Abstract
Osteoarthritis (OA) is the most common joint disease with an unsatisfactory therapy outcome and characterized by the degradation of articular cartilage and synovial inflammation. Here, we isolated bone marrow mesenchymal stem cells (BMSCs) from rat’s bone marrow and BMSC-derived exosome (BMSCs-Exo) from BMSCs successfully. MiR-135b was proved to be highly expressed in TGF-β1-stimulated BMSC-derived exosomes (BMSCs-ExoTGF-β1). Then, our results demonstrated that BMSCs-ExoTGF-β1 reduced OA-induced upregulation of pro-inflammatory factors in rat’s serum and damage in cartilage tissues, which was then reversed by miR-135b decreasing. Subsequently, we found that the OA-resulted M1 polarization of synovial macrophages (SMs) was repressed by BMSCs-ExoTGF-β1, this effect of BMSCs-ExoTGF-β1 was limited by miR-135b decreasing. We also proved that M2 polarization of SMs can be induced by miR-135b mimics. Furthermore, we found that the promotory effect of miR-135b and BMSCs-ExoTGF-β1 on M2 SMs polarization was reversed by increasing of MAPK6. Overall, our data showed that BMSCs-ExoTGF-β1 attenuated cartilage damage in OA rats through carrying highly expressed miR-135b. Mechanistically, miR-135b promoted M2 polarization of SMs through targeting MAPK6, thus improving cartilage damage. Our study provided a novel regulatory mechanism of BMSCs-Exo in OA development and revealed a new potential treatment target of OA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteoarthritis (OA) is a most common chronic and painful disorder in joints and characterized by the damage of cartilage, formation of osteophyte, infiltration of inflammatory cells, and remodeling in subchondral bone (Mianehsaz et al. 2019; Miller and Scanzello 2019). Currently, the prevalence of OA is approximately 15% globally and may be up to 35% in 2030 years, thus becomes a major cause of disability. However, the purpose of OA treatment in clinical is to attenuate the pain in patients as much as possible instead of repairing the damaged cartilage tissues, which mainly due to the lack of effective drugs (Kim and Yoo 2018; Oo et al. 2018). Therefore, it is necessary to explore the molecular mechanism of OA occurrence and development, thus providing a new therapy target of OA.
Although inflammation is a cause of OA or the result of OA remains unclear, the crucial role of it in OA development is ensured (Robinson et al. 2016). A large quantity of inflammatory cells infiltrate to synovial tissues in OA patients and animal model. Among these inflammatory cells, synovial macrophages (SMs) are an important component (Xie et al. 2019). It was confirmed that macrophage could differentiate into two types: classically activated M1 macrophage and alternatively activated M2 macrophage. These two types of macrophages have opposite function in the development of inflammation. M1 macrophage acted as pro-inflammatory cells, and M2 macrophage served as anti-inflammatory cells (Ma et al. 2017; Murray et al. 2014). Recently, Zhang et al. pointed out that M1 rather than M2 macrophages accumulate in synovial tissues of human and OA mouse and M1-polarized SMs facilitate the development of experimental OA. In addition, they also proved that M2-polarized SMs could prevent the development of OA (Zhang et al. 2018). Hence, exploring the regulatory mechanism of M1-polarized to M2-polarized SM conversion may offer a potential method to OA treatment.
Mesenchymal stem cells (MSCs), a type of multi-potent cells, were widely used to treat multiple disorders because of their capacity of self-renewal and the potential to differentiate into multiple lineages (Geng et al. 2020; Goradel et al. 2018; Mirzaei et al. 2018a, 2018b; Moradian Tehrani et al. 2018). It was revealed that the activity of chondrocytes and synoviocytes are decreased by SMs from OA patients, and human amniotic-derived MSCs could effectively reduce the damage in cartilage tissue mediated by SMs (Topoluk et al. 2018). Exosomes are the important messenger between different cells; a growing data demonstrated that MSCs could repress the development of disorders via secreting exosomes which carry proteins, miRNAs, long non-coding RNAs, or other small molecules (Fang et al. 2020). In our previous study, we found that transforming growth factor β1 (TGF-β1)-overexpressed MSCs could notably encourage the proliferation of chondrocyte through secreting exosomes which highly expressed miR-135b (Wang et al. 2018b). In addition, a recent study revealed that exosomes promote M1 macrophage to M2 macrophage conversion, thus suppressing the development of disease (Kim et al. 2019).
Mitogen-activated protein kinase 6 (MAPK6) is an important member of MAPK family, and participates in multiple cellular events (Hu et al. 2018). The results of bioinformatics analysis showed that there is a binding site to miR-135b on MAPK3 3′-UTR. Hence, in the present study, we further explored the role and mechanism of action of MSC-derived exosome in the degradation of cartilage tissues in OA and found that TGF-β1-stimulated bone marrow-derived MSC-secreted exosomes could effectively attenuate cartilage damage through promotion of M2 polarization of SMs via carrying highly expressed miR-135b and subsequent repressing MAPK6 expression. Our data confirmed a new regulatory mechanism of MSC-derived exosomes and miR-135b in the development of OA and indicated a potential therapy strategy.
Materials and methods
Animals
Twelve-week-old SD rats (300~330 g weight) were purchased from Shanghai experimental animal research center (Shanghai, China) and were raised in a standard environment with 12 h light/12 h dark cycle, and sufficient food and water. Three- to five-day-old newborn rats were used to prepare bone marrow-derived MSCs (BMSCs). All animal experiments were carried out according to the guidelines of the Care and Use of Laboratory Animals of the National Institutes of Health, and all experimental protocols were approved by the Animal Research Committee of First Affiliated Hospital of Anhui Medical University.
Culture of primary bone-derived MSCs
Primary BMSCs were collected from the bone marrow of newborn rats as previous study (Hou et al. 2018). In brief, rats’ femurs were removed under aseptic conditions, and then were washed using PBS solution for three times. Next, α-minimum essential medium (α-MEM, Gibco, Invitrogen, Carlsbad, CA, USA), which supplemented with 10% fetal bovine serum (FBS, Gibco), 100 U/ml penicillin (Sigma-Aldrich, St. Louis, MO), and 100 µg/ml streptomycin (Sigma-Aldrich), was used to rinse the bone cavity at longitudinal direction for 20 times. Then, the rinse solution was filtered through a 200-mesh sieve followed by culturing in an incubator with 37 °C and 5% CO2. The old medium was changed at 12 h later for incubation to remove the unattached cells. After that, the cell culture medium was updated once every 3 days. Finally, the cellular morphology of BMSCs was verified using a light microscope, and the expression of BMSC surface markers was identified using flow cytometry. BMSCs express high levels of CD29, CD44, and CD90 and low levels of CD11b, CD34, and CD45. According to the necessity of our study, 10 ng/ml TGF-β1 and equal volume of PBS were used to treat BMSCs for 24 h.
