Abstract
Ependymal cells located above the ventricular zone of the lateral, third, and fourth ventricles and the spinal cord are thought to form part of the adult neurogenic niche. Many studies have focused on ependymal cells as potential adult neural stem/progenitor cells. To investigate the functions of ependymal cells, a simple method to isolate subtypes is needed. Accordingly, in this study, we evaluated the expression of cluster of differentiation (CD) 9 in ependymal cells by in situ hybridization and immunohistochemistry. Our results showed that CD9-positive ependymal cells were also immunopositive for SRY-box 2, a stem/progenitor cell marker. We then isolated CD9-positive ependymal cells from the third ventricle using the pluriBead-cascade cell isolation system based on antibody-mediated binding of cells to beads of different sizes and their isolation with sieves of different mesh sizes. As a result, we succeeded in isolating CD9-positive populations with 86% purity of ependymal cells from the third ventricle. We next assayed whether isolated CD9-positive ependymal cells had neurospherogenic potential. Neurospheres were generated from CD9-positive ependymal cells of adult rats and were immunopositve for neuron, astrocyte, and oligodendrocyte markers after cultivation. Thus, based on these findings, we suggest that the isolated CD9-positive ependymal cells from the third ventricle included tanycytes, which are special ependymal cells in the ventricular zone of the third ventricle that form part of the adult neurogenic and gliogenic niche. These current findings improve our understanding of tanycytes in the adult third ventricle in vitro.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The ventricular system is a set of four communicating cavities (ventricles) within the brain. The ventricles are responsible for the production, transport, and removal of cerebrospinal fluid (CSF), which immerses the central nervous system (Del Bigio 2010). The ventricles are composed the lateral, third, and fourth ventricles and the central canal of the spinal cord. All ventricles are lined with ependymal cells, a specialized form of epithelium involved in the production of CSF (Del Bigio 2010). Ependymal cells are known to be composed of several populations that vary according to morphological and physiological characteristics, as follows: E1 cells with approximately 50 motile cilia; E2 cells with two motile cilia and very large basal bodies; and tanycytes with few cilia (Ohata and Alvarez-Buylla 2016). Ependymal cells in the central canal have 1–3 motile cilia (Millhouse 1971; Robins et al. 2013) and are commonly immunopositive for S100β, cluster of differentiation (CD) 24, forkhead box J1, and vimentin by immunohistochemistry (Pastrana et al. 2011). In addition, E1 cells and central canal ependymal cells are immunopositve for CD133 (Pfenninger et al. 2011). E2 cells express glial fibrillary acidic protein (GFAP), and tanycytes express GFAP and nestin (Robins et al. 2013).
The forebrain subventricular zone (SVZ) and the central canal ependymal zone are a well-characterized niche of neural stem cells (Pastrana et al. 2011; Silva-Vargas et al. 2013). A niche is a microenvironment that supports the self-renewal of stem cells and differentiation. Recent findings have suggested that tanycytes in the third ventricle also form the critical component of a hypothalamic stem cell niche (Robins et al. 2013). Morphological studies have mapped and defined subpopulations of tanycytes, i.e., β-, α2-, and α1-tanycytes, according to their position in the third ventricle (Robins et al. 2013; Rodriguez et al. 2005). β-Tanycytes line the median eminence, α2-tanycytes reside adjacent to the arcuate nucleus, and α1-tanycytes extend from the ventromedial nucleus to the dorsomedial nucleus (Robins et al. 2013). Many studies of lineage tracing analyses using transgenic (TG) mice have focused on tanycytes as potential adult neural stem cells. Robins et al. (2013) suggested that adult α2-tanycytes have the capacity to act as neural stem cells, and Haan et al. (2013) showed that β-tanycytes can proliferate in early adulthood, giving rise to new neurons.
To investigate the factors that trigger cell replacement or nervous tissue repair in tanycytes as neural stem/progenitor cells in the hypothalamus, it is essential to isolate pure primary cultures of β-, α2-, or α1-tanycytes from adult animals. Recently, we developed a method to separate particular cell populations from dispersed cells of the rat pituitary gland using antibodies against membrane proteins, such as CD proteins (Horiguchi et al. 2018; Horiguchi et al. 2016). Subsequently, we showed that the Cd9 gene was expressed in the majority of S100β/sex determining region of Y-box 2 (SOX2)-positive adult stem/progenitor cells in the rat anterior lobe of the pituitary gland and succeeded in the isolation of these cells from the anterior lobe using anti-CD9 antibodies together with the pluriBead-cascade cell isolation system (Horiguchi et al. 2018).
In this study, we characterized CD9-positive ependymal cells isolated using this pluriBead-cascade system. We analyzed whether ependymal cells in the lateral ventricle, third ventricle, and central canal of spinal cord also expressed Cd9. Finally, we evaluated whether isolated CD9-positive ependymal cells in the third ventricle had neurospherogenic potential and the capacity to differentiate into neurogenic cells.
Materials and methods
Animals
Adult Wistar rats were purchased from Japan SLC, Inc. (Shizuoka, Japan). Eight- to 10-week-old male rats weighing 200–250 g were given ad libitum access to food and water and housed under a 12-h light/dark cycle. Rats were killed by exsanguination from the right atrium under deep nembutal anesthesia and were then perfused with Hanks’ balanced salt solution (Life Technologies, Carlsbad, CA, USA) for isolation of CD9-positive cells from the third ventricle or with 4% paraformaldehyde in 0.05 M phosphate buffer (PB; pH 7.4) for in situ hybridization and immunohistochemistry. The current study was approved by the Committee on Animal Experiments of the School of Agriculture, Meiji University, and Kyorin University based on the NIH Guidelines for the Care and Use of Laboratory Animals.
