Abstract
Differentiation of mesenchymal stem cells (MSCs) into cardiomyocytes is a complex phenomenon, and attempts to find an effective inducing agent are still ongoing. We studied the effect of fibrin scaffold and sodium valproate (VPA, as a histone deacetylase inhibitor) on the differentiation of human adipose-derived stem cells (hADSCs) into cardiomyocyte-like cells. The cells were cultured in culture flask (2D) and in fibrin scaffold (3D), fabricated of human plasma fibrinogen, with and without VPA (1 mM). QRT-PCR, Western blot, and immunochemistry assays were used to evaluate the expression of cardiac markers at gene and protein levels. High levels of CD44, CD90, CD73, and CD105 were expressed on the surface of hADSCs. Treated encapsulated hADSCs (3D) presented significantly higher mRNA expression of HAND1 (1.54-fold), HAND2 (1.59-fold), cTnI (1.76-fold), MLC2v (1.4-fold), Cx43 (1.38-fold), βMHC (1.34-fold), GATA4 (1.48-fold), and NKX2.5 (1.66-fold) in comparison to 2D conditions at four weeks after induction. The protein expressions of NKX2.5 (0.78 vs 0.65), cTnI (1.04 vs 0.81), and Cx43 (1.11 vs 1.08) were observed in the differentiated cells both in 3D and 2D groups, while control cells were absolutely negative for these proteins. The frequency of cTnI and Cx43-positive cells was significantly higher in 3D (24.2 ± 15 and 12 ± 3%) than 2D conditions (19.8 ± 3 and 10 ± 2%). Overall, the results showed that VPA can increase cardiomyogenesis in hADSCs and that fibrin scaffold enhances the inductive effect of VPA. Results of this study may improve cell-based protocols for implementation of more successful cardiac repair strategies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Despite all efforts in scientific research and medical treatment, ischemic heart disease and myocardial infarction (MI) remain the leading causes of death worldwide (Mozaffarian et al. 2015). Although current interventions and pharmacologic treatments have been relatively successful, many patients are still suffering from irreversible cardiac damage due to lack of regenerative therapies of the cardiac muscle. The prevalence of congestive heart failure is increasing, which highlights the need for new treatment approaches to effectively regenerate cardiac injury (Choi et al. 2010; Lemcke et al. 2018). Among different strategies, cell therapy with mesenchymal stem cells (MSCs) created new hopes for the improvement of heart damages (Chen et al. 2015; Yang et al. 2013). In the meantime, human adipose-derived stem cells (hADSCs) are becoming one of the suitable sources of adult stem cells due to subcutaneous localization and easy collection in large quantities, easy extraction of stem cells and cultivation, and ability to combine with biomaterials and differentiate into cells of vital tissues, including cardiomyocyte-like cells, which have attracted considerable attention to the development of regenerative medicine (Grayson et al. 2008; Otsuki et al. 2018; Sukho et al. 2018; Tobita et al. 2011; Varghese et al. 2017). Meanwhile, some studies have shown that ADSCs are not differentiated well into cardiomyocyte-like cells in vivo (Gaustad et al. 2004; Planat-Benard et al. 2004; Rangappa et al. 2003). Use of ADSCs instead of cardiac-committed cells and lack of a suitable environment for supporting cell adhesion to keep the cells alive are among possible causes of this failure (Choi et al. 2010; Li et al. 2007; Xu et al. 2004; Zeng et al. 2011). Alternative approaches are using cardiac-inducing factors (chemical agents, hypoxia, and mechanical stimulation) and cardiac tissue engineering by biomaterials (Choi et al. 2010; Girão-Silva et al. 2014). The modification of structure and organization of chromatin is one of the regulatory mechanisms for differentiation of stem cells. The change in the histone structure can modify the accessibility and binding of transcription factors to DNA. Acetylation is one of the most important pro-differentiation changes of the histone that occurs by histone acetyltransferases (HATs) while the histone deacetylases (HDACs) cause condensation of the chromatin structure and reduction of transcriptional activity of the genes. Therefore, histone deacetylase inhibitors (HDACIs) may increase acetylation of the histone. VPA is a powerful HDACI, probably capable to mediate stem cell differentiation in the early stages (Li et al. 2014; Rice et al. 2007; Gurvich et al. 2004). In cardiac engineering in vitro, this differentiation can be induced by seeding the cells in scaffolds with a suitable composition, architecture, and mechanical properties, such as fibrin scaffold (Bagheri-Hosseinabadi et al. 2018). Albeit according to current knowledge, the exact mechanism and signaling pathways that mediate differentiation of stem cells into cardiomyocyte-like cells are not fully understood. Therefore, the purpose of this study was to investigate and compare the effect of VPA as an inducer, on cardiomyocyte genesis of hADSCs in 2D culture medium and 3D fibrin scaffold conditions.