Flow cytometry assay
Flow cytometry was carried out to identify the BMSCs and ensure the percentage of CD86- and CD163-positive SMs. Here, 100-μl cell suspension of BMSCs or SMs, which contain about 5.0 × 105 cells, was added into a new EP tube, and then incubated with antibodies (Santa Cruz Biotech, Santa Cruz, CA, USA) including fluorescein isothiocyanate (FITC)-labeled CD11b, CD34, CD45, or CD86, phycoerythrin (PE)-labeled CD29, CD44, CD90, or CD163 in the dark at 4 °C for 30 min. At last, the cells were analyzed using a BD FACSCalibur flow cytometer.
Exosome isolation and identification
Exosome extraction kit (Geneseed, Guangzhou, China) was utilized to collect exosomes from the cell culture supernatant of BMSCs following the manufacturer’s instruction. Transmission electron microscope (TEM) was used to detect the shape of exosome, nanoparticle tracking analysis (NTA) was utilized to analyze the diameter and concentration of exosomes, and Western blotting assay was performed to measure the marker protein expression of exosomes, which were used to identify exosomes.
Western blotting assay
Total RNA were extracted from cells and exosomes using RIPA lysis buffer (Beyotime, Beijing, China), and the concentration of protein was ensured using a Thermo Scientific NanoDrop 2000 Spectrophotometer. Next, equal amount of 20 μg proteins was separated by 12% SDS-PAGE gel and subsequently transferred onto polyvinylidene difluoride membranes (PVDF, Millipore, Bedford, MA). After incubation with 5% non-fat milk for 1 h at room temperature, the PVDF membranes were maintained with primary antibodies against CD9 (1:1000, Santa Cruz Biotech, USA), CD81 (1:1000, Santa Cruz Biotech), MAPK6 (1:2000, Abcam, Cambridge, MA, USA), SOX9 (1:2000, Santa Cruz Biotech), Aggrecan (1:2000, Santa Cruz Biotech), and β-actin (1:2000, Abcam) at 4 °C overnight. After that, the PVDF membranes were incubated with HRP-conjugated secondary antibody (Santa Cruz Biotech) for 1 h at room temperature. At last, the protein bands were visualized using an ECL kit (Santa Cruz Biotech), and then analyzed using ImageJ software.
QRT-PCR assay
Total RNA were isolated from cells and exosomes utilizing TRIzol reagent (Invitrogen) according to the manufacturer’s protocol followed by reverse transcripted into cDNA using the SuperScript First Strand Synthesis System (Invitrogen). After that, the qRT-PCR was carried out using SYBR® Premix Ex Taq™ II (TaKaRa Biotechnology, Dalian, China) on the CFX96 real-time qPCR instrument (Bio-Rad, Hercules, CA, USA) as the introductions. The relative expression of genes was calculated as the method of 2−ΔΔCt. GAPDH was served as the internal reference of mRNA, and the expression of miR-135b was normalized to U6. The primer sequences we used are as follows: miR-135b: forward: 5′-GGTATGGCTTTTCATTCCT-3′ and reverse: 5′-CAGTGCGTGTCGTGGAGT-3′; U6: forward: 5′-CTCGCTTCGGCAGC ACA-3′ and reverse: 5′-AACGCTTCACGAATTTGCGT-3′; Arg-1 mRNA: forward: 5′-TGGACTGGACCCAGTATTCA-3′ and reverse: 5′-CCCAAGAGTTGGGTTCA CTT-3′; iNOS mRNA: forward: 5′-GAGACAGGAAAGTCGGAAGC-3′ and 5′-GT GTTGAAGGCGTAGCTGAA-3′; MAPK6 mRNA: forward: 5′-TAAAGCCATTGA CATGTGGG-3′ and reverse: 5′-TCGTGCACAACAGGGATAGA-3′; GAPDH: forward: 5′-TCTCCCTCACAATTTCC ATCCC-3′ and reverse: 5′-TTTTTGTGGGT GCAGCGAAC-3’.
Cell transfection
Inhibitor negative control (INC), miR-135b inhibitor (miR-135bI), miR-135b mimic, and mimic negative control (mimic NC) were obtained from RiboBio. The full sequences of MAPK6 gene were inserted into the plasmid pcDNA to structure the overexpression system of MAPK6 (pcDNA-MAPK6), and then, pcDNA-MAPK6 was transfected into SMs together with miR-135b mimic or exosomes. MiR135b mimics: sense: 5′-ACAUAGGAAUGAAAAGCCAUATT-3′ and antisense: 5′-UAUGGCUU UUCAUUCCUAUGUGA-3′. In brief, the cells were planted into six-well plates with a density of 1 × 106 cells/well, and then, cell transfection was performed when the cells paved 70~80%. In brief, INC, miR-135bI, miR-135b mimic, mimic NC, and pcDNA-MAPK6 were mixed with Lipofectamine 2000 transfection reagent (Gibco Life Technology, Grand Island, NY) according to the manufacturer’s introduction. Next, these mixtures were incubated with the cells for 6 h at 37 °C. At 24 h later, the following experiments were done.
OA rat model and experimental group
The OA rat model was prepared as our previous study (Wang, Xu and Xu, 2018b). In brief, the medial collateral ligament and the medial meniscus were removed completely. Subsequently, under the premise that the tibial surface of rat not be damaged, we cut the meniscus at the narrowest point, and then broken the anterior cruciate ligament. At last, 0.05 mg/kg buprenorphine and 5 mg/kg gentamicin were injected into rats to attenuate the pain in animals.