Tissue preparation
Whole brains of adult rats were immediately immersed in a fixative consisting of 4% paraformaldehyde in 0.05 M PB (pH 7.4) for 20–24 h at 4 °C. The tissues were then immersed for more than 2 days in PB (pH 7.2) containing 30% sucrose at 4 °C, embedded in Tissue-Tek OCT compound (Sakura Finetek Japan, Tokyo, Japan), and frozen rapidly. Frozen frontal sections and horizontal sections (8 μm thick) were obtained using a cryostat (Tissue-Tek Polar DM; Sakura Finetek Japan) and mounted on slide glasses (Matsunami, Osaka, Japan).
In situ hybridization and immunohistochemistry
In situ hybridization was performed with digoxigenin (DIG)-labeled cRNA probes, as described in our previous report (Fujiwara et al. 2007). The following DNA fragments were amplified from the rat pituitary cDNA library by polymerase chain reaction (PCR) with specific primer sets: Cd9, 5′-GGCTATACCCACAAGGACGA-3′ and 5′-CCCGGATCCCTCTACTACAA-3′ (product length 528 bp). Amplified cDNAs were ligated into the pTA-2 vector (Toyobo Co., Ltd., Osaka, Japan) and were subcloned in a plasmid vector. Gene-specific antisense or sense DIG-labeled cRNA probes were generated using a Roche DIG RNA labeling kit (Roche Diagnostics, Basel, Switzerland). Cells were fixed with 4% paraformaldehyde in 0.025 M PB for 20 min at room temperature, and hybridization with DIG-labeled cRNA probes was performed at 58 °C for 16 h. Visualization of each type of mRNA was performed with alkaline phosphatase-conjugated anti-DIG antibodies (Roche Diagnostics) using 4-nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl phosphate (BCIP; Roche Diagnostics). Each observation was performed in triplicate.
For fluorescent double-labeling of Cd9 mRNA, in situ hybridization signals were visualized using a solution from an HNPP Fluorescent Detection Kit (Roche Diagnostics). After in situ hybridization, sections or cells were incubated in phosphate-buffered saline (PBS; pH 7.2) containing 2% normal goat or donkey serum for 20 min at 30 °C and then incubated with CD9-mouse monoclonal or other antibodies at room temperature. After washing with PBS, cells were incubated in PBS with secondary antibodies for 30 min at 30 °C. The antibodies used are detailed in Table 1. The absence of an observable nonspecific reaction was confirmed using normal mouse and rabbit sera. Nuclei were counterstained by incubation with Vectashield Mounting Medium containing 4,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA, USA). Sections and cells were scanned using a fluorescence microscope (cellSens Dimension System; Olympus, Tokyo, Japan). Each observation was performed at least three times.
For double immunofluorescence, frozen sections or cells were incubated overnight at room temperature in PBS with CD9 and Ki67 (a proliferation cell marker), SOX2 (a stem/progenitor cell marker), Neuronal nuclear antigen (NeuN; a neural cell marker), Glial fibrillary acidic protein (GFAP; an intermediate filament and a stem/progenitor cell and astrocyte marker), or receptor interacting protein (RIP; an oligodendrocyte marker). SOX2 is well known for its functions in embryonic stem cell pluripotency, maintenance, and self-renewal and adult tissue homeostasis of different tissues, particularly in the central nervous system (Feng and Wen 2015). Primary and secondary antibodies are detailed in Table 1. After washing with PBS, cells were incubated in PBS with secondary antibodies as described above. Sections and cells were scanned using a fluorescence microscope (cellSens Dimension System; Olympus). Each observation was performed in triplicate.
Two sets (A: Cd9, Nestin and Vimentin, B: CD9, SOX2 and GFAP) of multistaining were performed for the third ventricle. The number of positive cells in three different areas (157.5 × 210 μm2) was counted for each set using the cellSens Dimension system (Olympus), and the proportion compared with DAPI-positive cells was calculated.
Isolation of CD9-immunopositive cells from the third ventricle
The ventricular zone of the third ventricles of rats was excised, minced into small pieces, and then incubated in Hanks’ solution containing 1% trypsin (Invitrogen, Carlsbad, CA, USA) and 0.2% collagenase (Nitta Gelatin, Osaka, Japan) for 5 min at 37 °C. Thereafter, pieces were incubated in the same solution containing 5 μg/mL DNase I (Boehringer-Mannheim, Mannheim, Germany) for 5 min at 37 °C. After incubation in Hanks’ solution containing 0.3% ethylenediaminetetraacetic acid (EDTA; Wako Pure Chemicals, Osaka, Japan) for 5 min at 37 °C, the digest was washed with Hanks’ solution. Dispersed cells were separated from debris by centrifugation, rinsed, resuspended in Hanks’ solution by pipetting, and then filtered through nylon mesh (Becton Dickinson Labware, Franklin Lakes, NJ, USA). Dispersed cells were separated using the pluriBead-cascade cell isolation system (Universal Mouse pluriBeads kit; pluriSelect Corp., San Diego, CA, USA) as described previously (Pierzchalski et al. 2013) with mouse monoclonal anti-CD9 antibodies (BD Biosciences, Franklin Lakes, NJ, USA) according to the manufacturer’s instructions. Subsequently, CD9-immunopositive cells and negative cells were counted and used for quantitative PCR (qPCR) or plated onto slide glass for smear preparations or cultivation.
