Abstract
Biodiversity is important for ecosystem functioning and biotic interactions. In experimental grasslands, increasing plant species richness is known to increase the diversity of associated herbivores and their predators. If these interactions can also involve endosymbionts that reside within a plant or animal host is currently unknown. In plant-feeding aphids, secondary bacterial symbionts can have strong fitness effects on the host, e.g. resistance to natural enemies or fungal pathogens. We examined the secondary symbiont community in three species of aphid, each feeding on a unique host plant across experimental plots that varied in plant species richness. Aphids were collected in May and June, and the symbiont community identified using species-specific PCR assays. Aphis fabae aphids were found to host six different symbiont species with individual aphids co-hosting up to four symbionts. Uroleucon jaceae and Macrosiphum rosae hosted two and three symbiont species, respectively. We found that, at the aphid population level, increasing plant species richness increased the diversity of the aphid symbiont community, whereas at the individual aphid level, the opposite was found. These effects are potentially driven by varying selective pressures across different plant communities of varying diversities, mediated by defensive protection responses and a changing cost-benefit trade-off to the aphid for hosting multiple secondary symbionts. Our work extends documented effects of plant diversity beyond visible biotic interactions to changes in endosymbiont communities, with potentially far-reaching consequences to related ecosystem processes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biodiversity, i.e. the variety of life on earth, is not only of ethical and aesthetical value but also of functional importance (Hector et al. 2001). Low diversity in a community has now been consistently found to be associated with a lowered mean (and an increase in the variance) in many of the ecosystem functions investigated (Hooper et al. 2005; Allan et al. 2013). Most research on the relationship between plant diversity and ecosystem functioning has focused primarily on plant-related ecosystem variables, revealing positive effects of biodiversity on processes such as primary productivity, nutrient use efficiency and invasion resistance (Tilman et al. 1996; Ruijven et al. 2003), although the strength and shape of the relationship between plant diversity and ecosystem variables varies between the variables investigated (Roscher et al. 2004; Scherber et al. 2010; Allan et al. 2013).
More recently, studies of biodiversity effects have also incorporated trophic complexity (Duffy et al. 2007) focusing on variables relating to the interaction of plants with higher trophic levels, for example insect herbivores (Scherber et al. 2006; Borer et al. 2012; Loranger et al. 2014), fungal pathogens (Mitchell 2003), or on multi-trophic interactions (Haddad et al. 2009; Petermann et al. 2010b; Ebeling et al. 2012). These studies have shown that plant diversity affects interactions between plants and other trophic levels (Rzanny and Voigt 2012), as well as ecosystem functions mediated by these interactions (Scherber et al. 2010; Ebeling et al. 2014a). Again, the relationship between plant species richness and the abundances and diversities of other organisms differs between groups. For example, effects on the diversity of other organisms are stronger than effects on abundance (Allan et al. 2013), and insect herbivore diversity increases more strongly than predator diversity with increasing plant species richness (Scherber et al. 2010). The effect size of diversity can be high. In a long-term grassland experiment bottom–up plant-mediated diversity effects accounted for up to 60 % of explainable variation in the community of sucking herbivores whereas top–down forces (predation) explained only a further 25 % of the variation (Roscher et al. 2004; Rzanny et al. 2013). Resulting changes in the community of higher trophic level organisms can control rates of ecosystem functioning (Ebeling et al. 2014a; Loranger et al. 2014), and indeed cascading interaction effects may underlie many of the observed biodiversity effects on ecosystem processes for which there is currently no mechanistic explanation (Allan et al. 2013). However, analyses of plant diversity effects on interactions have been largely restricted to two-species interactions, while analyses of interactions of three or more partners are still rare (Haddad et al. 2009; Petermann et al. 2010b; Ebeling et al. 2012).
One example where interactions can span several trophic layers are plant-feeding aphids and their natural enemies (Bukovinszky et al. 2008). Plant species richness has been shown to affect the community composition, abundance and life-history traits of aphids and their associated parasitoids in experimental grassland plots (Petermann et al. 2010a, b). Aphids feed on the phloem sap of a restricted range of host plants, with only 5 % of species considered as polyphagous (Pettersson et al. 2007). While aphids can easily distinguish a host plant from a non-host plant, they are also sensitive to subtle physiological changes within a host plant species (Powell et al. 2006). Aphids are known to exhibit host choice for different plant genotypes, which can be context-dependent on environmental factors such as the presence of another aphid (Zytynska and Preziosi 2011; Zytynska et al. 2014). A combination of all these variables in concert with natural enemy pressure is likely to influence the interaction an aphid experiences with its host plant.
Interactions can also involve organisms that perhaps are often overlooked since they reside within a host itself, for example symbiotic microorganisms (Omacini et al. 2001). Many aphids are host to secondary (facultative) symbiont bacteria that have been shown to confer resistance to parasitic wasps and fungi (Oliver et al. 2003, 2014; Scarborough et al. 2005), mediate effects of heat stress (Montllor et al. 2002; Russell and Moran 2006) and be involved in host plant use (Tsuchida et al. 2004; Wagner et al. 2015). While certain symbiont–aphid combinations increased aphid survival on specific host plants, other studies have found little species-specific effects (McLean et al. 2011) or, rather, that the symbionts provide a general benefit, thus allowing an aphid colony to better survive on a sub-optimal host (Wulff and White 2015). Aphid symbionts are predominantly vertically transmitted, which means they are also a source of heritable variation which can lead to host-associated differentiation and population adaptation (Ferrari and Vavre 2011). They may also be horizontally transmitted (Russell et al. 2003), for example by parasitic wasps (Gehrer and Vorburger 2012) or potentially via the plant, as has been shown for Rickettsia in whitefly (Caspi-Fluger et al. 2012), although the extent to which this happens in aphids is unknown (Darby and Douglas 2003; Russell et al. 2003; Gehrer and Vorburger 2012; Henry et al. 2013).
Aside from the wide benefits of hosting certain symbionts, there can also be a fitness cost (reduced longevity and fecundity due to the energy demand of the symbiont) to the aphid (Chen et al. 2000; Vorburger and Gouskov 2011). Aphid symbionts are usually only hosted by a proportion of aphids in a population (Sandström et al. 2001), potentially due to a cost–benefit trade-off for hosting them. In some cases, symbiont species can be fixed in a population which suggests strong selection driving the association. For example, Arsenophonus was found to infect 100 % of Aphis glycines aphids in some soybean populations (Wulff et al. 2013). While Arsenophonus was not found to provide any defensive protection to natural enemies or a fungal pathogen, it was found to provide a more general benefit to the aphid, although this also indicates a reduced cost to hosting the symbiont and hence the larger number of aphids hosting it (Wulff et al. 2013; Wulff and White 2015). Further, Serratia, a symbiont with fitness effects on pea aphids and blue alfalfa aphids (Chen et al. 2000), was found to infect 95 % of Microlophium carnosum aphids on Urtica dioica (Haynes et al. 2003), perhaps suggesting either lower fitness costs or increased benefit is maintaining this symbiont.