Next step, according to the necessity of our study, the SD rats were randomly divided into six groups (n = 6 in each group): sham group, OA group, OA + BMSC-ExoPBS group, OA + BMSC-ExoTGF-β1 group, OA + INC-BMSC-ExoTGF-β1 group, and OA + miR-135bI-BMSC-ExoTGF-β1 group. In OA + BMSC-ExoPBS group, OA + BMSC-ExoTGF-β1 group, OA + INC-BMSC-ExoTGF-β1 group, and OA + miR-135bI-BMSC-ExoTGF-β1 group, the rats were injected with 100 μl exosomes through articular cavity, 1 × 1011 exosome particles/ml. Here, BMSC-Exo: BMSC-derived exosome; BMSC-ExoPBS: PBS-stimulated BMSC-derived exosome; BMSC-ExoTGF-β1: TGF-β1-treated BMSC-derived exosome; INC-BMSC-ExoTGF-β1: inhibitor NC together with TGF-β1-treated BMSC-derived exosome; miR-135bI-BMSC-ExoTGF-β1: miR-135b inhibitor together with TGF-β1-stimulated BMSC-derived exosome.
ELISA assay
The rats were anesthetized using 3% pelltobarbitalum natricum (50 mg/kg) and then abdominal aortic blood. The concentrations of IL-1β, PGE2, COX-2, COX-1, and NO in rat’s serum were determined using ELISA assay. Rat IL-1β, PGE2, COX-2, COX-1, and NO immunoassay kits were obtained from CusaBio (Wuhan, China), and the ELISA assay was carried out as the specific introduction. At last, the concentration of these factors was examined using an HT Microplate Reader (BioTek, Winooski, VT, USA) at the absorbance of 450 nm.
H&E staining and saffron-O-solid green staining
The pathological change in cartilage tissue of rats and the degradation of the tissues were ensured by H&E staining and saffron-O-solid green staining, respectively. Paraffin-embedded cartilage tissues were cut into 4-μm sections continuously, and then, the sections were dewaxed and subsequently hydrated. Then, H&E staining and saffron-O-solid green staining were completed using Hematoxylin-Eosin/HE Staining Kit (Solarbio) and Modified Safranine O-Fast Green FCF Cartilage Stain Kit (Solarbio) according to the manufacturer’s introductions, respectively.
Immunocytochemistry assay
Immunocytochemistry assay was performed to examine the cell number of CD86-positive and CD163-positive macrophages in synovial tissue. Paraffin sections were successively incubated with dewaxed, hydrated, EDTA antigen retrieval solution (Solarbio) for 10 min at 95 °C, 3% H2O2 for 15 min at room temperature, primary antibodies against CD86 (1:1000) and CD163 (1:1000) at 4 °C overnight, and HRP-conjugated secondary antibody for 30 min at 37 °C. Subsequently, the sections were stained with DAB solution (Solarbio) for 5 min, and then, hematoxylin solution was used to mark the nuclear of cells for 1 min. At last, the images of tissues were obtained using a light microscope.
Synovial macrophage preparation
Primary rat SMs were collected from the synovium of normal rats. Synovial tissues were removed and subsequently washed utilizing PBS solution for three times. Next, the tissues were cut into pieces of 2–3 mm2, and then maintained with 0.1% collagenase type II (Sigma-Aldrich) for 30 min at 37 °C temperature. After that, mononuclear cells were maintained with PBS buffer which contains the biotinylated anti-F4/80 antibody at 4 °C for 30 min. Followed by mixing with streptavidin-labeled magnetic particles (BD IMag Streptavidin Particles Plus-DM; BD Biosciences, Tokyo, Japan), the cells were added into an IMag separation system (BD Biosciences) for 30 min at 4 °C followed by 37 °C pre-warmed DMEM that was added into this system, and subsequently removed the tube and added DMEM into this tube to obtain F4/80-positive cells which were then centrifuged at 300g for 10 min. The precipitate was collected and cultured in a six-well plate. When testing, 1 μg/ml LPS (Sigma-Aldrich) was used to activate SM and induced its polarization.
Luciferase activity assay
The 3′-UTR fragments of MAPK6, which contain the wide-type (WT) or mutational (Mut) binding sites with miR-135b, were sub-cloned through PCR, and then, the gene sequences were inserted into a luciferase reporter gene vector PGL3 to form PGL3-MAPK6-WT and PGL3-MAPK6-Mut. Next step, the plasmid of PGL3-MAPK6-WT or PGL3-MAPK6-Mut was transfected into HEK293 cells together with miR-135b mimic or mimic NC using Lipofectamine 2000. At 48 h later, the luciferase activity of cells was measured using the Dual-Glo™ Luciferase Assay system (Promega, USA).
Statistical analysis
In this present study, all data were presented as mean ± standard deviation (SD). The software of SPSS 20.0 (SPSS, Chicago, USA) was utilized to analyze the results of our study. Student’s t test was performed to analyze the difference between two independent groups, and the differences between groups were ensured using one-way ANOVA method. Moreover, the differences were considered as statistically significant when the value of P was less than 0.05.
Results
Identification of BMSCs
On the beginning, primary BMSCs were isolated from the bone marrow of newborn rats. Flow cytometry was carried out to detect the expression of surface markers of BMSCs (CD11b, CD34, CD45, CD29, CD44, and CD90). As shown in supplementary Fig. 1, approximately 94% of cells expressed CD29 (f), CD44 (j), and CD90 (k), which were recognized as the markers of mesenchymal lineage. Approximately 0.5% of cells expressed CD11b (b), CD34 (c), and CD45 (d), which were recognized as the markers of hematopoietic lineage. Meantime, the fibroblast-like shape of BMSCs was observed under a light microscope (supplementary Fig. 1i). Subsequently, ability of BMSC differentiation into osteogenic (supplementary Fig. 1m) and adipogenic (supplementary Fig. 1n) lineages were ensured using alizarin red S staining and Oil red O staining, respectively. We obtained the BMSCs from rat’s bone marrow successfully.
MiR-135b was upregulated in BMSC-ExoTGF-β1. a The shape of exosomes was ensured using TEM. b The average diameter of exosomes was examined using NTA. c The expression of exosome markers, CD9 and CD81, was measured utilizing Western blotting assay. d qRT-PCR was carried out to detect the expression of miR-135b both in BMSC-ExoPBS and BMSC-ExoTGF-β1. **P < 0.01 contrasted to BMSC-ExoPBS group.