Measurement of the number of CD9 immunopositive cells
Four random fields were imaged per slide for smear preparations of isolated CD9-positive cell fractions by the pluriBead-cascade cell isolation system using a fluorescence microscope with a × 40 objective lens. The numbers of cells positive for CD9 and total numbers of cells stained by DAPI per area (157.5 × 210 μm2) were counted using the cellSens Dimension system (Olympus). Observations were performed three times for each experimental group.
Quantification of mRNA levels by qPCR
qPCR was performed as described previously (Horiguchi et al. 2016). Using TRIzol (Life Technologies), total RNA fractions were prepared from CD9-immunopositive cell fractions and negative cells and then incubated with RNase-free DNase I (1 U/tube; Promega, Madison, WI, USA) for 10 min at 37 °C. Next, cDNA was synthesized using a PrimeScript RT reagent kit (Takara Bio Inc., Shiga, Japan) with oligo-(dT)20 primers (Life Technologies). qPCR (Thermal Cycler Dice Real Time System II; Takara Bio Inc.) was performed using gene-specific primers and SYBR Premix Ex Taq (Takara) containing SYBR Green I. The sequences of the gene-specific primers were as follows: Cd9, 5′-GGCTATACCCACAAGGACGA-3′ and 5′-GCTATGCCACAGCAGTTCAA-3′ (product length: 140 bp); Sox2, 5′-CCATTTTCGTGGTCTTGTTT-3′ and 5′-TCAACCTGCATGGACATTTT-3′ (product length: 94 bp), and β-actin (b-actin), 5′-TGGCACCACACTTTCTACAATGAGC-3′ and 5′-GGGTCATCTTTTCACGGTTGG-3′ (product length: 106 bp). Quantification of b-actin was performed for normalization. The relative gene expression was calculated by comparing cycle times for each target qPCR. Cycle threshold values were converted to relative gene expression levels by using the 2-(ΔCt sample – ΔCt control) method. Each analysis was performed at least three times.
Neurosphere cultures
Isolated CD9-positive cells from the ventricular zone in the third ventricles of adult rats were plated on 35-mm untreated dishes (AGC Techno Glass, Shizuoka, Japan) at a density of 10,000 cells/mL in Dulbecco’s modified Eagle’s medium (DMEM)/F-12 containing B27 supplement (1:50; Thermo Fisher Scientific), N2 supplement (1:100; Wako, Osaka, Japan), bovine serum albumin (BSA; 0.5%; Sigma, St. Louis, MO, USA), basic fibroblast growth factor (bFGF; 20 ng/mL; R&D Systems, Minneapolis, MN, USA), and epidermal growth factor (EGF; 20 ng/mL; R&D Systems). Cultures were incubated in humidified chambers with 5% CO2 at 37 °C. Neurosphere growth was assessed 5 days after plating. Neurospheres were passaged by incubating for 20 min in 0.05% trypsin plus 5 mM EDTA in DMEM/F-12 containing BSA (0.5%), mechanically dissociated into a single-cell suspension, and cultured on 35-mm untreated dishes as described above for 5 days. Neurospheres were immediately collected manually using pipettes under a microscope and dispersed on Matrigel-coated 16-well chamber slides (0.4 cm2/well; Thermo Fisher Scientific). Differentiation of neurospheres was performed by the overlay three-dimensional culture method (Yoshida et al. 2016). Slides were coated with growth factor-reduced Matrigel diluted 1:10 in DMEM/F-12 containing 20 ng/mL bFGF and EGF without serum. Neurospheres were incubated for 7 days before fixation and processing. Neurospheres were incubated for 7 days before fixation, processing, and immunohistochemistry for NeuN, GFAP, or RIP together with CD9. The numbers of cells positive for CD9, NeuN, GFAP, and RIP in five random fields (157.5 × 210 μm2) from each well were counted using a fluorescence microscope with a × 40 objective lens, and the proportion compared with DAPI-positive cells was calculated. Observations were performed three times for each experimental group.
Statistical analysis
All data are presented as means ± standard errors of the means (n = 3). Significant differences between groups were determined by unpaired two-tailed Student’s t tests. Results with P values of less than 0.05 were considered statistically significant.
Results
Expression of the Cd9 gene in the lateral ventricle and spinal cord
First, we evaluated the expression of Cd9 in the lateral ventricle and central canal. Cd9 mRNA was detected in the lateral ventricle (Fig. 1a, b) and central canal of the spinal cord (Fig. 1c, d) by in situ hybridization with a DIG-labeled antisense cRNA probe. Cd9-expressing cells were located in the ventricular zone. No specific signal was detected in sections processed with the DIG-labeled sense RNA probe for Cd9 (data not shown). Immunohistochemical analysis revealed that CD9-immunopositive (CD9-positive) cells were detected in the cell layer facing the lateral ventricle (Fig. 1e, f) and central canal of the spinal cord (Fig. 1g, h). To identify CD9-positive cells, we performed double-immunohistochemistry for CD9 and SOX2 (nuclear protein), NeuN (nuclear protein), GFAP (cytoplasmic protein), or RIP (cytoplasmic protein). Some CD9-positive cells were immunopositive for SOX2 (Fig. 1i, k). In addition, we also performed double staining by in situ hybridization for Cd9 and immunohistochemistry for vimentin (an ependymal cell marker). Vimentin-positive cells among the ventricular zone expressed Cd9 (Fig. 1j, l). However, CD9-positive cells were immunonegative for NeuN (Fig. 1m, o), GFAP (Fig. 1n, p), and RIP (data not shown).