There is growing evidence for aphids to host multiple species of symbiont (Zytynska and Weisser 2015). In field surveys, aphids host on average 1–2 symbiont species (Ferrari et al. 2012; Russell et al. 2013; Smith et al. 2015). Multiple infection may benefit the aphids through increased resistance (Oliver et al. 2006), but there could also be an increased cost (Oliver et al. 2014). Multiple infections are also linked to an increased chance of transmission failures leading to more stable single infections and thus fewer than expected multiple infections in natural populations (Oliver et al. 2014). Further, within the aphid symbiont community, positive and negative interactions between symbiont species could additionally influence the frequency of multiple infections, either through impacts on fitness or through transmission failures (Frantz et al. 2009; Ferrari et al. 2012; Oliver et al. 2014).
Despite the growing area of aphid–symbiont research, we still know little of the range of effects that different symbiont species can have on the aphid host. It is increasingly becoming clear that the effect of the symbiont can be context-dependent on aphid species, aphid clone, symbiont strain, host plant species and other abiotic factors (Montllor et al. 2002; Tsuchida et al. 2002; Leonardo and Muiru 2003; Brady and White 2013; Russell et al. 2013). Many of these factors could also be mediated by the surrounding interacting community to which an aphid, and its symbiont community, could respond. Aphid fitness is linked to plant quality, and plant diversity has been shown to change the availability of carbon and nitrogen in a plant, with cascading effects on associated grasshopper herbivores and pollinating bees (Abbas et al. 2014). However, the effect of this on the aphid or its symbionts is unknown, although some symbionts may have the ability to benefit the aphid if there is a nutritional imbalance in the plant, potentially leading to symbiont-mediated aphid–plant interactions. For example, Serratia is known to be able to act as a nutritional symbiont in Cinara cedri aphids (Pérez-Brocal et al. 2006), and Arsenophonus has been found to provide an overall general benefit to A. glycines aphids, perhaps providing a nutritional advantage (Wulff and White 2015).
The potentially large community of symbionts associated with aphids, along with the large food web in which aphids are embedded, make them ideal new study system for investigating plant diversity effects on interaction webs (Omacini et al. 2001; Bukovinszky et al. 2008; Petermann et al. 2010b; Hackett et al. 2013). Within an aphid population, there are two levels at which to consider the effect of the plant community on the symbiont community: at the individual aphid level and at the population level. At the individual aphid level, the benefit and costs associated with hosting symbionts are important, as these can directly determine the fitness of the individual aphid. As the species richness of symbionts in an individual aphid increases, the resistance to different selection pressures could increase but so also can the associated costs (Vorburger et al. 2013; Oliver et al. 2014). At the population level, the resulting frequency will be determined by the collective effect on all the aphids. For example, if one particular symbiont combination is by far the most beneficial to host, then it is likely that it will be found in the majority of aphids in a population; in this case, the diversity of symbionts across the population would be low. However, if there is a higher diversity of selection pressures, then many different symbionts could confer resistance benefits, and perhaps one symbiont combination is not the overall optimal population-level strategy. Without a single optimal strategy, different aphids will likely host different symbiont communities.
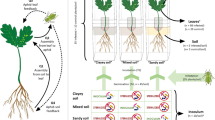
It is not yet known whether the surrounding plant community can mediate changes in the symbiont community of aphids, and we study this in an experimental grassland on plots with varying levels of plant species richness. Given the plausibility that higher plant species richness leads to higher diversity of selection pressures on the aphids and symbionts, we might expect to see an increase in symbiont diversity across the population with increasing plant species richness. In this study, we examined the symbiont community both within individual aphids and at the aphid population level in order to determine if the species richness (SR) of the plant community would influence the symbiont community structure (Fig. 1). We collected three species of aphids from host plants growing in experimental plots that differed in plant species richness. The main hypotheses we tested were:
-
1.
There are non-random associations of different symbiont species in an aphid.
-
2.
Plant species richness influences:
-
(a)
Symbiont species richness at the aphid level.
-
(b)
Symbiont species richness at the population (plot) level.
-
(c)
Symbiont diversity at the population (plot) level.
Fig. 1 Graphical illustration of the study system. We collected three species of aphids each feeding on a unique host plant across experimental plots with varying plant species richness. The aphids were tested for eight common secondary bacterial symbionts and the resulting effect of differing plant species richness on the symbiont community was analysed. The analyses were performed at two different levels, the first at the individual aphid which considered the species richness of symbionts within an aphid, and the second at the population (or plot) level which looked at symbiont species richness and diversity across all aphids of a single species in each experimental plot
-
(a)
Materials and methods
Study site
This study was undertaken within the framework of the Jena Experiment (Germany), a large grassland biodiversity experiment where both the species richness and functional diversity of plant communities are manipulated. We used existing plots within the trait-based diversity experiment (Ebeling et al. 2014b), which contain 20 different species of grasses and non-legume herbs separated into three pools of eight species each, with some species occurring in two pools. The experiment comprises experimental communities differing in species richness (SR) manipulated with four levels (1–4 species) in plots of 3.5 × 3.5 m. Due to the design of the experiment, each host plant species occurs in 13–15 experimental plots of the 46 plots per species pool. Plots are located in three spatial blocks to account for differences in surrounding vegetation.