MiR-135b was increased in TGF-β1-modified BMSC-derived exosomes
BMSCs were stimulated with PBS and TGF-β1, respectively. At 24 h later, PBS-stimulated BMSC-derived exosome (BMSC-ExoPBS) and TGF-β1-stimulated BMSC-derived exosome (BMSC-ExoTGF-β1) were collected from cell supernatants, and then, the cup- or round-shaped morphology of the exosomes was identified using TEM (Fig. 1a). At the same time, the average diameter of exosomes at 100 nm was ensured by NTA (Fig. 1b). Besides, we also found that the characteristic markers of exosomes, CD9 and CD81, expressed in BMSC-ExoPBS and BMSC-ExoTGF-β1 (Fig. 1c). After that, we examined the expression of miR-135b. QRT-PCR results indicated that miR-135b was increased in BMSC-ExoTGF-β1 (Fig. 1d). We successfully obtained the BMSCs-Exo, and proved the overexpressed miR-135b in BMSC-ExoTGF-β1.
BMSC-ExoTGF-β1 attenuated injury and promoted repair of cartilage tissues in OA rats via secreting miR-145b
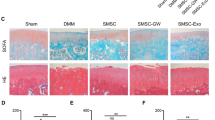
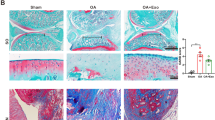
To investigate the effect of BMSC-ExoTGF-β1 on cartilage injury and repair in OA, we established the OA rat model, which was then treated with BMSC-ExoPBS, BMSC-ExoTGF-β1, INC-BMSC-ExoTGF-β1, or miR-135bI-BMSC-ExoTGF-β1. Firstly, our data showed that the expression of miR-135b was significantly downregulated in miR-135bI-BMSC-ExoTGF-β1 group due to miR-135b inhibitor treatment (Fig. 3a). Next, ELISA results demonstrated that the production of inflammatory factors (IL-1β, PGE2, COX-2, COX-1, and NO) in serum of OA rats was upregulated, which were then reduced by BMSC-ExoTGF-β1. However, the inhibition of BMSC-ExoTGF-β1 to these inflammatory factors was partly reversed by miR-135bI-BMSC-ExoTGF-β1 (Fig. 3b–f). After that, we further detect the pathological change in cartilage tissues from each rat, and our results showed that compared with sham group, in OA group, the cartilage tissues displayed lower chondrocyte number and degeneration and rough surface of the tissues. However, OA-induced injury in cartilage tissues can be mildly improved by BMSC-ExoPBS, but greatly improved by BMSC-ExoTGF-β1, while the improvement of BMSC-ExoTGF-β1 to OA-induced cartilage injury was partly reversed by miR-135b knockdown (Fig. 2a–l). Moreover, the results of saffron-O-solid green staining proved that OA caused the loss of cartilage tissues, while a small percentage of cartilage tissue was rescued by BMSC-ExoPBS treatment and a large percentage of cartilage tissue was rescued by BMSC-ExoTGF-β1. But, the inhibitory effect of BMSC-ExoTGF-β1 on OA-induced cartilage damage partly reduced after miR-135b decreasing (Fig. 2m–r). Subsequently, we further detected the expression of osteogenesis-related proteins, SOX9 and aggrecan, and our data displayed that the expression of these two proteins was significantly downregulated in OA group, while this effect of OA was repressed by BMSC-ExoTGF-β1. Meantime, the inhibition of BMSC-ExoTGF-β1 to OA-induced downregulation of SOX9 and aggrecan was partly limited by miR-135b decreasing (Fig. 3 g and h). Taken together, BMSC-ExoTGF-β1 could effectively attenuate OA-induced inflammatory production and cartilage damage through carrying highly expressed miR-135b.
BMSC-ExoTGF-β1 improved cartilage injury via carrying miR-135b. The exosomes from PBS-treated BMSC, TGF-β1-treated BMSCs, inhibitor NC-treated BMSC following TGF-β1, and miR-135b inhibitor-treated BMSC following TGF-β1 were individually injected into OA rats. (a–l) The pathological changes in cartilage tissues were confirmed using H&E staining. (m–r) Saffron-O-solid green staining was performed to measure the cartilage damage.
BMSC-ExoTGF-β1 improved inflammation via carrying miR-135b. (a) The expression level of miR-135b in exosomes from inhibitor NC-treated BMSC and miR-135b inhibitor-treated BMSC was measured by using qRT-PCR. **P < 0.01 contrasted to INC-BMSC-ExoTGF-β1. Then, the exosomes produced by PBS-treated BMSC, TGF-β1-treated BMSCs, inhibitor NC-treated BMSC following TGF-β1, and miR-135b inhibitor-treated BMSC following TGF-β1 were individually injected into OA rats. (b–f) The concentration of inflammatory factors (IL-1β, PGE2, COX-2, COX-1, and NO) in serum was ensured by ELISA assay. (g, h) The expression of SOX and aggrecan in protein level were detected using Western blotting assay and analyzed. **P < 0.01 contrasted to sham group, #P < 0.05 and ##P < 0.01 compared with OA group, and &P < 0.05 and &&P < 0.01 compared with BMSC-ExoTGF-β1 group
BMSC-ExoTGF-β1 promoted M2 polarization of SMs via carrying miR-135b
It was demonstrated that the infiltration of inflammatory cells plays a crucial role in OA development (Shen et al. 2013). Here, we detected the percentages of M1- and M2-polarized SMs in synovial tissues of rats, and found that OA induced M1 polarization of SMs. The percentage of M2-polarized SMs in OA rats was upregulated following BMSC-ExoTGF-β1 treatment. However, the upregulation of BMSC-ExoTGF-β1 to M2-polarized SMs was partly limited by miR-135b decreasing (Fig. 4a–o). Furthermore, the qRT-PCR results displayed that expression of miR-135b was remarkably reduced in synovial tissues from OA rats, while BMSC-ExoTGF-β1 could notably upregulate the expression of miR-135b, which was then inhibited by miR-135b inhibitor (Fig. 4p). In summary, BMSC-ExoTGF-β1 promoted M2 polarization of SMs via carrying highly expressed miR-135b.
BMSC-ExoTGF-β1 boosted M2 polarization of SMs by secreting miR-135b. SD rats were randomly divided into six groups: sham, OA, OA + BMSC-ExoPBS, OA + BMSC-ExoTGF-β1, OA + INC-BMSC-ExoTGF-β1, OA + miR-135bI-BMSC-ExoTGF-β1.(a–o) Immunohistochemistry assay was performed to detect the CD86- (M1) and CD163-positive (M2) cell numbers in synovial tissues from each rat. The ratio of M1/M2 macrophages was analyzed. (p) The expression of miR-135b in synovial tissues was examined using qRT-PCR. **P < 0.01 contrasted with sham group, #P < 0.05 contrasted with OA group and &&P < 0.01 compared with BMSC-ExoTGF-β1 group.