Materials and methods
Ethical approval was obtained from the Institutional Ethical Review Board at Kerman University of Medical Sciences, Kerman, Iran (approval no IR.KMU.REC.1395-961).
Materials
Many of the materials were prepared from Sigma Company (Sigma-Aldrich, St. Louis, MO, USA), but the FITC-conjugated antibodies against CD45, CD44, and CD34 were purchased from Chemicon Co., USA, and PE-conjugated CD105, CD90, and CD73 were from BD Biosciences. Chondrogenic differentiation kit was purchased from GIBCO, Invitrogen, USA; LDH kit from Pars-Azma, Iran; RNX-Plus kit total RNA isolation from CinnaGen, Iran; RevertAid cDNA synthesis kit and SYBR Green from Thermo Fisher Scientific Inc., MA, USA; protein marker E2912 and chemiluminescence reagent (ECL) from Santa Cruz, USA; rabbit polyclonal antibody against cTnI, mouse monoclonal antibodies against NK2 homeobox 5 (NKX2.5) and connexin 43, goat anti-rabbit IgG H&L (HRP), and goat anti-mouse IgG H&L (HRP) from Abcam Company; and β-actin (rabbit polyclonal) from Biorbyt.
Methods
Preparation of hADSCs
Isolation and culture of hADSCs were performed as previously reported (Bagheri-Hosseinabadi et al. 2018). Briefly, the paravertebral adipose tissues were obtained from three healthy volunteer women (aged 30–50 years) who underwent elective liposuction surgery. A written consent form was obtained to use their lipoaspirate tissues for research purposes. The tissues were washed with phosphate-buffered saline (PBS) solution containing penicillin (300 U/ml), streptomycin (100 μg/ml), and amphotericin-B (5 μg/ml). Tissues were gently shaken for 60 min in DMEM/F12 containing 1% collagenase type IV at 37 °C. The enzyme activity was neutralized by PBS containing 20% fetal bovine serum (FBS). After centrifuging and separation, hADSCs were cultured in DMEM/F12 supplemented with 10% (v/v) FBS, penicillin (100 U/ml), streptomycin (100 μg/ml), and amphotericin-B (2.5 μg/ml) in a humidified incubator at 37 °C in an atmosphere of 5% CO2. On the next day, the medium was removed and the cells were washed by PBS and replenished with fresh completed medium every 72 h until they reached confluence. For future experiments, the hADSCs were expanded until passage 4.
Phenotypic characteristics of hADSCs
Flow cytometric analysis of the cells was used for the detection of hADSC-specific surface markers (CD90, CD73, CD105, CD44, CD34, and CD45). A total of 1.0 × 105 hADSCs/ml were blocked with a 1:9 dilution of normal goat serum in PBS at 4 °C for 15 min, and then the cells were washed and re-suspended in cold PBS. In the following step, the blocked hADSCs were incubated with fluorochrome-labeled marker-specific antibodies for 60 min at 4 °C in darkness. Fluorochrome-labeled isotype antibodies (mouse IgG monoclonal isotype standards) were also used for staining of the control cells. After washing with PBS (containing 2% FBS), flow cytometry analyses using BD FACS Calibur (Becton-Dickinson, USA) were used for data acquisition, and WinMDI software (BD Biosciences, USA) was used for further data analyses (Latifpour et al. 2011).
Induction of osteogenic, adipogenic, and chondrogenic differentiation
Differentiation potential of the hADSCs into adipocytes, osteoblasts, and chondrocytes was examined by the following methods for confirming their stemness potential.