Study species and collection
We collected aphids from the plots in May and July 2012. In May, over the 14 days preceding collection, 2 days reached temperatures above 25 °C, 7 above 20 °C; there were 8 rainy days with a total of 23.6 mm rain. In July, over the 14 days preceding collection, 5 days reached temperatures above 25 °C, 12 above 20 °C and there were 9 rain days with a total of 39.3 mm rain (data from the Max-Planck-Institute for Biogeochemistry, Jena, Germany; responsibility of Olaf Kolle; http://www.bgc-jena.mpg.de/wetter/). Overall aphid abundance was found to be low across the experimental plots. Therefore, we first identified plant species that were hosting aphids and then carefully screened each plot containing the host plant species in order to count all aphid colonies. In May 2012, only the black bean aphid (Aphis fabae, Scop.) occurred on the field site and was present only on Anthriscus sylvestris (L.) Hoffm. (Apiaceae); this host plant was present in 15 experimental plots (pool 3). In July 2012, we collected Uroleucon jaceae (L.) from the host plant Centaurea jacea (L.), (Asteraceae) and the rose aphid (Macrosiphum rosae, L.) on Knautia arvensis (L.) (Dipsacaceae); C. jacea and K. arvensis are present in 13 and 14 experimental plots, respectively (both in pool 1). For symbiont analysis, one aphid from each colony was collected and stored in 100 % ethanol at −20 °C until DNA extraction. We only collected wingless aphids with a preference for adults or 4th instar, in order to use only those aphids which have been born on the plants from which they were collected and to reduce the chance of collecting an opportunistic aphid visitor. As covariates for our data, we counted the number of aphid colonies per plot as our measure of aphid population density, which we expected to have a positive relationship with both plant and symbiont species richness. We assumed that aphids in a colony (a discrete group of aphids) were all offspring from the same mother aphid and as such would all belong to the same clone, since during the summer months aphids reproduce asexually. We also estimated the cover of the host plant (%, arcsine-transformed), as this could also be influenced by plant species richness and might affect aphid density. Since the plant carbon and nitrogen content may influence aphid fitness through increased nutrition, and could influence or be influenced by the secondary symbionts, potentially through changing the aphid carbon and nitrogen content, we also collected data on this. For the carbon (C) and nitrogen (N) analysis, two additional aphids were collected from ten separate colonies per plot and pooled for analysis (total of 20 aphids per plot). Plant stem material was collected from where the ten separate aphid colonies had been feeding. The aphids and plant samples were dried and analysed for C and N content (Flash EA 112 Thermo). The aphids and plants collected in this study are not protected under any national or international guidelines, thus no special permissions were required.
Symbiont community identification
DNA was extracted from the aphids using the ‘salting-out’ method (Sunnucks and Hales 1996). Up to ten aphids per plot (i.e. ten colonies) were tested for symbiont presence/absence. When more than ten aphids had been collected, we used the ten largest aphids to maximise DNA yield recovered. We used species-specific PCR primers to detect eight different bacteria symbionts, following the methods in Thao and Baumann (2004), Tsuchida et al. (2010) and Ferrari et al. (2012). The bacterial symbionts we detected in the samples were Hamiltonella defensa, Regiella insecticola, Serratia symbiotica, Rickettsia, Spiroplasma and Arsenophonus; for brevity, these symbionts are referred to as Hamiltonella, Regiella, Serratia, Rickettsia, Spiroplasma and Arsenophonus, respectively. ‘X-type’ (a γ Proteobacteria) and Rickettsiella were also examined for but no positive infections were detected. The presence of the bacteria was visualised using agarose gel electrophoresis, with a successful amplification of bacterial DNA shown by a band on the gel. We repeated the analysis for a smaller number of samples for quality control, and used both positive and negative controls in every PCR run to ensure the absence of a band was informative and not due to a failed amplification. Additional validation of these methods was performed on a subset of samples, which were sequenced to confirm that the species-specific primers were amplifying only from the correct symbiont. Samples were chosen with respect to single and multiple infection detection, and showed that the bands we detected on the agarose gel were from the correct symbiont.
The presence–absence data were used to create a community matrix with each individual aphid in a row and each bacterial species in a column. This data were prepared in three ways to benefit the analysis: (1) the species richness (number of species in a plot) of symbiotic bacteria was calculated for each individual aphid, giving one data point for every aphid analysed; (2) the species richness of symbiotic bacteria was calculated at the plot level, giving one data point per plot analysed; and (3) the Shannon–Weaver diversity of the symbiont community at the plot level was calculated, giving one diversity value for each plot. Species richness and Shannon diversity were calculated in the vegan package in R (Oksanen et al. 2015). The plant and aphid C:N samples were pooled to produce one data point per plot; all other explanatory variables were also calculated per plot (host plant cover and aphid density).
Data analysis
The symbiont communities differed among aphid species, and therefore we analysed them separately. To test the association between different symbiont species, we first used a Chi-square test on the presence and absence of the different species across all aphids within each species, using the Yate’s correction for small numbers of observations. This was calculated in the Community Analysis Package 4 (Pisces Conservation). We then calculated the expected number of aphids that would host each different symbiont combination, based on the frequency of the symbionts (all plots combined; Table S1). There was no spatial structuring of symbiont presence (i.e. each symbiont was found in every block), and so in theory every symbiont could be found in every plot. A G-test {likelihood ratio test for goodness-of-fit; G = 2∑ [Observed × ln (Observed/Expected)]} was used to test the expected and observed number of aphids hosting the different combinations. Fisher’s exact tests were used to test for the influence of plant diversity (low or high) on the association between specific symbionts.
At the aphid level, we conducted linear mixed effect models using plot as a random factor to control for non-independence of sampling several aphids within each plot. Host plant cover was arc-sin-transformed. Models with normal errors were used since these gave better model fit (diagnostic plots showed these better fitted the underlying assumptions) than using an alternative error distribution (e.g. Poisson for count data with many zeros). Each model was first run with all variables (block, aphid density, plant cover, plant C:N, aphid C:N and plant SR). We conducted model simplification using the R package lme4 (Bates et al. 2014) to assess the BIC (Bayesian information criterion) using the function anova to compare different models and the AIC (Akaike information criterion) through using the function drop1. Once the best model was found (lowest BIC and AIC), we used the nlme package in R (Pinheiro et al. 2015) for significant testing. At the plot level, we used linear models, with the same predictor variables as above, because each data point was from a single plot. A backwards stepwise removal of non-significant terms was again used to obtain the minimum adequate models. Interactions were tested each in turn but none were retained. Statistical models were run in R v.3.2.0 (R Core Development Team 2015) using R-studio v.0.98.977 (http://www.rstudio.org/).
Confounding variables: plant diversity effects on host plant cover, aphid density and nutrient content
Plant diversity differences can drive many changes in an experimental plot, such as effects on host plant cover which will likely, in turn, affect aphid density. Before conducting our main analyses, we considered the effect of plant SR on host plant cover, aphid density and plant and aphid nutrient contents. Across all plots, we found little effect of plant SR on these covariates. The only significant associations (P < 0.05) were (1) plant SR influenced the cover of Knautia arvensis, where host plant cover decreased with increasing plant SR (F 1,10 = 5.87, P = 0.036); (2) Macrosiphum rosae aphid C:N was lower when aphid density was higher (F 1,10 = 5.17, P = 0.046); and (3) there was a positive relationship between plant and aphid C:N for the Aphis fabae aphid and Anthriscus sylvestris plant combination (F 1,10 = 5.42, P = 0.040). Thus, due to the low number of correlations between potentially confounding effects, we show that any influence of plant SR on the aphid symbiont community is not driven by a confounding effect of these covariates and we can safely include them in the statistical models.