Exosomal miR-135b accelerated M2 polarization of SMs
To further explore the molecular mechanism of BMSC-ExoTGF-β1 facilitates M2 polarization of SMs, we cultured the primary SMs from OA and normal rats. Then, LPS was used to activate macrophage, and the cells were divided into six groups: control, LPS, LPS + BMSC-ExoPBS, LPS + BMSC-ExoTGF-β1, LPS + INC-BMSC-ExoTGF-β1, and LPS + miR135bI-BMSC-ExoTGF-β1. Firstly, the expression of miR-135b in cells was measured. Our data showed that LPS decreased the expression of miR-135b, while BMSC-ExoTGF-β1 effectively increased the expression of miR-135b in LPS-stimulated SMs, which was then inhibited by miR-135b decreasing (Fig. 5a). Subsequently, the qRT-PCR results revealed that LPS significantly increased the expression of iNOS, a marker of M1 macrophage. In LPS-treated SMs, BMSC-ExoTGF-β1 could notably decrease the expression of iNOS and increase the expression of Arg-1 that a marker of M2 macrophage, while these effects of BMSC-ExoTGF-β1 were partly repressed by miR-135b decreasing (Fig. 5b). In addition, the promotion of BMSC-ExoTGF-β1 to M2 polarization of SMs through carrying highly expressed miR-135b in LPS-treated SMs also was ensured by the results of flow cytometry (Fig. 5c–g). Next, we used miR-135b mimic instead of TGF-β1-stimulated BMSC-derived exosomal miR-135b to investigate the role of miR-135b in SM polarization. As shown in Fig. 5h, the expression of miR-135b was significantly upregulated by its mimics. Besides, the flow cytometry results indicated that upregulation of miR-135b could remarkably reduce the percentage of CD86-positive (M1) SMs and increase the percentage of CD163-positive (M2) SMs (Fig. 5i–m). Moreover, upregulation of miR-135b repressed the expression of iNOS and facilitated Arg-1 that was found (Fig. 5n). Our data proved that BMSC-ExoTGF-β1 could promote M2 polarization of SMs through carrying highly expressed miR-135b in cellular level.
Exosomal miR-135b induced SMs to M2 polarization. Exosomes, from different BMSCs, were incubated with SMs following LPS for 24 h. a The expression of miR-135b was measured by qRT-PCR. b The expression of iNOS and Arg-1 mRNAs also weredetected using qRT-PCR assay. iNOS is a marker of M1 macrophage, and Arg-1 is a marker of M2 macrophage. c–g Flow cytometry was carried out to detect the percentage of CD86- and CD163-positive SMs. **P < 0.01 contrasted to control group, ##P < 0.01 compared with LPS group, and &&P < 0.01 compared with BMSC-ExoTGF-β1 group. Subsequently, miR-135b mimic and mimic NC were transfected into SMs. h qRT-PCR was performed to detect the expression of miR-135b. i–m The percentage of CD86- and CD163-positive SMs and the ratio of M1 and M2 SMs were ensured using flow cytometry. n The expression of iNOS and Arg-1 mRNAs were examined using qRT-PCR. **P < 0.01 compared with mimic NC group
MiR-135b promoted M2 macrophage polarization via inhibiting MAPK6
Next, we predicated the potential downstream target of miR-135b through bioinformatics analysis, and found that MAPK6 is a target of miR-135b. Our data demonstrated that the expression of MAPK6 both in gene (Fig. 6a) and protein (Fig. 6 b and b’) levels were upregulated in cartilage tissues of OA rat, while MAPK6 expression was repressed by BMSC-ExoTGF-β1. The effect of BMSC-ExoTGF-β1 was partly reversed after miR-135b decreasing. Meantime, the expression of MAPK6 mRNA (Fig. 6c) and protein (Fig. 6 d and d’) also were increased in LPS-stimulated SMs, and then were decreased in BMSC-ExoTGF-β1-co-treated SMs, while the effect of BMSC-ExoTGF-β1 was limited by miR-135b knockdown. According to these results, we asked a question that whether miR-135b inhibits the expression of MAPK6 in OA progression, and then regulates the polarization of SMs. In this present study, we firstly proved the relationship between MAPK6 3′-UTR and miR-135b using luciferase reporter assay. MAPK6 mRNA was indeed the target of miR-135b (Fig. 6 e and e’). In addition, pcDNA-MAPK6 and miR-135b mimic were co-transfected into SMs to explore whether miR-135b facilitates M2 polarization of SMs through inhibiting the expression of MAPK6. Our data showed that increasing of miR-135b significantly promoted M2 polarization of SMs, while it was suppressed by MAPK6 overexpression (Fig. 7a–e). At the same time, increasing of miR-135b-induced upregulation of Arg-1 mRNA and downregulation of iNOS mRNA were partly repressed by MAPK6 upregulation (Fig. 7f). Overall, these data confirmed that MAPK6 as a target of miR-135b mediated the regulation of miR-135b to M2 polarization of SMs.
MAPK6 acted as a target of miR-135b. (a) The expression of MAPK6 mRNA in cartilage tissues from each rat was measured using qRT-PCR. **P < 0.01 contrasted to cham group, ##P < 0.01 contrasted to OA group, and &&P < 0.01 contrasted to BMSC-ExoTGF-β1 group. (b and b’) The expression of MAPK6 protein in cartilage tissues from each rat was measured using Western blotting assay. **P < 0.01 contrasted to sham group, ##P < 0.01 contrasted to OA group, and &&P < 0.01 contrasted to BMSC-ExoTGF-β1 group. (c) The expression of MAPK6 mRNA in SMs was examined using qRT-PCR. **P < 0.01 compared with control group, ##P < 0.01 compared with LPS group and &&P< compared with BMSC-ExoTGF-β1 group. (d and d’) Western blotting assay was performed to ensure the expression of MAPK6 in SMs. **P < 0.01 compared with control group, ##P < 0.01 compared with LPS group, and &&P < 0.01 compared with BMSC-ExoTGF-β1 group. (e and e’) The relationship between miR-135b and MAPK6 3′-UTR was ensured using luciferase activity assay.
MiR-135b promoted M2 polarization of SMs through inhibition of MAPK6 expression. (a–e) Flow cytometry assay was carried out to detect the percentages of CD86- and CD163-positive SMs, and the ratio of M1 and M2 were analyzed. (f) The expression of iNOS and Arg-1 mRNAs were measured by qRT-PCR. **P < 0.01 compared with mimic NC group, and ##P < 0.01 compared with miR-135b mimic group.