Identification of Cd9-expressing and CD9-positive cells in the rat ventricular zone. a, b, e, f, i, j, m, and n Lateral ventricle. c, d, g, h, k, l, o, and p Central canal of the spinal cord. a–d In situ hybridization for Cd9. e–h Immunohistochemistry for CD9. i, k Double-immunostaining for CD9 (magenta) and SOX2 (green). j, l Double staining for Cd9 (magenta) by in situ hybridization and vimentin (green) by immunohistochemistry. m, o Double-immunostaining for CD9 (magenta) and NeuN (green). n, p Double-immunostaining for CD9 (magenta) and GFAP (green). White arrowheads indicate double-positive cells. Open arrowheads indicate single positive cells. LV, lateral ventricle. CC, central canal. Bars, 200 μm (e), 100 μm (g), and 10 μm (f, h, and p)
Expression of Cd9 in the third ventricle
Hypothalamic tanycytes are unique ependymal cells located the ventricular zone of the third ventricle and are thought to form part of the adult neurogenic and gliogenic niche. Tanycytes are largely absent in the anterior hypothalamus and in dorsal ventricular regions of the third ventricle (Lee et al. 2012). The central hypothalamic slices were accurately subdissected (Fig. 2a, b). Tanycytes constitute heterogeneous cell populations and exhibit different characteristics according to their location, spatial relationships, and morphology. α1-Tanycytes line the region of the ventromedial nucleus and part of the dorsomedial nucleus, and α2-tanycytes line the area of the arcuate nucleus. β-Tanycytes line the infundibular recess and median eminence (Fig. 2b) (Goodman and Hajihosseini 2015; Rizzoti and Lovell-Badge 2017; Robins et al. 2013). Hematoxylin and eosin (HE) staining at the third ventricle of Wistar rats is shown in Fig. 2c. The immunoreactivities for nestin and vimentin were strong in the regions containing α2-tanycytes and β-tanycytes, respectively (Fig. 2d, e). Cd9 mRNA was detected in the third ventricle by in situ hybridization with a DIG-labeled antisense cRNA probe (Fig. 2f–j). Cd9-expressing cells were detected in the regions containing ependymocytes (Fig. 2g), α1-tanycytes (Fig. 2h), and α2-tanycytes (Fig. 2i), but were rare in β-tanycytes (Fig. 2j), indicating that CD9-positive cells were composed of α-tanycytes and ependymocytes. The proportions of Cd9-expressing cells per total cells were 98.0% ± 0.7% (ependymocyte area), 96.3% ± 1.5% (α-tanycyte area), and 8.5% ± 1.0% (β-tanycyte area; Fig. 4). By double staining for Cd9 mRNA with in situ hybridization and the neural stem/progenitor cell marker nestin or ependymal cell marker vimentin with immunohistochemistry, we observed double-positive cells (Fig. 2k, l). Almost all Cd9-expressing cells were immunopositive for Nestin and Vimentin (Fig. 4).
Identification of Cd9-expressing cells in the third ventricle. a, b Illustration of subdissection of the rat brain (a) and mediolateral/dorsoventral subdissections (b). HT, hypothalamus. VMN, ventromedial nucleus. ARC, arcuate nucleus. ME, median eminence. c HE staining of the third ventricle. d, e Immunohistochemistry for nestin and vimentin. f, In situ hybridization for Cd9. g–j High-magnification images of the boxed area in f. k, l Double staining by in situ hybridization for Cd9 (magenta) and by immunohistochemistry for nestin (k) or vimentin (l) (green) with DAPI (blue). Bars, 200 μm (c, e, and f), 50 μm (j), and 10 μm (l)
Characterization of Cd9-expressing cells in the third ventricle
To characterize Cd9-expressing cells in the third ventricle, we performed double staining by immunohistochemistry for CD9 and Ki67 (a proliferation marker), SOX2, NeuN, GFAP, or RIP. CD9 immunopositivity was detected in Cd9-expressing cells by in situ hybridization in the third ventricle (Fig. 3a, b). These locations coincided with the results of RNA and protein analyses, suggesting that this immunoreactivity represented specific staining. Cd9-expressing cells were immunopositve for Ki67 (Fig. 3c, d). SOX2-positive cells were observed in the ventricular zone, and some were immunopositive for CD9 (Fig. 3e, f). The proportions of CD9 and SOX2 double-positive cells per total cells in the ventricular zone were 97.0% ± 0.4% (ependymocyte area), 95.6% ± 0.6% (α-tanycyte area), and 9.5% ± 0.8% (β-tanycyte area; Fig. 4). In addition, GFAP-positive cells in the ventricular zone were immunopositive for CD9 (Fig. 3g, h), whereas NeuN- (Fig. 3i, j) or RIP-positive cells (Fig. 3k, l) were not. The GFAP-positive ependymal cells were thought to be tanycytes. In the α-tanycyte area of the ventricular zone, almost all GFAP-positive cells (26.6%, per total cells) were immunopositive for CD9 (25.1% ± 3.3%, per total cells; Fig. 4).