Results
Aphid and symbiont occurrence
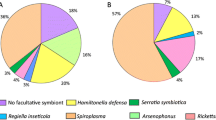
In May 2012, we sampled 118 Aphis fabae aphids from the host plant Anthriscus sylvestris in all 15 plots where A. sylvestris occurred. Aphis fabae was found to host six species of secondary symbionts (average 2 symbionts per aphid) with various infection rates; Arsenophonus (57.6 %), Rickettsia (44.9 %), Regiella (35.6 %), Spiroplasma (24.6 %), Hamiltonella (14.4 %) and Serratia (11.0 %). We found only five aphids (4.2 %) that hosted none of the tested symbionts, and 29.7 % hosting only one, predominantly Arsenophonus. Co-infections occurred with 42.4 % of aphids hosting two species, 21.2 % hosting three and 2.5 % hosting four symbionts (Hamiltonella, Serratia, Rickettsia and Spiroplasma). Based on the population frequency of the symbionts (Table S1), some symbionts and combinations of symbionts were found more often than expected (Table S2). For example, more than the expected number of aphids co-hosted Hamiltonella and Serratia, both as double- and triple-infections alongside Rickettsia or Spiroplasma, despite these two symbionts having the lowest frequencies in the population. A Chi-square association test at the community level, also indicated a positive significant association between Hamiltonella and Serratia (X 2 = 61.95, P < 0.05).
Two additional aphid species occurred in the experimental plots in July 2012. We sampled 104 Uroleucon jaceae aphids from the host plant Centaurea jacea in all 13 plots with this host plant. These aphids hosted only two species of secondary symbiont (average per aphid 1.34): Serratia and Rickettsia, with 97.1 and 36.5 % infection rate, respectively. The majority of aphids (60.6 %) hosted one symbiont (Serratia), with 36.5 % co-hosting both the symbionts and three aphids (2.9 %) which did not host any symbiont. Here, all aphids hosting Rickettsia also hosted Serratia and, thus, no more aphids than expected were found to host these due to the high frequency of Serratia in this species (Tables S1, S3). Finally, we also sampled 97 Macrosiphum rosae aphids from the host plant Knautia arvensis in all 14 plots with this host plant. This aphid species hosted three species of secondary symbiont (average per aphid 1.89): Hamiltonella, Regiella and Serratia with infection rates of 8.3, 82.5, and 98.9 %, respectively. All aphids were found to host at least one species of symbiont, with 14.4 % hosting one (mostly Serratia), 81.5 % hosting two (mostly Regiella and Serratia) and 4.1 % hosting all three species. Similarly to A. fabae, we found more than the expected number of aphids hosting Hamiltonella and Serratia together (Table S4), despite the low frequency of Hamiltonella in this aphid species (Table S1).
Diversity effects on symbiont richness and diversity
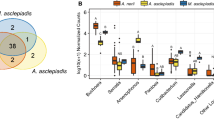
At the level of individual aphids within plots, the bacterial symbiont species richness was found to decrease with increasing plant SR for two of the three aphid species studied (Table 1; Fig. 2). On the contrary, at the plot level, the symbiont species richness and symbiont Shannon diversity was found to increase with increasing plant SR (Table 1; Fig. 2). The plots with four plant species hosted aphids with a greater number of different combinations of symbionts than did the other plots (Fig. S1). Aphis fabae aphids collected from the plot with a single plant species hosted only five different symbiont combinations, while from the two, three and four plant species richness plots, the aphids hosted 13, 10, and 17 combinations, respectively. For M. rosae, all aphids hosted the Regiella–Serratia symbiont combination in the plot where plant species richness equalled one, whereas those in plots with four plant species hosted three different combinations of symbionts.
Symbiont community changes across a plant species richness gradient. The effect of plant species richness on the species richness of secondary bacterial symbionts in aphids (at the aphid level and at the plot level) and on symbiont diversity (Shannon, at the plot level), across three combinations of aphid and host plant species. Both axes show ‘jittered’ data points in order to show the full extent of the data. We used linear mixed effects models at the individual aphid level, to control for plot (as a random effect) and we used linear models at the plot level. Minimum adequate model results are presented in Table 1
The positive association found between the Hamiltonella and Serratia symbionts in A. fabae was also influenced by plant species richness. We found that, in the low species richness plots (with one or two plant species), 17.8 % (8/45) of aphids co-hosted these two symbionts, whereas at the higher species richness plots (with three or four plant species), only 5.3 % (4/71) of aphids co-hosted them (Fishers exact test P = 0.031). Therefore, the association was driven in part by the aphid–symbiont communities within the lower plant species richness plots. In M. rosae aphids, all aphids with Hamiltonella also hosted Serratia, so there was no effect of plant species richness (Fishers exact test P = 0.136). By comparing the frequency of symbiont presence across the plant species richness gradient, we find that Regiella tends to increase in A. fabae but decrease in M. rosae with increasing plant species richness (Fig. S2). Similarly, Rickettsia shows a decrease in A. fabae and no effect in U. jaceae with increasing plant species richness, suggesting different mechanisms acting on this symbiont across the species despite the general patterns being the same (see also Fig. S3, S4).
At the individual aphid level, there was little effect of covariates on symbiont SR (Table 1): within U. jaceae aphids, an increase in plant C:N resulted in a decrease in the SR of symbionts (F 1,9 = 6.79, P = 0.029) and this result could not be explained by changes in plant C:N across plant SR levels (F 1,9 = 3.73, P = 0.090; Table 2). At the plot level, more variables significantly influenced the symbiont SR and Shannon diversity (Table 1). In particular, the cover of the host plant influenced the symbiont SR and diversity, yet in contradictory ways for the different aphid species. For A. fabae and U. jaceae, there was a decrease in symbiont SR and diversity with increasing host plant cover, but for M. rosae there was the opposite result, where there was an increase in symbiont SR and diversity with increasing host plant cover (Table 1). Increasing aphid C:N of A. fabae was found to decrease the symbiont SR and Shannon diversity at the plot level, but not at the individual aphid level (Table 1); this corresponds to the weak negative relationship between aphid C:N and plant SR (F 1,11 = 4.76, P = 0.052; Table 2). Lastly, an increase in aphid density was found to be weakly associated with an increase in symbiont species richness and Shannon diversity, but only for A. fabae (Table 1).