BMSC-ExoTGF-β1 promoted M2 polarization of SMs via inhibiting MAPK6
Furthermore, we used BMSC-ExoTGF-β1 instead of miR-135b mimic to treat SMs together with pcDNA-MAPK6 or empty vector following LPS. Here, we found that the percentage of CD86-positive cells was notably upregulated by MAPK6 increasing, and CD163-positive cells were downregulated in BMSC-ExoTGF-β1 and LPS co-treated SMs following MAPK6 increasing (Fig. 8a–e). Combined with the above experimental results, we can know that BMSC-ExoTGF-β1 promoted M2 polarization of SMs through increasing MAPK6 expression. This conclusion also was proved by the results of Western blotting assay. As shown in Fig. 8f, the expression of iNOS was increased, while Arg-1 was decreased by MAPK6 overexpression in BMSC-ExoTGF-β1 and LPS co-treated SMs.
Increasing of MAPK6 repressed BMSC-ExoTGF-β1-induced M2 polarization of SMs. BMSC-ExoTGF-β1 and LPS were used to treat SMs which were then transfected with pcDNA-MAPK6 or empty vector. At 24 h later, (a–e) the percentages of CD86- and CD163-positive SMs were ensured by flow cytometry, and the ratio of M1/M2 SMs was analyzed. (f) The expression of iNOS and Arg-1 mRNAs were examined using Western blotting assay. **P < 0.01 compared with BMSC-ExoTGF-β1 group.
Discussion
MSCs can be isolated from multiple tissues like adipose, umbilical cord, umbilical cord blood and bone marrow, and have the potential to differentiate into osteoblasts, muscle cells, adipocytes, cardiomyocytes and chondrocytes (Mirzaei et al. 2016; Mohammadi et al. 2016). Among these MSC origins, BMSCs are the most common source of MSCs (Darvish and Payandeh 2019; Vahidinia et al. 2019). Positively expressed CD29, CD44, CD90, CD105, and CD73 and negatively expressed CD45, CD34, CD14, and CD11b can be acted as the membrane markers of BMSC identification (Zakirova et al. 2019). At present, it was demonstrated that MSCs play a crucial role in the treatment of multiple disorders, such as malignant tumors, neurological diseases, tissue fibrosis, as well as degenerative joint disease (Cortes-Dericks and Galetta 2019; Mendes-Pinheiro et al. 2019; Park et al. 2019; Yoshida and Nakashima 2018). Liu et al. revealed that small ubiquitin-like modifier-modified MSC differentiate toward chondrocytes and repress inflammation, thus facilitate the repair of articular cartilage (Liu et al. 2020). Tong et al. demonstrated that umbilical cord-derived MSCs could effectively improve the development of experimental OA in rats through enhancing cartilage superficial layer cells and repressing synovial inflammation (Tong et al. 2020). More and more studies confirmed the potential of MSCs in OA treatment. Traditionally, MSCs were used to treat OA mainly due to the fact that they have the ability to differentiation into chondrocytes, while MSCs may be a feasible therapy strategy for cartilage repair; their clinical use still has many questions. Hence, more and more researchers put their sight on the ability of paracrine secretory of MSCs (Colombini et al. 2019). It was confirmed that MSCs could be against the development of disease through secreting exosomes which carry miRNAs; these miRNAs repressed the progression of the disease via inhibiting the expression of their downstream target (Li et al. 2020). Here, we obtained the primary BMSCs from rat’s bone marrow successfully, and then, our study indicated that the expression of miR-135b was increased in exosomes from TGF-β1-stimulated BMSCs. Importantly, BMSCs-ExoTGF-β1 notably reduced the damage of cartilage tissues and promoted its repair through carrying highly expressed miR-135b.
Macrophages are important mediator of inflammation response via production of cytokines, such as M1-polarized macrophage-derived IL-1β and TGF-β, and M2-polarized macrophage-derived IL-10, and inhibition of inflammation response in macrophage could ameliorate experimental arthritis (Wang et al. 2018a). Recently, many studies proved the imbalance between M1- and M2-polarized macrophage closely associated with the development of OA. Liu et al. revealed that the ratio of M1- and M2-polarized macrophage in the knee of OA patients higher than the normal group and the higher ratio means higher degree of lesions (Liu et al. 2018). Pro-inflammatory M1-polarized macrophage could boost the inflammation and degradation of cartilage, while enhancement of M2 macrophage polarization could remarkably attenuate cartilage damage and facilitate its repair (Utomo et al. 2016; Zhou et al. 2019). Furthermore, BMSCs-Exo was proved to induce the conversion of M1-polarized macrophage toward M2-polarized macrophage through submitting exosomal miRNA (Deng et al. 2020; Zhao et al. 2019). Here, our data demonstrated that BMSCs-ExoTGF-β1 promoted M2 polarization of SMs in OA rats and LPS-induced SMs through carrying miR-135b.
Subsequently, to investigate the regulatory mechanism of miR-135b in M2 SM polarization, we predicted the downstream target of it. We found that MAPK6, also named as ERK3, involves in series of biological processes (Huang et al. 2019). Nevertheless, the role of MAPK6 in cartilage injury remains unclear. In this present study, we proved that TGF-β1-stimulated BMSCs promoted M2 SM polarization through inhibition the expression of MAPK6 via submitting exosomal miR-135b.
Here, our data revealed a new action mechanism of BMSC-Exo to attenuate the damage of cartilage tissues in OA rats through section of exosomal miR-135b. Specifically, miR-135b inhibited the degradation of cartilage tissues via facilitating M2 polarization of SMs by repressing the expression of MAPK6.