Double staining for CD9 and several cell markers. a–d Double staining by in situ hybridization for Cd9 (magenta) and by immunohistochemistry for CD9 (green) (a and b) and Ki67 (green) (c and d). a, c Merged image of DAPI staining and CD9 or Ki67. e–l Double-immunostaining for CD9 and SOX2 (e and f), GFAP (g and h), NeuN (i and j), or RIP (k and l). e, g, i, and k Merged images of DAPI staining and SOX2, GFAP, NueN, and RIP. Bar, 10 μm
The population of Cd9-expressing cells and cells positive for CD9, SOX2, GFAP, Nestin, and Vimentin in ependymocyte-, α-tanycyte-, and β-tanycyte-rich areas of the third ventricles. The total number of cells in each area was counted as the number of DAPI-stained nuclei. Populations of Cd9-expressing cells and Nestin-positive cells (a), Cd9-expressing cells and Vimentin-positive cells (b), CD9- and Nestin-positive cells (c), and CD9- and Vimentin-positive cells (d) are shown. Null cells indicates negative cells positive for only DAPI
Isolation of CD9-positive cells from the third ventricle
Next, we attempted to purify CD9-positive ependymal cells from the third ventricle using rat monoclonal anti-CD9 antibodies combined with the pluriBead-cascade cell isolation system. The brains were transversally cut between the optic chiasm and pituitary gland as a reference point, leaving the rostral and caudal portion of the brain (Fig. 5a). As shown in Fig. 5b, c, the brain around the third ventricle was cuboidally excised. The cells were dispersed and separated using a Universal pluriBeads kit as described previously (Horiguchi et al. 2018; Horiguchi et al. 2016). One drop of suspension for the CD9-positive fraction was used for smear preparation and immunocytochemistry. We observed that most of the cells were immunopositive for CD9 (Fig. 5d–f). The proportion of CD9-positive cells in the CD9-positive fraction was 86.1% (Fig. 5g). The remaining 14% of cells in the CD9-positve cell fraction were presumably contaminated with neural cells, glial cells, and endothelial cells (data not shown). Cd9 mRNA levels were 2.0-fold higher in the CD9-positive cell fraction than in the CD9-negative cell fraction (Fig. 5h). Moreover Sox2 gene expression was also significantly higher in the CD9-positive cell fraction than in the CD9-negative cell fraction (Fig. 5h).
Isolation of CD9-positive cells from the third ventricle. a Ventral view of the rat brain. The dotted lines indicate the plane between the optic chiasm and the pituitary gland. b Caudal view of the rostral fragment obtained after transversal sectioning of the rat brain, as indicated in a. c Tissue fragment containing the third ventricles (area enclosed by the dotted line in b). The area enclosed by the dotted line was utilized for cell dispersion. d–f Immunocytochemistry for CD9 on smear preparations of the CD9-positive fraction. DAPI staining (blue, d), immunocytochemistry for CD9 (magenta, e), and the merged image (f) are shown. g The ratio of CD9-immunopositive cells in the smear preparation of the CD9-positive fraction. h mRNA levels in CD9-negative cell fraction (open bars) and CD9-positive cell fraction (closed bars), as determined by qPCR (mean ± standard error of the mean, n = 3), followed by normalization with an internal control (b-actin). The graph represents Cd9 and Sox2. **P < 0.01. Bars, 5 mm (c), 50 μm (f)
Neurospherogenic properties and differentiation capacities of CD9-positive ependymal cells of the third ventricle
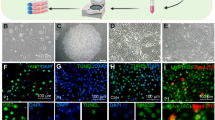
To investigate whether adult CD9-positive ependymal cells isolated from the third ventricle had neurospherogenic properties, we performed floating neurosphere culture. We observed that CD9-positive cells showed neurosphere-forming ability (Fig. 6a), whereas the CD9-negative cell fraction did not (Fig. 6b). The numbers of neurospheres per well are shown in Fig. 6c. Neurospheres formed by CD9-positive ependymal cells could propagate to form first- (Fig. 6d), second- (Fig. 6e), and third-passaged (Fig. 6f) spheres. To confirm whether an undifferentiated state was maintained, we performed immunocytochemistry for CD9 plus SOX2, NeuN, GFAP, or RIP after cultivation of neurospheres on Matrigel for 1 day. We observed that most cells in neurospheres were positive for CD9 and SOX2 (Fig. 6g–i), and some cells were positive for GFAP (Fig. 6j–l), but not NeuN and RIP (data not shown). After withdrawing the growth factors from the culture medium on Matrigel, the cells in a part of neurospheres showed morphological changes (Fig. 7a, b) and were able to differentiate into all three main neural lineages, including cells positive for NeuN (neurons; Fig. 7c), GFAP (astrocytes; Fig. 7d), and RIP (oligodendrocytes; Fig. 7e). Most NeuN-, GFAP-, and RIP-positive cells after differentiation were immunonegative for CD9 on their membrane (data not shown). The proportions of NeuN-, GFAP-, and RIP-positive cells were 15.6%, 20.6%, and 12.5%, respectively (Fig. 7f).