Discussion
Our work is the first to study symbiont communities of aphids across different plant species communities. Our results suggest that the community structure of aphid secondary bacterial symbionts is affected by the species richness of the plant community in which the host plant of the aphid resides. The effect of the plant community was apparent even though each aphid species occurred on its own unique host plant along the diversity gradient, indicating there is variation for some trait, or traits, underlying this effect which we have not studied here. We found higher symbiont species richness at the plot level than the individual aphid level, with non-random associations between certain symbiont species. The average number of symbionts hosted by an aphid was 1.34–2.00 depending on the species, which is consistent with previous studies that found pea aphids to host on average 1–2 symbionts (Ferrari et al. 2012; Russell et al. 2013). Even though correlation does not necessarily imply causation, our results suggest that plant biodiversity, in particular plant species richness, causes directed changes in the symbiont communities.
In all three aphid species studied, the symbiont species richness and diversity at the level of the plot increased with increasing plant species richness. The average increase in symbionts across the plant species richness gradient (1–4 plant species) was around 0.5–0.8 symbiont species at the plot level. The apparently small increase in the number of symbionts is biologically significant, due to the strong effects that even a single symbiont can have on the aphid host, for example on aphid survival. Interestingly, at the level of the individual aphid, we found the opposite effect where a decrease in symbiont species richness was found with increasing plant species richness. There are a number of potential explanations for the effect of plant species richness on the symbiont community. While our results are only suggestive as to which hypothesis is most likely to be supported, i.e. they cannot clearly rule out one or the other hypotheses as our study was not designed to test a particular underlying hypothesis, they clearly show that it is worthwhile to study aphid–symbiont interactions in a wider ecological context. Future studies are needed to put the individual hypotheses we discuss to particular tests.
The first important observation is that there were differences in the plant diversity effects on the within- and between-host symbiont community. This shows that there are potentially different mechanisms driving the presence and abundance of symbionts at these two levels. One scenario which explains these patterns is a potential trade-off between the number and strength of selection pressures, with more diverse, but individually weaker, selection pressures within the plots with higher plant species richness. An earlier paper on the same field site showed that, at high plant species richness parasitoid diversity is increased but parasitism rate is decreased compared to plots with lower plant species richness (Petermann et al. 2010b). Further, Smith et al. (2015) found that an increase in the number of pea aphids hosting multiple symbionts (Hamiltonella, Serratia, and Regiella), linked to increased parasitoid-induced mortality (i.e. stronger selection pressure), was later followed by a dramatic drop in symbiont frequency when the parasitism rate reduced (i.e. reduced selection pressure). Thus, temporal variation in selection pressures should also be considered in future work.
At the individual aphid level, we found higher symbiont species richness at low plant species richness, contrary to the pattern at the plot level. This might be surprising, since one may expect that higher symbiont species richness would be found in the plots with higher plant species richness, as aphids with multiple symbionts would be favoured if selection pressures are diverse. If there was little cost to hosting a symbiont, it would be expected to be maintained in a population. Thus, we suggest that costs must also mediate the interactions to some extent and could explain the reduction in the numbers of aphids hosting multiple symbionts at higher plant species richness (Clay 2014). The cost of hosting symbionts is expected to increase with multiple infections (Oliver et al. 2006), and although this can vary with aphid genotype and symbiont combination (Lukasik et al. 2013) it is not expected that an aphid could host a wide variety of symbionts unless the benefit of multiple hosting outweighs any increased costs, but this is still to be fully explored. Under this scenario in our study, if, in the higher plant species richness plots selection pressures were indeed more diverse, then the impact of each selection pressure on each different symbiont must not have been sufficient to maintain it in our populations. We would then see only those symbionts retained that either infer little cost, or are sufficiently beneficial, to the host. To test if this is true would require exposing aphids with different combinations of symbionts into plots of different diversities and measuring aphid survival (and fecundity) in the presence and absence of challenges such as parasitoids. In the low plant species richness plots, potentially with a lower diversity of selection pressures, then fewer combinations of symbionts present the optimal strategy. This leads to many aphids hosting the same symbiont communities (i.e. lower symbiont diversity across the plot). If hosting multiple symbionts presents the optimal strategy, then more aphids will host multiple symbionts (i.e. higher symbiont species richness within individual aphids). For example, we found an association between Hamiltonella and Serratia in A. fabae, which were more often found together in aphids from lower plant species richness plots. In laboratory studies, this combination has been shown to increase aphid protective defence (Oliver et al. 2006): the protection offered by Hamiltonella against parasitoid wasps is less effective at higher temperatures but Serratia can protect against heat shock (above 25 °C, which was reached in all our sampling periods) (Oliver et al. 2003, 2006; Bensadia et al. 2006; Russell and Moran 2006).
Importantly, it may not just simply be the taxonomic diversity of symbionts that is relevant to the aphid hosts but their functional diversity. The association between Hamiltonella and Serratia in A. fabae might indicate functional complementarity within a host, with Hamiltonella conferring parasitoid resistance and Serratia enabling the protection to continue under high temperatures. Thus, the relative benefit of this combination is higher than co-hosting two symbionts that provide the same service to the host (redundancy). While a number of studies have quantified the effect of particular combinations of symbionts on aphid–natural enemy interactions (e.g. Nyabuga et al. 2010), generally little is known about which symbionts are functionally complementary and in what respects. Further species-specific experiments on the aphid and symbiont species tested here need to be conducted, both in the laboratory and in the field.
At low plant species richness, aphids hosted relatively similar symbiont combinations (Fig. S1), resulting in an overall low symbiont species richness at the overall plot level. Conversely, at higher plant species richness, the aphids host more combinations of symbionts (Fig. S1), leading to an increased symbiont species richness and diversity at the plot level. Such a pattern is again consistent with the hypothesis that there are changes in selection pressures across the plant species richness gradient with more diverse challenges at the more diverse plots. For A. fabae, at plant species richness of one, the aphids host only five different combinations, whereas at plant species richness of four, they host 17 different combinations. One potential caveat is that the pattern may be affected by sampling effort because more aphids on more plots were able to be examined for the higher plant species richness. However, the statistical methods used should, to some extent, account for this variation, and we limited our number of aphids sampled to each plot to ten so as not to overestimate any diversity effect of the higher plant species richness plots where we expected to collect more aphids.