Abbreviations
- BMSCs:
-
Bone marrow-derived MSCs
- BMSC-Exo:
-
BMSCs-derived exosome
- BMSC-ExoPBS :
-
PBS-stimulated BMSC-derived exosome
- BMSC-ExoTGF - β1 :
-
TGF-β1-treated BMSC-derived exosome
- INC-BMSC-ExoTGF - β1 :
-
Inhibitor NC together with TGF-β1-treated BMSC-derived exosome
- MiR-135bI-BMSC-ExoTGF - β1 :
-
MiR-135b inhibitor together with TGF-β1-stimulated BMSC-derived exosome
- MSCs:
-
Mesenchymal stem cells
- NTA:
-
Nanoparticle tracking analysis
- OA:
-
Osteoarthritis
- SMs:
-
Synovial macrophages
- TEM:
-
Transmission electron microscope
- TGF-β1:
-
Transforming growth factor β1
References
Colombini A, Perucca Orfei C, Kouroupis D, Ragni E, De Luca P, ViganÒ M, Correa D, de Girolamo L (2019) Mesenchymal stem cells in the treatment of articular cartilage degeneration: New biological insights for an old-timer cell. Cytotherapy 21:1179–1197
Cortes-Dericks L, Galetta D (2019) The therapeutic potential of mesenchymal stem cells in lung cancer: benefits, risks and challenges. Cell Oncol 42:727–738
Darvish M, Payandeh Z (2019) Umbilical cord blood mesenchymal stem cells application in hematopoietic stem cells expansion on nanofiber three-dimensional scaffold. J Cell Biochem. https://doi.org/10.1002/jcb.28487
Deng H, Wu L, Liu M, Zhu L, Chen Y, Zhou H, Shi X, Wei J, Zheng L, Hu X, Wang M, He Z, Lv X, Yang H (2020) Bone Marrow Mesenchymal Stem Cell-Derived Exosomes Attenuate LPS-Induced ARDS by Modulating Macrophage Polarization Through Inhibiting Glycolysis in Macrophages. Shock (Augusta, Ga) https://doi.org/10.1097/SHK.0000000000001549.
Fang SB, Zhang HY, Wang C, He BX, Liu XQ, Meng XC, Peng YQ, Xu ZB, Fan XL, Wu ZJ, Chen D, Zheng L, Zheng SG (2020) Small extracellular vesicles derived from human mesenchymal stromal cells prevent group 2 innate lymphoid cell-dominant allergic airway inflammation through delivery of miR-146a-5p. J Extracell Vesicles 9:1723260
Geng Y, Chen J, Alahdal M, Chang C, Duan L, Zhu W, Mou L, Xiong J, Wang M, Wang D (2020) Intra-articular injection of hUC-MSCs expressing miR-140-5p induces cartilage self-repairing in the rat osteoarthritis. J Bone Miner Metab 38:277–288
Goradel NH, Hour FG, Negahdari B, Malekshahi ZV, Hashemzehi M, Masoudifar A, Mirzaei H (2018) Stem cell therapy: A new therapeutic option for cardiovascular diseases. J Cell Biochem 119:95–104
Hou B, Ye Z, Ji W, Cai M, Ling C, Chen C, Guo Y (2018) Comparison of the effects of BMSC-derived Schwann cells and autologous Schwann cells on remyelination using a rat sciatic nerve defect model. Int J Biol Sci 14:1910–1922
Hu C, Huang S, Wu F, Ding H (2018) miR-98 inhibits cell proliferation and induces cell apoptosis by targeting MAPK6 in HUVECs. Exp Ther Med 15:2755–2760
Huang ZQ, Xu W, Wu JL, Lu X, Chen XM (2019) MicroRNA-374a protects against myocardial ischemia-reperfusion injury in mice by targeting the MAPK6 pathway. Life Sci 232:116619
Kim H, Wang SY, Kwak G, Yang Y, Kwon IC, Kim SH (2019) Exosome-Guided Phenotypic Switch of M1 to M2 Macrophages for Cutaneous Wound Healing. 6:1900513
Kim JR, Yoo JJ (2018) Therapeutics in Osteoarthritis Based on an Understanding of Its Molecular Pathogenesis. 19:
Li Y, Zhou J, Zhang O, Wu X, Guan X, Xue Y, Li S, Zhuang X, Zhou B, Miao G, Zhang L (2020) Bone marrow mesenchymal stem cells-derived exosomal microRNA-185 represses ventricular remolding of mice with myocardial infarction by inhibiting SOCS2. Int Immunopharmacol 80:106156
Liu B, Zhang M, Zhao J, Zheng M, Yang H (2018) Imbalance of M1/M2 macrophages is linked to severity level of knee osteoarthritis. Experimental and therapeutic medicine 16:5009–5014
Liu Y, Li M, Yin Z, Zhou S, Qiu Y (2020) SUMO-modified bone marrow mesenchymal stem cells promoted the repair of articular cartilage in rats. Cell Biol Int 44:560–568
Ma PF, Gao CC, Yi J, Zhao JL, Liang SQ, Zhao Y, Ye YC, Bai J, Zheng QJ, Dou KF, Han H, Qin HY (2017) Cytotherapy with M1-polarized macrophages ameliorates liver fibrosis by modulating immune microenvironment in mice. J Hepatol 67:770–779
Mendes-Pinheiro B, Anjo SI, Manadas B, Da Silva JD, Marote A, Behie LA, Teixeira FG, Salgado AJ (2019) Bone marrow mesenchymal stem cells’ secretome exerts neuroprotective effects in a Parkinson’s disease rat model. Frontiers in bioengineering and biotechnology 7:294
Mianehsaz E, Mirzaei HR, Mahjoubin-Tehran M, Rezaee A, Sahebnasagh R, Pourhanifeh MH, Mirzaei H, Hamblin MR (2019) Mesenchymal stem cell-derived exosomes: A new therapeutic approach to osteoarthritis? Stem cell research & therapy 10:340
Miller RE, Scanzello CR (2019) An emerging role for Toll-like receptors at the neuroimmune interface in osteoarthritis 41:583–594
Mirzaei H, Sahebkar A, Avan A, Jaafari MR, Salehi R, Salehi H, Baharvand H, Rezaei A, Hadjati J, Pawelek JM, Mirzaei HR (2016) Application of mesenchymal stem cells in melanoma: a potential therapeutic strategy for delivery of targeted agents. Curr Med Chem 23:455–463
Mirzaei H, Sahebkar A, Sichani LS, Moridikia A, Nazari S, Sadri Nahand J, Salehi H, Stenvang J, Masoudifar A, Mirzaei HR, Jaafari MR (2018) Therapeutic application of multipotent stem cells. J Cell Physiol 233:2815–2823
Mirzaei H, Salehi H, Oskuee RK, Mohammadpour A, Mirzaei HR, Sharifi MR, Salarinia R, Darani HY, Mokhtari M, Masoudifar A, Sahebkar A, Salehi R, Jaafari MR (2018) The therapeutic potential of human adipose-derived mesenchymal stem cells producing CXCL10 in a mouse melanoma lung metastasis model. Cancer Lett 419:30–39