Neurospherogenic potential in CD9-positve cells. a, b Bright-field images of CD9-positive (a) and -negative (b) cell fractions on floating neurosphere culture. Arrowheads indicate neurospheres. c The number of neuropheres in a well of CD9-positive (CD9+) and -negative (CD9-) cell fractions. **P < 0.01. d–f Bright-field images of neurospheres. The first- (d), second- (e), and third-passaged (f) neurospheres formed by CD9-positive cells are shown. g–i Double-immunostaining for CD9 (g) and SOX2 (h) in neurospheres. i Merged images of DAPI staining (blue) and double-immunostaining for CD9 (magenta, g) and SOX2 (green, h). j–l Double-immunostaining for CD9 (g) and GFAP (h). i Merged image of DAPI staining (blue) and double-immunostaining for CD9 (magenta, j) and GFAP (green, k)
Differentiation capacities of CD9-positive ependymal cells. a Bright-field images of neurospheres with morphological changes after cultivation on Matrigel for 7 days. Bar, 50 μm. b The proportion of neurospheres with morphological changes among all neurospheres (mean ± standard error of the mean, n = 4). c–e Merged image of DAPI staining and immunostaining for NeuN (c), GFAP (d), or RIP (e) after cultivation on 3D Matrigel. Arrowheads indicate the immunopositive cells. Bar, 50 μm. f The proportion of NeuN-, GFAP-, and RIP-positive cells after cultivation on 3D Matrigel (mean ± standard error of the mean, n = 3)
Discussion
In this study, we showed that the ependymal cells in the ventricle expressed Cd9 and succeeded in isolation of these cells using anti-CD9 monoclonal antibodies. Then, using isolated CD9-positive ependymal cells from the third ventricle of adult rats, we confirmed that the isolated CD9-positive ependymal cells, but not the CD9-negative cells, were neurospherogenic and had the capacity to differentiate into neurons, astrocytes, and oligodendrocytes.
Several groups have developed various methods to isolate ependymal cells from rodents. Moreover, the development of transgenic animals has allowed us to distinguish living cells in the ventricular zone and separate these cells from the brain by fluorescent-activated cell sorting (FACS) (Robins et al. 2013). In many cases, isolation of ependymal cells from the spinal cord and lateral ventricle wall of adult rodents has been carried out by cell labeling with magnetic antibodies for application in magnetic cell separation with beads or by cell labeling with fluorescent antibodies for application in FACS using antibodies against CD133 and CD24 (Coskun et al. 2008; Pfenninger et al. 2007; Pfenninger et al. 2011). In the current study, we described a new marker and simple method using the pluriBead-cascade cell isolation system to obtain ependymal cells from the hypothalamus of adult rats. This isolation system is based on antibody-mediated binding of cells to beads of different sizes and their isolation with sieves of different mesh sizes (Horiguchi et al. 2016; Pierzchalski et al. 2013). Additionally, this isolation strategy does not require specialized equipment, as is needed for FACS analysis. Thus, this method is simpler than other previously described methods.
CD9 is a member of the tetraspanin family; accordingly, CD9 has four putative membrane spanning domains and functions as a key player in many physiological and pathological processes by associating with a various cell-surface molecules (Le Naour et al. 2000). Through these interactions, CD9 modifies multiple cellular events, including fertilization, cellular adhesion, motility, and tumor invasion (Berditchevski 2001; Boucheix and Rubinstein 2001; Hemler 2003; Maecker et al. 1997). In our previous study, we showed that CD9 in S100β-positive cells of the anterior pituitary gland contributes to sustaining proliferation activity (Horiguchi et al. 2018). Moreover, some ependymal cells are also known to have proliferative activity (Hamilton et al. 2009; Robins et al. 2013). In this study, we showed that CD9-positive cells in the third ventricle were immunopositive for Ki67. Similar to our observation in this study that D9-positive cells in the third ventricle were immunopositive for Ki67, Hemler et al. reported that the related tetraspanin, CD81, regulates cell motility and proliferation by facilitating the organization of multimolecular membrane complexes, including CD9 and integrins (Hemler 2005). CD9/CD81 double-knockout mice exhibit reduced proliferation, elevated apoptosis, and reduced lung, bone, spleen, testis, and pituitary development (Jin et al. 2018). CD9 may also play a role in maintaining the proliferative activity of ependymal cells during nervous system development. Although Cd9 was not expressed in the region containing β-tanycytes in the current study, β-tanycytes have been shown to proliferate well and are neurogenic, generating new neurons in the hypothalamic nuclei during the postnatal/juvenile period (Lee et al. 2012). Robins et al. (2013) demonstrated that FGF signaling is necessary for α-tanycyte proliferation and neurosphere formation, whereas β-tanycytes are unable to form neurospheres. Recently, CD9 was shown to be a new binding partner for junctional adhesion molecules and integrins, mediating FGF-regulated mitogen-activated protein kinase activation (Peddibhotla et al. 2013). Based on these findings, we suggest that CD9 expression in α1- and α2-tanycytes may be correlated with self-renewing neurospheres through FGF signaling. β-Tanycytes line the median eminence, α2-tanycytes reside adjacent to the arcuate nucleus, and α1-tanycytes extend from the ventromedial nucleus to the dorsomedial nucleus (Robins et al. 2013). Recent studies have reported that adult α2-tanycytes residing adjacent to the arcuate nucleus have the capacity to act as neural stem cells (Robins et al. 2013) and β-tanycytes lining the median eminence can proliferate in early adulthood and differentiate into new neurons (Haan et al. 2013). Thus, pure primary cultures of β-, α2-, and α1-tanycytes must be isolated from the hypothalamus of adult animals. However, at this time, we have not been able to examine neural function. Our novel method to enrich α-tanycytes described in the current study may encourage further in vitro studies and provide insights into the functions of these cells.
