann and Agrawal 2011; Nakadai and Murakami 2015). During outbreaks, this cost may be a better option than the risk of facing starvation, due to either induced resistance or intra-specific competition (Svanbäck and Bolnick 2007; Plath et al. 2011; Branco et al. 2014). Alternatively, as PPM abundance increased, the increase in host range at the population level may have simply resulted from a greater number of individuals being able to exploit new hosts, due to larger intra-specific variability in individual insect preferences (Bolnick et al. 2011).
Contrary to our expectations, the marked preferences for some particular species did not result from the avoidance of introduced species: both native and introduced species were equally attacked by PPM. Even if they escape natural enemies from their native range (“the enemy release hypothesis”, Keane and Crawley 2002), non-native plant species can face novel herbivores in the area of introduction (Parker et al. 2012), particularly when species from the same genus are present in the introduction area (Branco et al. 2015). The likelihood for introduced plants to recruit new herbivore species was shown to increase with phylogenetic or trait similarity with native plant species (Ness et al. 2011), even within the same genus (da Ros et al. 1993; Pearse et al. 2013). Phylogenetic relatedness among pine species was, therefore, a stronger driver of host use pattern than tree region of origin. Likewise, plant traits predicted host use better than introduction status or region of origin as previously observed for oaks (Pearse 2011).
We found that needle width was a key predictor of PPM attack probability on a particular pine species. Démolin (1969) observed that the ability of female moth to firmly ‘hook’ pine needles for oviposition was greatest for needles between 1.6 and 2 mm wide, which corresponded to the length of PPM’s tarsi. Accordingly, we showed that the probability of attack was greatest for needles with a width approaching 2 mm, irrespective of species’ introduction status. Yet, this trait displayed a phylogenetic signal in pines. Therefore, PPM may have been ecologically fitted to lay eggs on non-native pines having a particular combination of needle traits (Forister and Wilson 2013). This confirms that phylogenetic signals can be detected when considering plant–insect interactions, but this needs not be the result of coevolution (Agosta 2006).
We showed that PPM attacked significantly fewer pine species than it would be expected by chance. This is consistent with previous studies reporting PPM female preferences for alternative host species (Stastny et al. 2006; Paiva et al. 2011), but it conflicts with the view that PPM females are rather unselective regarding oviposition sites (Hódar et al. 2002). In this study, we counted the number of nests, and not actual oviposition events. It is common that offspring survive or perform better on plants that their mother preferred, and that females prefer plants (for oviposition) that are more suitable to the offspring (i.e., “mother knows best”; Gripenberg et al. 2010). Whether or not this is the case in PPM remains to be tested properly, using behavioral experiments. Although we interpreted host preference in terms of female choice, it cannot be excluded that the observed distribution of PPM nests resulted (in part or entirely) from differential survival of eggs or young larvae. For instance, Hódar et al. (2002) found that females were unselective regarding oviposition, while the survival of early instar larvae varied among pine species. By contrast Stastny et al. (2006) reported that host preference based on number of nests in the field was consistent with oviposition preference in controlled experiments. Such discrepancies may result from local adaptations of PPM populations (Zovi et al. 2008), which makes it difficult to generalize findings about host preference in PPM. However, the fact that a certain host trait particularly relevant to female moth behavior (correspondence between needle width and moth tarsi length) was found to explain host preference should provide confidence in our interpretation of host range expansion driven by female’s choice.
Our study was conducted in an arboretum at the northern edge of PPM’s geographic range (Battisti et al. 2005). It is possible that the hierarchy in host use by herbivores in marginal populations may not reflect preferences in more central populations. For instance, choice behavior may be less conservative if normally preferred hosts are scarce or missing (Carrasco et al. 2015), which was not the case in the arboretum. However, PPM was shown to act conservatively regarding host preferences (Stastny et al. 2006) and despite the unbalanced design, no less than 41 pine species were available in the arboretum. PPM was, therefore, given the choice between different potential host species so that we can reject the possibility that the location of this study biased PPM’s choice towards non-native species. In addition, the location of the arboretum at the front edge of PPM geographical range expansion provided further reassurance that no coevolution processes were behind host range expansion.
Conclusion
PPM showed clear preference for particular pine species, those which have wider needles. This choice was independent of pine introduction status indicating that non-native species were neither more nor less likely to be attacked by the PPM than native species. Importantly, the host range increased with PPM population density in a non-random way. Host range expansion occurred on pines closely related to regular hosts, the latter being ‘ecologically fitted’ to be attacked by the PPM as a result of needle traits that displayed a phylogenetic signal. Regardless of the mechanisms underlying the observed patterns, the density dependence of host use may have profound implications for the population dynamics not only of PPM, but also of other, co-occurring pests on the same host trees (Bolnick et al. 2011). From an applied point of view, our results allow the identification of pine species that would be at higher risk of PPM attack, should it be accidentally introduced outside its natural geographic range or should exotic pines be planted in the native range of PPM (Lombardero et al. 2012).