Plant species richness may affect aphid–symbiont interactions in different ways. The scenarios presented above discuss patterns of symbiont distribution driven by plant diversity effects on selection pressures influencing aphids, such as pathogens or parasitoids. It is worth noting that such an effect on aphid natural enemies may be caused by increasing plant species richness in many different ways, for example by an increased diversity of flowers that aphid parasitoids may use for feeding (Ebeling et al. 2008), by providing more hosts for entomopathogens (Scherber et al. 2010) or by affecting microclimatic conditions. In addition, there may also be direct plant effects on particular aphid–symbiont combinations. We tested for the effect of plant chemistry (carbon and nitrogen) and found little effect. In addition, we also found inconsistent effects of aphid density, plant cover and aphid C:N on the symbiont community of the three focal aphid species. These results show that the forces driving the effects we detected cannot be captured by these simple traits. In the Jena Experiment, it has been shown that plant species richness affects a multitude of other variables (Allan et al. 2013) that need to be systematically explored to unravel the mechanisms underlying aphid symbiont interactions in differently diverse plots.
It may also be worth speculating about how the patterns of aphid–symbiont combinations in different plots will manifest. Variation in symbiont presence among an aphid population will be driven, in part, by colonisation–extinction scenarios. In grasslands, host plants will be colonised by a number of different founding aphid mothers that host varying symbiont communities, and often will come from neighbouring plots or from outside the experimental field. Upon settling on a host plant, there will be selection for particular symbiont combinations that confer a higher fitness (Oliver et al. 2008). Dispersal morphs are present only at certain times in the season, as has been shown for many aphid species, for example when moving from a winter to a summer host or due to a reduction in host plant quality (Müller et al. 2001). During the summer months (May–September), the generation time of an aphid can be as little as 7 days, due to the telescopic reproductive strategy of the summer asexual aphids. Thus, while the founding mother symbiont status will certainly influence the symbiont community structure, the following unwinged generations will be more influenced by selective dynamics within each plot rather than by new migrants. In our study, we collected only unwinged aphids that had been produced and developed on the host plant, i.e. we collected aphids after at least one but probably several generations of clonal selection had taken place. In addition to clonal selection, transmission rates of aphid symbionts from mother to offspring (or paternal inheritance if in sexual generations; Moran and Dunbar 2006) are not always 100 % efficient, even in laboratory populations where external factors are controlled (Chen and Purcell 1997; Dykstra et al. 2014), and so may be lost across multiple generations. Although horizontal transmission is not considered to be a frequent occurrence in aphids, the parthenogenetic reproduction of aphids coupled with a high-fitness advantage would nevertheless quickly increase the frequency of a new highly advantageous symbiont in a population. In whitefly, Bemesia tabaci, horizontal transmission of Arsenophonus is thought to be relatively common, as shown through a study looking at phylogenetic incongruence between these symbionts and the different B. tabaci cryptic species (Ahmed et al. 2013).
Our study also suggests that it may be worth studying the effects of the plant community on insect–microorganism interactions more widely, beyond aphids, and also consider wider implications of such interactions for the ecosystem as a whole. For example, in an experimental system, Hamiltonella infection status of aphids explained some variation in the relative allocation of plant biomass to the roots (Hackett et al. 2013), and in whitefly, this symbiont was found to suppress plant defences (Su et al. 2015). Further, symbionts could mediate dietary breadth of an aphid (Wagner et al. 2015), which could, in theory, enhance aphid invasiveness or host-switching abilities (Feldhaar 2011). Despite limited current research on the community level effects of bacterial secondary symbionts in insects, such studies show promising evidence, from laboratory, field and modelling work, to indicate that these symbionts play a much stronger and wider role than previously acknowledged (Oliver et al. 2014). Future work should follow up this line of research using manipulative approaches with the aphid–symbiont community. For example, one could create an experimental system with diverse plant species (or even plant genotypes) and, using a variety of aphids with different symbiont communities, test the outcome of different cost–benefit trade-offs, with varying selection pressures such as natural enemies or nutrient limitation.
In conclusion, we present evidence from a field study that shows an association between plant species richness and the secondary symbiont bacterial community in aphids, indicating that plant diversity effects can cascade across multiple trophic levels. The patterns we found were consistent across three different aphid–plant–symbiont communities, showing a general pattern not restricted to specific species combinations. It is possible that there are similar effects of plant diversity on other as yet unstudied sub-webs of the entire organismic community. Functional biodiversity research has so far described a large number of effects of plant species richness on a wide range of processes at the ecosystem level (Allan et al. 2013). Our work extends documented effects of plant diversity beyond visible biotic interactions to changes in endosymbiont communities, with potentially far-reaching consequences for related ecosystem processes.
References
Abbas M et al (2014) Plant diversity effects on pollinating and herbivorous insects can be linked to plant stoichiometry. Basic Appl Ecol 15:169–178
Ahmed MZ, De Barro PJ, Ren S-X, Greeff JM, Qiu B-L (2013) Evidence for horizontal transmission of secondary endosymbionts in the Bemisia tabaci cryptic species complex. PLoS ONE 8:e53084
Allan E et al (2013) A comparison of the strength of biodiversity effects across multiple functions. Oecologia 173:223–237. doi:10.1007/s00442-012-2589-0