Osteogenic differentiation
hADSCs were seeded onto plates at a density of 3.0 × 103 cells/cm2 in the complete medium. On the next day, osteogenic medium containing DMEM-LG supplemented with 10% FBS, penicillin (100 U/ml), streptomycin (100 μg/ml), amphotericin-B (2.5 μg/ml), glycerophosphate 10 mM, dexamethasone 10 nM, and 50 μg/ml ascorbic acid was added to the cells. For Alizarin Red staining, the osteogenic medium was removed after three weeks, and the cells were washed with PBS and then fixed with 4% paraformaldehyde. Alizarin Red staining was performed according to the established protocol reported in the published paper (Bagheri-Hosseinabadi et al. 2018). The stained cells were observed by an inverted optical microscope (Olympus, Japan), and the images were recorded for further analysis.
Adipogenic differentiation
To direct hADSCs towards adipogenic differentiation, 3.0 × 104 hADSCs/ml were seeded in plates. After 24 h, the medium was replaced by adipogenic differentiation medium (DMEM/F12 containing 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 2.5 μg/ml amphotericin-B, 100 nM dexamethasone, and 200 nM indomethacin). After three weeks, the induction medium was aspirated and the cells were fixed in 4% paraformaldehyde and stained with Oil Red O solution (Bagheri-Hosseinabadi et al. 2018). The formation of lipid droplets was observed by a phase-contrast microscope.
Chondrogenic differentiation
A total of 3.0 × 104 hADSCs/ml were seeded in the plates. On the next day, the cultured cells were exposed to chondrogenic differentiation kit (according to the manual). After three weeks, Alcian blue staining was performed for quantification of sulfated proteoglycans (Bagheri-Hosseinabadi et al. 2018). The images were prepared using an invert microscope (Olympus, Japan).
Fabrication of fibrin scaffold, cell encapsulation, and analysis of hADSC morphology
The protocol for fabrication of fibrin scaffold has been reported in our previous works (Bagheri-Hosseinabadi et al. 2018, 2017). In brief, fibrinogen was extracted from human fresh-frozen plasma (FFP) by adding equal volume of protamine sulfate. After centrifuging, the precipitated fibrinogen was dissolved using 0.1 M sodium citrate. In order to use the fibrinogen solution in cell culture, it had to be filtered through a 0.2-μm mesh filter. For seeding cells in fibrin scaffold, the suspension composed of 1.0 × 105 hADSCs, 50 μl CaCl2 (50 mM), and 50 μl thrombin solution (40 IU/ml) was transferred to a 5 ml syringe containing 200 μl of fibrinogen solution and allowed to polymerize in CO2 incubator at 37 °C for 10 min. After ensuring that the gel had been formed, each of the scaffold was put in a single well of a 6-well plate containing 2 ml DMEM/F12 supplemented with 10% FBS, 1% of the aforementioned antibiotic, and 2% tranexamic acid (as an anti-fibrinolytic agent), which were incubated in a humidified incubator at 37 °C in an atmosphere of 5% CO2 for appropriate times.
Fibrin scaffold microstructure was evaluated by a scanning electron microscope (SEM, AIS 2100, Seron Technology, South Korea) at an accelerating voltage of 20 kV. At first, the samples were washed by PBS and then fixed by 3% glutaraldehyde. Subsequently, the samples were immediately immersed in liquid nitrogen and dried for 24 h in a freeze dryer. Dried fibrin scaffolds were mounted onto gold using a sputter-coated (SC7620, Emitech, UK) for 160 s. Images of fibrin scaffold were captured, and the fiber diameter was determined by ImageJ software (NIH, USA).
For analysis of hADSCs encapsulated into the fibrin scaffold, after seven days of culture of 2.0 × 105 cell/ml in the fibrin scaffold, the samples were stained with Hoechst staining to observe hADSC attachment and grow in the hydrogel. For 250 μl Hoechst solution (5 μg/ml, 33258) was added to the scaffolds and incubated for 30–45 min at 37 °C. Next, the staining solution was removed from the scaffolds, and, after washing by PBS, the hADSCs were observed using a phase-contrast fluorescence microscope.