References
Agosta SJ (2006) On ecological fitting, plant–insect associations, herbivore host shifts, and host plant selection. Oikos 114:556–565. doi:10.1111/j.2006.0030-1299.15025.x
Agrawal AA, Fishbein M (2006) Plant defense syndromes. Ecology 87:S132–S149
Araújo MS, Bolnick DI, Layman CA (2011) The ecological causes of individual specialisation. Ecol Lett 14:948–958. doi:10.1111/j.1461-0248.2011.01662.x
Bartoń K (2016) MuMIn: multi-model inference. R package version 1.15.6. https://cran.r-project.org/package=MuMIn
Bates D, Maechler M, Bolker B, Walker S (2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67(1):1–48. doi:10.18637/jss.v067.i01
Battisti A, Stastny M, Nether̀er S et al (2005) Expansion of geographic range in the pine processionary moth caused by increased winter temperatures. Ecol Appl 15:2084–2096
Benson DA, Karsch-Mizrachi I, Lipman DJ et al (2011) GenBank. Nucleic Acids Res 39:D32–D37. doi:10.1093/nar/gkq1079
Bertheau C, Brockerhoff EG, Roux-Morabito G et al (2010) Novel insect-tree associations resulting from accidental and intentional biological “invasions”: a meta-analysis of effects on insect fitness. Ecol Lett 13:506–515
Bezemer TM, Harvey JA, Cronin JT (2014) Response of native insect communities to invasive plants. Annu Rev Entomol 59:119–141. doi:10.1146/annurev-ento-011613-162104
Bolnick DI, Amarasekare P, Araújo MS et al (2011) Why intraspecific trait variation matters in community ecology. Trends Ecol Evol 26:183–192. doi:10.1016/j.tree.2011.01.009
Branco M, Dhahri S, Santos M, Ben Jamaa ML (2014) Biological control reduces herbivore’s host range. Biol Control 69:59–64. doi:10.1016/j.biocontrol.2013.11.001
Branco M, Brockerhoff EG, Castagneyrol B et al (2015) Host range expansion of native insects to exotic trees increases with area of introduction and the presence of congeneric native trees. J Appl Ecol 52:69–77. doi:10.1111/1365-2664.12362
Cappuccino N, Carpenter D (2005) Invasive exotic plants suffer less herbivory than non-invasive exotic plants. Biol Lett 1:435–438. doi:10.1098/rsbl.2005.0341
Carrasco D, Larsson MC, Anderson P (2015) Insect host plant selection in complex environments. Curr Opin Insect Sci 8:1–7. doi:10.1016/j.cois.2015.01.014
Castagneyrol B, Régolini M, Jactel H (2014) Tree species composition rather than diversity triggers associational resistance to the pine processionary moth. Basic Appl Ecol 15:516–523. doi:10.1016/j.baae.2014.06.008
D’Costa L, Simmonds MSJ, Straw N et al (2014) Leaf traits influencing oviposition preference and larval performance of Cameraria ohridella on native and novel host plants. Entomol Exp Appl 152:157–164. doi:10.1111/eea.12211
da Ros N, Ostermeyer R, Roques A, Raimbault JP (1993) Insect damage to cones of exotic conifer species introduced in arboreta. J Appl Entomol 115:113–133. doi:10.1111/j.1439-0418.1993.tb00371.x
Démolin G (1969) Comportement des adultes de Thaumetopoea pityocampa Schiff.: dispersion spatiale, importance écologique. Ann For Sci 26:81–102
Dequeiroz A, Gatesy J (2007) The supermatrix approach to systematics. Trends Ecol Evol 22:34–41. doi:10.1016/j.tree.2006.10.002
Desurmont GA, Donoghue MJ, Clement WL, Agrawal AA (2011) Evolutionary history predicts plant defense against an invasive pest. Proc Natl Acad Sci 108:7070–7074. doi:10.1073/pnas.1102891108
Forister ML, Wilson JS (2013) The population ecology of novel plant–herbivore interactions. Oikos 122:657–666. doi:10.1111/j.1600-0706.2013.00251.x
Gilbert GS, Briggs HM, Magarey R (2015) The impact of plant enemies shows a phylogenetic signal. PLoS One 10:e0123758. doi:10.1371/journal.pone.0123758
Gripenberg S, Mayhew PJ, Parnell M, Roslin T (2010) A meta-analysis of preference–performance relationships in phytophagous insects. Ecol Lett 13:383–393. doi:10.1111/j.1461-0248.2009.01433.x
Harvey JA, Biere A, Fortuna T et al (2010) Ecological fits, mis-fits and lotteries involving insect herbivores on the invasive plant, Bunias orientalis. Biol Invasions 12:3045–3059. doi:10.1007/s10530-010-9696-9
Hódar JA, Zamora R, Castro J (2002) Host utilisation by moth and larval survival of pine processionary caterpillar Thaumetopoea pityocampa in relation to food quality in three Pinus species. Ecol Entomol 27:292–301. doi:10.1046/j.1365-2311.2002.00415.x