Bates D, Maechler M, Bolker B, Walker S (2014) lme4: Linear mixed-effects models using Eigen and S4. R package version 1.1-7. http://CRAN.R-project.org/package=lme4
Bensadia F, Boudreault S, Guay JF, Michaud D, Cloutier C (2006) Aphid clonal resistance to a parasitoid fails under heat stress. J Insect Physiol 52:146–157
Borer ET, Seabloom EW, Tilman D (2012) Plant diversity controls arthropod biomass and temporal stability. Ecol Lett 15:1457–1464. doi:10.1111/ele.12006
Brady CM, White JA (2013) Cowpea aphid (Aphis craccivora) associated with different host plants has different facultative endosymbionts. Ecol Entomol 38:433–437. doi:10.1111/een.12020
Bukovinszky T, van Veen F, Jongema Y, Dicke M (2008) Direct and indirect effects of resource quality on food web structure. Science 319:804
Caspi-Fluger A et al (2012) Horizontal transmission of the insect symbiont Rickettsia is plant-mediated. Proc R Soc Lond B 279:1791–1796
Chen DQ, Purcell AH (1997) Occurrence and transmission of facultative endosymbionts in aphids. Curr Microbiol 34:220–225
Chen DQ, Montllor CB, Purcell AH (2000) Fitness effects of two facultative endosymbiotic bacteria on the pea aphid, Acyrthosiphon pisum, and the blue alfalfa aphid A. kondoi. Entomol Exper Appl 95:315–323. doi:10.1046/j.1570-7458.2000.00670.x
Clay K (2014) Defensive symbiosis: a microbial perspective. Funct Ecol 28:293–298. doi:10.1111/1365-2435.12258
Darby A, Douglas A (2003) Elucidation of the transmission patterns of an insect-borne bacterium. Appl Environ Microbiol 69:4403–4407
Duffy JE, Cardinale BJ, France KE, McIntyre PB, Thebault E, Loreau M (2007) The functional role of biodiversity in ecosystems: incorporating trophic complexity. Ecol Lett 10:522–538
Dykstra HR et al (2014) Factors limiting the spread of the protective symbiont Hamiltonella defensa in Aphis craccivora aphids. Appl Environ Microbiol 80:5818–5827
Ebeling A, Klein AM, Schumacher J, Weisser WW, Tscharntke T (2008) How does plant richness affect pollinator richness and temporal stability of flower visits? Oikos 117:1808–1815. doi:10.1111/j.1600-0706.2008.16819.x
Ebeling A, Klein AM, Weisser WW, Tscharntke T (2012) Multitrophic effects of experimental changes in plant diversity on cavity-nesting bees, wasps, and their parasitoids. Oecologia 169:453–465. doi:10.1007/s00442-011-2205-8
Ebeling A et al (2014a) Plant diversity impacts decomposition and herbivory via changes in aboveground arthropods. PLoS ONE 9:e106529. doi:10.1371/journal.pone.0106529
Ebeling A et al (2014b) A trait-based experimental approach to understand the mechanisms underlying biodiversity–ecosystem functioning relationships. Basic Appl Ecol 15:229–240
Feldhaar H (2011) Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol Entomol 36:533–543
Ferrari J, Vavre F (2011) Bacterial symbionts in insects or the story of communities affecting communities. Philos Trans R Soc Lond B 366:1389–1400
Ferrari J, West JA, Via S, Godfray HCJ (2012) Population genetic structure and secondary symbionts in host-associated populations of the pea aphid complex. Evolution 66:375–390. doi:10.1111/j.1558-5646.2011.01436.x
Frantz A, Calcagno V, Mieutzet L, Plantgenest M, Simon JC (2009) Complex trait differentiation between host-populations of the pea aphid Acyrthosiphon pisum (Harris): implications for the evolution of ecological specialisation. Biol J Linn Soc 97:718–727
Gehrer L, Vorburger C (2012) Parasitoids as vectors of facultative bacterial endosymbionts in aphids. Biol Lett 8:613–615. doi:10.1098/rsbl.2012.0144
Hackett SC, Karley AJ, Bennett AE (2013) Unpredicted impacts of insect endosymbionts on interactions between soil organisms, plants and aphids. Proc R Soc Lond B 280:2013. doi:10.1098/rspb.2013.1275
Haddad NM, Crutsinger GM, Gross K, Haarstad J, Knops JMH, Tilman D (2009) Plant species loss decreases arthropod diversity and shifts trophic structure. Ecol Lett 12:1029–1039
Haynes S et al (2003) Diversity of bacteria associated with natural aphid populations. Appl Environ Microbiol 69:7216–7223. doi:10.1128/aem.69.12.7216-7223.2003
Hector A, Joshi J, Lawler S, Spehn E, Wilby A (2001) Conservation implications of the link between biodiversity and ecosystem functioning. Oecologia 129:624–628
Henry LM et al (2013) Horizontally transmitted symbionts and host colonization of ecological niches. Curr Biol 23:1713–1717
Hooper D et al (2005) Effects of biodiversity on ecosystem functioning: a consensus of current knowledge. Ecol Monogr 75:3–35
Leonardo TE, Muiru GT (2003) Facultative symbionts are associated with host plant specialization in pea aphid populations. Proc R Soc Lond B 270:S209
Loranger H et al (2014) Invertebrate herbivory increases along an experimental gradient in grassland plant diversity. Oecologia 174:183–193
Lukasik P, Guo H, van Asch M, Ferrari J, Godfray HC (2013) Protection against a fungal pathogen conferred by the aphid facultative endosymbionts Rickettsia and Spiroplasma is expressed in multiple host genotypes and species and is not influenced by co-infection with another symbiont. J Evol Biol 26:2654–2661. doi:10.1111/jeb.12260
McLean A, van Asch M, Ferrari J, Godfray H (2011) Effects of bacterial secondary symbionts on host plant use in pea aphids. Proc R Soc Lond B 278:760
Mitchell CE (2003) Trophic control of grassland production and biomass by pathogens. Ecol Lett 6:147–155. doi:10.1046/j.1461-0248.2003.00408.x
Montllor CB, Maxmen A, Purcell AH (2002) Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol 27:189–195
Moran NA, Dunbar HE (2006) Sexual acquisition of beneficial symbionts in aphids. Proc Natl Acad Sci USA 103:12803–12806
Müller CB, Williams IS, Hardie J (2001) The role of nutrition, crowding and interspecific interactions in the development of winged aphids. Ecol Entomol 26:330–340
Nyabuga FN, Outreman Y, Simon JC, Heckel DG, Weisser WW (2010) Effects of pea aphid secondary endosymbionts on aphid resistance and development of the aphid parasitoid Aphidius ervi: a correlative study. Entomol Exp Appl 136:243–253. doi:10.1111/j.1570-7458.2010.01021.x
Oksanen J, Guillaume Blanchet F, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2015). vegan: Community Ecology Package. R package version 2.3-0. http://CRAN.R-project.org/package=vegan
Oliver KM, Russell JA, Moran NA, Hunter MS (2003) Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci USA 100:1803
Oliver KM, Moran NA, Hunter MS (2006) Costs and benefits of a superinfection of facultative symbionts in aphids. Proc R Soc Lond B 273:1273–1280
Oliver KM, Campos J, Moran NA, Hunter MS (2008) Population dynamics of defensive symbionts in aphids. Proc R Soc Lond B 275:293