In conclusion, we found that Cd9 was expressed in cells in the region containing α2- and α1-tanycytes in the third ventricle. In addition, we succeeded in isolating CD9-positive ependymal cells with 86% purity utilizing anti-CD9 antibodies and the pluriBead-cascade cell isolation system. Finally, we confirmed that neurogenic and gliogenic stem/progenitor cells were included in CD9-positive ependymal cells. These findings improve our understanding of the adult neural niche in the third ventricles.
References
Berditchevski F (2001) Complexes of tetraspanins with integrins: more than meets the eye. J Cell Sci 114(Pt 23):4143–4151
Boucheix C, Rubinstein E (2001) Tetraspanins. Cellular and molecular life sciences : CMLS 58(9):1189–1205. https://doi.org/10.1007/pl00000933
Coskun V, Wu H, Blanchi B, Tsao S, Kim K, Zhao J, Biancotti JC, Hutnick L, Krueger RC Jr, Fan G, de Vellis J, Sun YE (2008) CD133+ neural stem cells in the ependyma of mammalian postnatal forebrain. Proc Natl Acad Sci U S A 105(3):1026–1031. https://doi.org/10.1073/pnas.0710000105
Del Bigio MR (2010) Ependymal cells: biology and pathology. Acta Neuropathol 119(1):55–73. https://doi.org/10.1007/s00401-009-0624-y
Feng R, Wen J (2015) Overview of the roles of Sox2 in stem cell and development. Biol Chem 396(8):883–891. https://doi.org/10.1515/hsz-2014-0317
Fujiwara K, Maekawa F, Kikuchi M, Takigami S, Yada T, Yashiro T (2007) Expression of retinaldehyde dehydrogenase (RALDH)2 and RALDH3 but not RALDH1 in the developing anterior pituitary glands of rats. Cell Tissue Res 328(1):129–135. https://doi.org/10.1007/s00441-006-0345-7
Goodman T, Hajihosseini MK (2015) Hypothalamic tanycytes-masters and servants of metabolic, neuroendocrine, and neurogenic functions. Front Neurosci 9:387. https://doi.org/10.3389/fnins.2015.00387
Haan N, Goodman T, Najdi-Samiei A, Stratford CM, Rice R, El Agha E, Bellusci S, Hajihosseini MK (2013) Fgf10-expressing tanycytes add new neurons to the appetite/energy-balance regulating centers of the postnatal and adult hypothalamus. J Neurosci 3(14):6170–80. https://doi.org/10.1523/JNEUROSCI.2437-12.2013.
Hamilton LK, Truong MK, Bednarczyk MR, Aumont A, Fernandes KJ (2009) Cellular organization of the central canal ependymal zone, a niche of latent neural stem cells in the adult mammalian spinal cord. Neuroscience 164(3):1044–1056. https://doi.org/10.1016/j.neuroscience.2009.09.006
Hemler ME (2003) Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu Rev Cell Dev Biol 19:397–422. https://doi.org/10.1146/annurev.cellbio.19.111301.153609
Hemler ME (2005) Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol 6(10):801–811. https://doi.org/10.1038/nrm1736
Horiguchi K, Nakakura T, Yoshida S, Tsukada T, Kanno N, Hasegawa R, Takigami S, Ohsako S, Kato T, Kato Y (2016) Identification of THY1 as a novel thyrotrope marker and THY1 antibody-mediated thyrotrope isolation in the rat anterior pituitary gland. Biochem Biophys Res Commun 480(2):273–279. https://doi.org/10.1016/j.bbrc.2016.10.049
Horiguchi K, Fujiwara K, Yoshida S, Nakakura T, Arae K, Tsukada T, Hasegawa R, Takigami S, Ohsako S, Yashiro T, Kato T, Kato Y (2018) Isolation and characterisation of CD9-positive pituitary adult stem/progenitor cells in rats. Sci Rep 8(1):5533. https://doi.org/10.1038/s41598-018-23923-0
Jin Y, Takeda Y, Kondo Y, Tripathi LP, Kang S, Takeshita H, Kuhara H, Maeda Y, Higashiguchi M, Miyake K, Morimura O, Koba T, Hayama Y, Koyama S, Nakanishi K, Iwasaki T, Tetsumoto S, Tsujino K, Kuroyama M, Iwahori K, Hirata H, Takimoto T, Suzuki M, Nagatomo I, Sugimoto K, Fujii Y, Kida H, Mizuguchi K, Ito M, Kijima T, Rakugi H, Mekada E, Tachibana I, Kumanogoh A (2018) Double deletion of tetraspanins CD9 and CD81 in mice leads to a syndrome resembling accelerated aging. Sci Rep 8(1):5145. https://doi.org/10.1038/s41598-018-23338-x
Le Naour F, Rubinstein E, Jasmin C, Prenant M, Boucheix C (2000) Severely reduced female fertility in CD9-deficient mice. Science (New York, NY) 287(5451):319–321
Lee DA, Bedont JL, Pak T, Wang H, Song J, Miranda-Angulo A, Takiar V, Charubhumi V, Balordi F, Takebayashi H, Aja S, Ford E, Fishell G, Blackshaw S (2012) Tanycytes of the hypothalamic median eminence form a diet-responsive neurogenic niche. Nat Neurosci 15(5):700–702. https://doi.org/10.1038/nn.3079