Jactel H, Barbaro L, Battisti A et al (2015) Insect–tree interactions in Thaumetopoea pityocampa. In: Roques A (ed) Processionary moths and climate change: an update. Springer, Netherlands, pp 265–310
Jorge LR, Prado PI, Almeida-Neto M, Lewinsohn TM (2014) An integrated framework to improve the concept of resource specialisation. Ecol Lett 17:1341–1350. doi:10.1111/ele.12347
Keane RM, Crawley MJ (2002) Exotic plant invasions and the enemy release hypothesis. Trends Ecol Evol 17:164–170. doi:10.1016/S0169-5347(02)02499-0
Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO (2010) Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26(11):1463–1464. doi:10.1093/bioinformatics/btq166
Leslie AB, Beaulieu JM, Rai HS et al (2012) Hemisphere-scale differences in conifer evolutionary dynamics. Proc Natl Acad Sci 109:16217–16221. doi:10.1073/pnas.1213621109
Li S, Daudin JJ, Piou D et al (2015) Periodicity and synchrony of pine processionary moth outbreaks in France. For Ecol Manag 354:309–317. doi:10.1016/j.foreco.2015.05.023
Lombardero MJ, Alonso-Rodríguez M, Roca-Posada EP (2012) Tree insects and pathogens display opposite tendencies to attack native vs. non-native pines. For Ecol Manag 281:121–129. doi:10.1016/j.foreco.2012.06.036
Münkemüller T, Lavergne S, Bzeznik B et al (2012) How to measure and test phylogenetic signal: how to measure and test phylogenetic signal. Methods Ecol Evol 3:743–756. doi:10.1111/j.2041-210X.2012.00196.x
Nakadai R, Murakami M (2015) Patterns of host utilisation by herbivore assemblages of the genus Caloptilia (Lepidoptera; Gracillariidae) on congeneric maple tree (Acer) species. Ecol Entomol 40:14–21. doi:10.1111/een.12148
Nakadai R, Murakami M, Hirao T (2014) Effects of phylogeny, leaf traits, and the altitudinal distribution of host plants on herbivore assemblages on congeneric Acer species. Oecologia 175:1237–1245. doi:10.1007/s00442-014-2964-0
Nakagawa S, Schielzeth H (2013) A general and simple method for obtaining R 2 from generalized linear mixed-effects models. Methods Ecol Evol 4:133–142. doi:10.1111/j.2041-210x.2012.00261.x
Nakládal O, Uhlíková H (2015) Review of historical outbreaks of the nun moth with respect to host tree species. J For Sci 61:18–26. doi:10.17221/94/2014-JFS
Ness JH, Rollinson EJ, Whitney KD (2011) Phylogenetic distance can predict susceptibility to attack by natural enemies. Oikos 120:1327–1334. doi: 10.1111/j.1600-0706.2011.19119.x
Pagel M (1999) Inferring the historical patterns of biological evolution. Nature 401:877–884. doi:10.1038/44766
Paiva MR, Mateus E, Santos MH, Branco MR (2011) Pine volatiles mediate host selection for oviposition by Thaumetopoea pityocampa (Lep., Notodontidae). J Appl Entomol 135:195–203. doi:10.1111/j.1439-0418.2010.01550.x
Paradis E (2012) Analysis of phylogenetics and evolution with R. Springer New York, New York
Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290
Parker JD, Burkepile DE, Lajeunesse MJ, Lind EM (2012) Phylogenetic isolation increases plant success despite increasing susceptibility to generalist herbivores: phylogenetic relatedness and invasion success. Divers Distrib 18:1–9. doi:10.1111/j.1472-4642.2011.00806.x
Parker IM, Saunders M, Bontrager M et al (2015) Phylogenetic structure and host abundance drive disease pressure in communities. Nature 520:542–544. doi:10.1038/nature14372
Pearse IS (2011) The role of leaf defensive traits in oaks on the preference and performance of a polyphagous herbivore, Orgyia vetusta. Ecol Entomol 36:635–642. doi:10.1111/j.1365-2311.2011.01308.x
Pearse IS, Hipp AL (2009) Phylogenetic and trait similarity to a native species predict herbivory on non-native oaks. Proc Natl Acad Sci 106:18097–18102. doi:10.1073/pnas.0904867106
Pearse IS, Harris DJ, Karban R, Sih A (2013) Predicting novel herbivore–plant interactions. Oikos 122:1554–1564. doi:10.1111/j.1600-0706.2013.00527.x
Pérez-Contreras T, Soler JJ, Soler M (2014) Host selection by the pine processionary moth enhances larval performance: an experiment. Acta Oecol 55:15–22. doi:10.1016/j.actao.2013.10.006
Pinheiro J, Bates D, DebRoy S, Sarkar D and R Core Team (2016) nlme: Linear and Nonlinear Mixed Effects Models. R package version 3.1-128. Bioinformatics 26:1463–1464