Oliver KM, Smith AH, Russell JA (2014) Defensive symbiosis in the real world—advancing ecological studies of heritable, protective bacteria in aphids and beyond. Funct Ecol 28:341–355. doi:10.1111/1365-2435.12133
Omacini M, Chaneton EJ, Ghersa CM, Müller CB (2001) Symbiotic fungal endophytes control insect host–parasite interaction webs. Nature 409:78–81
Pérez-Brocal V et al (2006) A small microbial genome: the end of a long symbiotic relationship? Science 314:312–313
Petermann JS, Muller CB, Roscher C, Weigelt A, Weisser WW, Schmid B (2010a) Plant species loss affects life-history traits of aphids and their parasitoids. PLoS ONE 5:e12053. doi:10.1371/journal.pone.0012053
Petermann JS, Müller CB, Weigelt A, Weisser WW, Schmid B (2010b) Effect of plant species loss on aphid–parasitoid communities. J Anim Ecol 79:709–720. doi:10.1111/j.1365-2656.2010.01674.x
Pettersson J, Tjallingii WF, Hardie J (2007) Host plant selection and feeding. In: VanEmden HF, Harrington R (eds) Aphids as crop pests. CABI, Wallingford, pp 87–113
Pinheiro J, Bates D, DebRoy S, Sarkar D, Team RC (2015) nlme: linear and nonlinear mixed effects models. R package version 3:1–120
Powell G, Tosh CR, Hardie J (2006) Host plant selection by aphids: behavioral, evolutionary, and applied perspectives. Annu Rev Entomol 51:309–330
R Core Development Team (2015) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Roscher C et al (2004) The role of biodiversity for element cycling and trophic interactions: an experimental approach in a grassland community. Basic Appl Ecol 5:107–121
Ruijven J, De Deyn GB, Berendse F (2003) Diversity reduces invasibility in experimental plant communities: the role of plant species. Ecol Lett 6:910–918
Russell JA, Moran NA (2006) Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proc R Soc Lond B 273:603–610
Russell J, Latorre A, Sabater-Muñoz B, Moya A, Moran N (2003) Side-stepping secondary symbionts: widespread horizontal transfer across and beyond the Aphidoidea. Mol Ecol 12:1061–1075
Russell JA et al (2013) Uncovering symbiont-driven genetic diversity across North American pea aphids. Mol Ecol 22:2045–2059. doi:10.1111/mec.12211
Rzanny M, Voigt W (2012) Complexity of multitrophic interactions in a grassland ecosystem depends on plant species diversity. J Anim Ecol 81:614–627. doi:10.1111/j.1365-2656.2012.01951.x
Rzanny M, Kuu A, Voigt W (2013) Bottom–up and top–down forces structuring consumer communities in an experimental grassland. Oikos 122:967–976. doi:10.1111/j.1600-0706.2012.00114.x
Sandström JP, Russell JA, White JP, Moran NA (2001) Independent origins and horizontal transfer of bacterial symbionts of aphids. Mol Ecol 10:217–228
Scarborough CL, Ferrari J, Godfray H (2005) Aphid protected from pathogen by endosymbiont. Science 310:1781
Scherber C et al (2006) Effects of plant diversity on invertebrate herbivory in experimental grassland. Oecologia 147:489–500
Scherber C et al (2010) Bottom-up effects of plant diversity on multitrophic interactions in a biodiversity experiment. Nature 468:553–556. doi:10.1038/nature09492
Smith AH et al (2015) Patterns, causes and consequences of defensive microbiome dynamics across multiple scales. Mol Ecol 24:1135–1149. doi:10.1111/mec.13095
Su Q, Oliver KM, Xie W, Wu Q, Wang S, Zhang Y (2015) The whitefly-associated facultative symbiont Hamiltonella defensa suppresses induced plant defenses in tomato. Funct Ecol. doi:10.1111/1365-2435.12405
Sunnucks P, Hales DF (1996) Numerous transposed sequences of mitochondrial cytochrome oxidase I-II in aphids of the genus Sitobion (Hemiptera: Aphididae). Mol Biol Evol 13:510–524
Thao ML, Baumann P (2004) Evidence for multiple acquisition of Arsenophonus by whitefly species (Sternorrhyncha: Aleyrodidae). Curr Microbiol 48:140–144
Tilman D, Wedin D, Knops J (1996) Productivity and sustainability influenced by biodiversity in grassland ecosystems. Nature 379:718–720
Tsuchida T, Koga R, Shibao H, Matsumoto T, Fukatsu T (2002) Diversity and geographic distribution of secondary endosymbiotic bacteria in natural populations of the pea aphid, Acyrthosiphon pisum. Mol Ecol 11:2123–2135. doi:10.1046/j.1365-294X.2002.01606.x
Tsuchida T, Koga R, Fukatsu T (2004) Host plant specialization governed by facultative symbiont. Science 303:1989
Tsuchida T et al (2010) Symbiotic bacterium modifies aphid body color. Science 330:1102–1104
Vorburger C, Gouskov A (2011) Only helpful when required: a longevity cost of harbouring defensive symbionts. J Evol Biol 24:1611–1617. doi:10.1111/j.1420-9101.2011.02292.x
Vorburger C, Ganesanandamoorthy P, Kwiatkowski M (2013) Comparing constitutive and induced costs of symbiont-conferred resistance to parasitoids in aphids. Ecol Evol 3:706–713. doi:10.1002/ece3.491
Wagner SM et al (2015) Facultative endosymbionts mediate dietary breadth in a polyphagous herbivore. Funct Ecol. doi:10.1111/1365-2435.12459
Wulff JA, White JA (2015) The endosymbiont Arsenophonus provides a general benefit to soybean aphid (Hemiptera: Aphididae) regardless of host plant resistance (Rag). Environ Entomol 44:574–581
Wulff JA, Buckman KA, Wu K, Heimpel GE, White JA (2013) The endosymbiont Arsenophonus is widespread in soybean aphid, Aphis glycines, but does not provide protection from parasitoids or a fungal pathogen. PLoS ONE 8:e62145. doi:10.1371/journal.pone.0062145
Zytynska SE, Preziosi RF (2011) Genetic interactions influence host preference and performance in a plant-insect system. Evol Ecol 25:1321–1333
Zytynska SE, Weisser W (2015) The natural occurrence of secondary bacterial symbionts in aphids. Ecol Entomol (in press)
Zytynska SE, Franz L, Hurst B, Johnson A, Preziosi RF, Rowntree J (2014) Host plant genotypic diversity and community genetic interactions mediate aphid spatial distribution. Ecol Evol 4:121–131
Acknowledgments
This work was funded by the DFG through the Jena Experiment (FOR 1451, WE 3081). We thank Anne Ebeling and the Jena Experiment gardeners for managing and maintaining the experimental plots.
Author contribution statement
SEZ, WWW and STM designed the experiment, SEZ, MM and WU conducted fieldwork, SS performed the molecular work, WU performed the chemical analyses. SEZ analysed the data and wrote the first draft, with STM and WWW contributing substantially to revisions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Merijn Kant.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zytynska, S.E., Meyer, S.T., Sturm, S. et al. Secondary bacterial symbiont community in aphids responds to plant diversity. Oecologia 180, 735–747 (2016). https://doi.org/10.1007/s00442-015-3488-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00442-015-3488-y