Maecker HT, Todd SC, Levy S (1997) The tetraspanin superfamily: molecular facilitators. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 11(6):428–442
Millhouse OE (1971) A Golgi study of third ventricle tanycytes in the adult rodent brain. Zeitschrift fur Zellforschung und mikroskopische Anatomie (Vienna, Austria : 1948) 121 (1):1–13
Ohata S, Alvarez-Buylla A (2016) Planar organization of multiciliated ependymal (E1) cells in the brain ventricular epithelium. Trends Neurosci 39(8):543–551. https://doi.org/10.1016/j.tins.2016.05.004
Pastrana E, Silva-Vargas V, Doetsch F (2011) Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell 8(5):486–498. https://doi.org/10.1016/j.stem.2011.04.007
Peddibhotla SS, Brinkmann BF, Kummer D, Tuncay H, Nakayama M, Adams RH, Gerke V, Ebnet K (2013) Tetraspanin CD9 links junctional adhesion molecule-A to alphavbeta3 integrin to mediate basic fibroblast growth factor-specific angiogenic signaling. Mol Biol Cell 24(7):933–944. https://doi.org/10.1091/mbc.E12-06-0481
Pfenninger CV, Roschupkina T, Hertwig F, Kottwitz D, Englund E, Bengzon J, Jacobsen SE, Nuber UA (2007) CD133 is not present on neurogenic astrocytes in the adult subventricular zone, but on embryonic neural stem cells, ependymal cells, and glioblastoma cells. Cancer Res 67(12):5727–5736. https://doi.org/10.1158/0008-5472.can-07-0183
Pfenninger CV, Steinhoff C, Hertwig F, Nuber UA (2011) Prospectively isolated CD133/CD24-positive ependymal cells from the adult spinal cord and lateral ventricle wall differ in their long-term in vitro self-renewal and in vivo gene expression. Glia 59(1):68–81. https://doi.org/10.1002/glia.21077
Pierzchalski A, Mittag A, Bocsi J, Tarnok A (2013) An innovative cascade system for simultaneous separation of multiple cell types. PLoS One 8(9):e74745. https://doi.org/10.1371/journal.pone.0074745
Rizzoti K, Lovell-Badge R (2017) Pivotal role of median eminence tanycytes for hypothalamic function and neurogenesis. Mol Cell Endocrinol 445:7–13. https://doi.org/10.1016/j.mce.2016.08.020
Robins SC, Stewart I, McNay DE, Taylor V, Giachino C, Goetz M, Ninkovic J, Briancon N, Maratos-Flier E, Flier JS, Kokoeva MV, Placzek M (2013) alpha-Tanycytes of the adult hypothalamic third ventricle include distinct populations of FGF-responsive neural progenitors. Nature communications 4:2049. doi:https://doi.org/10.1038/ncomms3049
Rodriguez EM, Blazquez JL, Pastor FE, Pelaez B, Pena P, Peruzzo B, Amat P (2005) Hypothalamic tanycytes: a key component of brain-endocrine interaction. Int Rev Cytol 247:89–164. https://doi.org/10.1016/s0074-7696(05)47003-5
Silva-Vargas V, Crouch EE, Doetsch F (2013) Adult neural stem cells and their niche: a dynamic duo during homeostasis, regeneration, and aging. Curr Opin Neurobiol 23(6):935–942. https://doi.org/10.1016/j.conb.2013.09.004
Yoshida S, Nishimura N, Ueharu H, Kanno N, Higuchi M, Horiguchi K, Kato T, Kato Y (2016) Isolation of adult pituitary stem/progenitor cell clusters located in the parenchyma of the rat anterior lobe. Stem Cell Res 17(2):318–329. https://doi.org/10.1016/j.scr.2016.08.016
Acknowledgments
We would like to thank Editage (www.editage.jp) for English language editing.
Funding
This work was supported by the JSPS KAKENHI (grant nos. 16K08475 to K.H., 21380184 to Y.K., and 24580435 to T.K.), by a MEXT-supported Program for the Strategic Research Foundation at Private Universities (2014–2018), and by the Meiji University International Institute for BioResource Research (MUIIR).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
The current study was approved by the Committee on Animal Experiments of the School of Agriculture, Meiji University, and Kyorin University based on the NIH Guidelines for the Care and Use of Laboratory Animals. This article does not contain any studies with human participants.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Horiguchi, K., Yoshida, S., Hasegawa, R. et al. Isolation and characterization of cluster of differentiation 9-positive ependymal cells as potential adult neural stem/progenitor cells in the third ventricle of adult rats. Cell Tissue Res 379, 497–509 (2020). https://doi.org/10.1007/s00441-019-03132-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-019-03132-5