Plath M, Dorn S, Riedel J et al (2011) Associational resistance and associational susceptibility: specialist herbivores show contrasting responses to tree stand diversification. Oecologia 169:477–487. doi:10.1007/s00442-011-2215-6
Rasmann S, Agrawal AA (2011) Evolution of specialization: a phylogenetic study of host range in the red milkweed beetle (Tetraopes tetraophthalmus). Am Nat 177:728–737. doi:10.1086/659948
R Core Team (2016) R: a language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria. https://www.R-project.org/
Régolini M, Castagneyrol B, Dulaurent-Mercadal A-M et al (2014) Effect of host tree density and apparency on the probability of attack by the pine processionary moth. For Ecol Manag 334:185–192. doi:10.1016/j.foreco.2014.08.038
Revell LJ (2010) Phylogenetic signal and linear regression on species data. Methods Ecol Evol 1:319–329. doi:10.1111/j.2041-210X.2010.00044.x
Revell LJ (2012) phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol 3:217–223. doi:10.1111/j.2041-210X.2011.00169.x
Richardson DM (ed) (2000) Ecology and biogeography of Pinus. Cambridge Univ. Press, Cambridge
Schielzeth H (2010) Simple means to improve the interpretability of regression coefficients: interpretation of regression coefficients. Methods Ecol Evol 1:103–113. doi:10.1111/j.2041-210X.2010.00012.x
Schlaepfer MA, Runge MC, Sherman PW (2002) Ecological and evolutionary traps. Trends Ecol Evol 17:474–480. doi:10.1016/S0169-5347(02)02580-6
Smith SA, Beaulieu JM, Donoghue MJ (2009) Mega-phylogeny approach for comparative biology: an alternative to supertree and supermatrix approaches. BMC Evol Biol 9:37. doi:10.1186/1471-2148-9-37
Srivastava DS, Cadotte MW, MacDonald AAM et al (2012) Phylogenetic diversity and the functioning of ecosystems. Ecol Lett 15:637–648. doi:10.1111/j.1461-0248.2012.01795.x
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi:10.1093/bioinformatics/btl446
Stastny M, Battisti A, Petrucco-Toffolo E et al (2006) Host-plant use in the range expansion of the pine processionary moth, Thaumetopoea pityocampa. Ecol Entomol 31:481–490. doi:10.1111/j.1365-2311.2006.00807.x
Svanbäck R, Bolnick DI (2007) Intraspecific competition drives increased resource use diversity within a natural population. Proc R Soc Lond B Biol Sci 274:839–844. doi:10.1098/rspb.2006.0198
Underwood N (2010) Density dependence in insect performance within individual plants: induced resistance to Spodoptera exigua in tomato. Oikos 119:1993–1999. doi:10.1111/j.1600-0706.2010.18578.x
White JA, Whitham TG (2000) Associational susceptibility of cottonwood to a box elder herbivore. Ecology 81:1795–1803
Whitfeld TJS, Novotny V, Miller SE et al (2012) Predicting tropical insect herbivore abundance from host plant traits and phylogeny. Ecology 93:S211–S222. doi:10.1890/11-0503.1
Zovi D, Stastny M, Battisti A, Larsson S (2008) Ecological costs on local adaptation of an insect herbivore imposed by host plants and enemies. Ecology 89:1388–1398. doi:10.1890/07-0883.1
Acknowledgments
We thank Manuela Branco for providing important ideas and must-read papers. We also thank Ian S. Pearse and anonymous reviewers for their helpful comments on original version of the present manuscript. Original analyses were done by Colleen Carlson and EB. Contributions by EB were supported by MBIE core funding to Scion and the Better Border Biosecurity collaboration (http://www.b3nz.org).
Data accessibility
Data used in this study are uploaded as supplementary material (SM2 and SM3). SM2 corresponds to raw data. SM3 is the phylogenetic tree (Newick format) of pine species planted in Arboretum National des Barres.
Author contribution statement
DP, EB and HJ developed the original idea. SD and ML provided the phylogenetic tree. DP and NP collected data. BC analysed the data and drafted the first version of the MS. All authors provided comments and contributed to the final version.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Raphael Didham.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Castagneyrol, B., Jactel, H., Brockerhoff, E.G. et al. Host range expansion is density dependent. Oecologia 182, 779–788 (2016). https://doi.org/10.1007/s00442-016-3711-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00442-016-3711-5