Mohammadi M, Jaafari MR, Mirzaei HR, Mirzaei H (2016) Mesenchymal stem cell: a new horizon in cancer gene therapy. Cancer Gene Ther 23:285–286
Moradian Tehrani R, Verdi J, Noureddini M, Salehi R, Salarinia R, Mosalaei M, Simonian M, Alani B, Ghiasi MR, Jaafari MR, Mirzaei HR, Mirzaei H (2018) Mesenchymal stem cells: A new platform for targeting suicide genes in cancer 233:3831–3845
Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, Locati M, Mantovani A, Martinez FO, Mege JL, Mosser DM, Natoli G, Saeij JP, Schultze JL, Shirey KA, Sica A, Suttles J, Udalova I, van Ginderachter JA, Vogel SN, Wynn TA (2014) Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 41:14–20
Oo WM, Yu SP, Daniel MS, Hunter DJ (2018) Disease-modifying drugs in osteoarthritis: Current understanding and future therapeutics. Expert opinion on emerging drugs 23:331–347
Park MJ, Moon SJ, Baek JA, Lee EJ, Jung KA, Kim EK, Kim DS, Lee JH, Kwok SK, Min JK, Kim SJ (2019) Metformin augments anti-inflammatory and chondroprotective properties of mesenchymal stem cells in experimental osteoarthritis 203:127–136
Robinson WH, Lepus CM, Wang Q, Raghu H, Mao R, Lindstrom TM, Sokolove J (2016) Low-grade inflammation as a key mediator of the pathogenesis of osteoarthritis. Nat Rev Rheumatol 12:580–592
Shen PC, Lu CS, Shiau AL, Lee CH, Jou IM, Hsieh JL (2013) Lentiviral small hairpin RNA knockdown of macrophage inflammatory protein-1γ ameliorates experimentally induced osteoarthritis in mice. Hum Gene Ther 24:871–882
Tong W, Zhang X, Zhang Q, Fang J, Liu Y, Shao Z, Yang S, Wu D, Sheng X, Zhang Y, Tian H (2020) Multiple umbilical cord-derived MSCs administrations attenuate rat osteoarthritis progression via preserving articular cartilage superficial layer cells and inhibiting synovitis. Journal of orthopaedic translation 23:21–28
Topoluk N, Steckbeck K, Siatkowski S, Burnikel B, Tokish J, Mercuri J (2018) Amniotic mesenchymal stem cells mitigate osteoarthritis progression in a synovial macrophage-mediated in vitro explant coculture model 12:1097–1110
Utomo L, Bastiaansen-Jenniskens YM, Verhaar JA, van Osch GJ (2016) Cartilage inflammation and degeneration is enhanced by pro-inflammatory (M1) macrophages in vitro, but not inhibited directly by anti-inflammatory (M2) macrophages. Osteoarthritis and cartilage 24:2162–2170
Vahidinia Z, Azami Tameh A, Nejati M, Beyer C, Talaei SA, Etehadi Moghadam S, Atlasi MA (2019) The protective effect of bone marrow mesenchymal stem cells in a rat model of ischemic stroke via reducing the C-Jun N-terminal kinase expression. Pathol Res Pract 215:152519
Wang Q, Zhou X, Zhao Y, Xiao J, Lu Y, Shi Q, Wang Y, Wang H, Liang Q (2018) Polyphyllin I ameliorates collagen-induced arthritis by suppressing the inflammation response in macrophages through the NF-κB pathway. Frontiers in immunology 9:2091
Wang R, Xu B, Xu H (2018) TGF-β1 promoted chondrocyte proliferation by regulating Sp1 through MSC-exosomes derived miR-135b. Cell cycle (Georgetown, Tex) 17:2756–2765
Xie J, Huang Z, Yu X, Zhou L, Pei F (2019) Clinical implications of macrophage dysfunction in the development of osteoarthritis of the knee. Cytokine Growth Factor Rev 46:36–44
Yoshida K, Nakashima A (2018) Serum-free medium enhances the immunosuppressive and antifibrotic abilities of mesenchymal stem cells utilized in experimental renal fibrosis 7:893–905
Zakirova EY, Valeeva AN, Aimaletdinov AM, Nefedovskaya LV, Akhmetshin RF, Rutland CS, Rizvanov AA (2019) Potential therapeutic application of mesenchymal stem cells in ophthalmology. Exp Eye Res 189:107863
Zhang H, Lin C, Zeng C, Wang Z, Wang H, Lu J, Liu X, Shao Y, Zhao C, Pan J, Xu S, Zhang Y, Xie D, Cai D, Bai X (2018) Synovial macrophage M1 polarisation exacerbates experimental osteoarthritis partially through R-spondin-2. Ann Rheum Dis 77:1524–1534
Zhao J, Li X, Hu J, Chen F, Qiao S, Sun X, Gao L, Xie J, Xu B (2019) Mesenchymal stromal cell-derived exosomes attenuate myocardial ischaemia-reperfusion injury through miR-182-regulated macrophage polarization. Cardiovasc Res 115:1205–1216
Zhou F, Mei J, Han X, Li H, Yang S, Wang M, Chu L, Qiao H, Tang T (2019) Kinsenoside attenuates osteoarthritis by repolarizing macrophages through inactivating NF-κB/MAPK signaling and protecting chondrocytes. Acta pharmaceutica Sinica B 9:973–985
Funding
This study was supported by Anhui Province Natural Science Youth Funding Projects (grant no. 1908085QH318).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
This study was approved by the Animal Research Committee of First Affiliated Hospital of Anhui Medical University.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
441_2020_3319_MOESM1_ESM.tif
Supplementary file1: Supplementary figure 1. BMSCs identification. (a-k) Flow cytometry was performed to verify the expression of BMSCs markers (CD11b, CD34 and CD45 Low; CD29, CD44 and CD90 High). (l) The shape of BMSCs was identified using light microscope. (m) The ability of BMSCs differentiation into osteogenic lineage was ensured using alizarin red staining. (n) The ability of BMSCs differentiation into adipogenic lineage was confirmed using Oil red O staining. (TIF 1.76 MB)
Rights and permissions
About this article
Cite this article
Wang, R., Xu, B. TGF-β1-modified MSC-derived exosomal miR-135b attenuates cartilage injury via promoting M2 synovial macrophage polarization by targeting MAPK6. Cell Tissue Res 384, 113–127 (2021). https://doi.org/10.1007/s00441-020-03319-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-020-03319-1