Effect of sodium valproate on hADSCs
Literature review revealed dose-dependent cytotoxic effects of VPA on mesenchymal stem cells (Kwiecińska et al. 2012; Grabarska et al. 2014). To investigate the optimum dose, hADSCs were cultured in the presence of six concentrations of VPA (0, 0.5, 1, 2, 5, 10 mM) for 24 h. At the end of the incubation period, the metabolic activity (as index of cell viability) was measured using Water-Soluble Tetrazolium salt (WST-1) assay. The absorbance of wells was measured at 450 nm with 630 nm as a reference wavelength. Acellular wells were used as blanks. Cell viability rate in each well was exhibited as a percent to the control well (without VPA) absorbance (see Bagheri-Hosseinabadi et al. 2018 for details). In the similar culture conditions, the rate of cell injury due to 24 h VPA exposure was measured in the cell culture media using LDH release assay. LDH activity was measured as an index of LDH release according to the details published in Bagheri-Hosseinabadi et al. (2018).
Induction of cardiac differentiation
For cardiac differentiation of hADSCs, 1.0 × 105 cells/ml were first seeded in both cell culture flask and fibrin scaffolds. In the 2D group, one day after seeding and in 3D group, 7 days after encapsulating of the cells, completed medium was removed and the cells were directed towards cardiomyogenic phenotype using specific media containing DMEM/F12 without FBS, supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml), amphotericin-B (2.5 μg/ml), and two selected concentrations of sodium valproate (0.5 and 1 mM), based on the results of WST-1 cell viability test and LDH release assay (see Fig. 4). In the following day in both groups, the cardiomyogenic medium was replaced by DMEM/F12 containing 10% FBS and 1% antibiotics, while in the 3D group, 2% tranexamic acid, an anti-fibrinolytic agent, was also added to the medium. Thereafter, the medium was renewed every three days until day 28. In the 2D group, morphological changes were evaluated using an inverted phase-contrast microscope regularly. In the fibrin hydrogel scaffolds (3D), this could not be performed due to the opacity of the fibrin gel, which prevented the cell visibility through the inverted microscope. Independent negative control groups (VPA untreated cells) were used for 2D and 3D cultures.
Analysis of cardiogenic gene expressions by qRT-PCR
Relative mRNA expressions of HAND1, HAND2, GATA4, NKX2.5, MLC2v, βMHC, cardiac troponin I (cTnI), and connexin 43 (Cx43) genes in hADSCs treated with 1 mM VPA and untreated hADSC samples in 2D and 3D groups at the fourth week past induction were assessed by qRT-PCR in comparison to the housekeeping gene, GAPDH. QRT-PCR was performed as reported previously (Bagheri-Hosseinabadi et al. 2018). Simply, the total RNA was isolated by a precipitation method using RNX-Plus total RNA isolation kit, according to the manufacturer’s protocol. For evaluation of quantity and quality of total RNA extracts, NanoDrop and agarose gel electrophoresis were used, respectively. Then, 100 ng/μl DNase-treated total RNA was reverse-transcribed into cDNA using RevertAid cDNA synthesis kit. Quantitative RT-PCR was performed with SYBR Green according to the manufacturer’s instructions; using a Rotor-Gene™ 6000 (Corbett Research, Australia). Untreated hADSCs, cultured in the absence of VPA, incubated in cell culture flask or in fibrin scaffold, were considered as negative control groups. Human embryonic heart tissue was used as positive control (with parental permission). The primer sequences used for qRT-PCR are indicated in Table 1. GAPDH cDNA was also used as internal control. All experiments in 2D and 3D groups were repeated independently at least three times. Raw data were analyzed with REST Relative Quantification Software version 2.2.3 (Qiagen, Inc.). The fold changes in expression of the target genes among the treated, positive, and negative control groups were calculated using the 2−ΔΔCT equation.
Analysis of cardiogenic protein expressions by Western blot
For analyzing cardiac differentiation rate of hADSCs, the expression of cardiac-specific proteins, including NKX2.5, cTnI, and Cx43, was determined by Western blot at the end of the fourth week past induction. Total protein was extracted from untreated hADSCs (negative controls), human embryonic tissue (positive control), and treated hADSCs (2D and 3D groups initially treated with 1 mM VPA) by a RIPA lysis buffer containing protease inhibitor cocktail. The protein content of the supernatants was determined using Bradford’s protein assay. Then, gel electrophoresis was carried out. For this purpose, approximately 25 μg of total protein from cell lysates and protein marker were loaded on the 12% polyacrylamide gel and transferred onto a PVDF membrane. In the next step, PVDF membranes were washed and blocking solution (5% BSA in PBST) was added. Then, the membranes were subjected to the primary antibodies, including, rabbit polyclonal antibody against cTnI (diluted at 1:10,000) and mouse monoclonal antibodies against NKX2.5 and connexin 43 (diluted at 1:10,000) for 12 h. Then, the blot membranes were washed and reacted with the specific secondary antibodies (goat anti-rabbit IgG H&L (HRP) and goat anti-mouse IgG H&L (HRP) (diluted at 1:10,000) for 60 min. Afterwards, the blot membranes were rinsed by PBST, and, subsequently, immunoreactive bands (the antigen–antibody complex) were visualized via chemiluminescence reagent. For imaging the blots, an appropriate fluorescence scanner (BioRad, Germany) was used. Finally, the Western blots were quantified by ImageJ software. β-actin (rabbit polyclonal, 1:200) was applied for normalization of all the proteins.
Analysis of cardiogenic proteins by immunocytochemistry
The expression of cTnI and connexin 43 proteins in 3D and 2D cultured cells was determined by immunocytochemistry. At the end of the fourth week, fibrin scaffold encapsulated and plate-cultured cells from treated and untreated groups were fixed using 10% neutral buffered formalin for 24 h and 4% paraformaldehyde solution for 30 min, respectively. In the 3D group, additional steps, including embedding culture slices in paraffin blocks, cutting into sections, mounting on glass slides, and rehydration of the sections were performed. All cells were permeabilized using 0.2% Triton X-100 in PBS at RT for 20 min and then incubated with blocking buffer (5% goat serum, 0.01% Triton X-100, 2% BSA in PBS) at RT for 45 min. Primary antibody (rabbit polyclonal antibody against cardiac troponin I and mouse monoclonal antibody against connexin 43) diluted in PBS (1:100) was added, and the cells were incubated overnight at 4 °C. After washing (3 times for 2D and 6 times for 3D samples), the cells were incubated with secondary antibody (goat anti-rabbit IgG, diluted in PBS to 1:200) at RT for 1 h. Washing step was repeated, and, finally, the samples were stained with Hoechst for 10 min and observed by an invert fluorescence microscope (Olympus, Japan). The percentage of cTnI and connexin 43-positive cells was determined in three sections for each group.
Statistical analysis
The results are presented as mean ± standard deviation (SD) of at least three replicated measurements. Two independent-sample t test and one-way ANOVA followed by Tukey’s post-hoc test were used to compare the results after normality assumption was checked. Statistically significant level was set to P < 0.05.
Results
Phenotypic characteristics of hADSCs
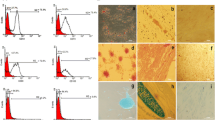
Figure 1 shows the results of flow cytometric analyses of the cell surface markers. High levels of mesenchymal-specific cell surface markers, CD44, CD90, CD73, and CD105 (Fig. 1a–d), were expressed on the surface of the hADSCs, whereas, hematopoietic cell markers, CD34 (Fig. 1e) and CD45 (Fig. 1f), were absent.
Induction of osteogenic, adipogenic, and chondrogenic differentiation
Adipose-derived stem cells usually have the ability to differentiate into multiple lineages. Differentiation of hADSCs into osteocytes (Fig. 2a), adipocytes (Fig. 2b), and chondrocytes (Fig. 2c) was induced using lineage-specific differentiation culture conditions.
(a) Alizarin Red, (b) Oil Red O, and (c) Alcian blue staining implied the presence of (a) calcium deposits, (b) intracellular lipid droplets, and (c) sulfated proteoglycans in the differentiated cells, indicating osteogenic, adipogenic, and chondrogenic potentials of hADSCs, respectively, after implementing proper inducing processes. Scale bars are 40 μm in (a) and 100 μm in (b) and (c)
Structure of fibrin scaffold
Detailed surface morphology of the cell-free fibrin scaffold was provided using scanning electron microscopy in which the fibrin scaffold had an interconnected porous structure with a pore size of 70–145 μm (Fig. 3a).
(a) SEM image of fabricated fibrin scaffold showing an interconnected porous structure with pore sizes ranging from 70 to 145 μm. (b–d) Localization of hADSCs on fibrin scaffold. (b) Light microscopy of fibrin scaffolds with phase-contrast microscope. (c) Human adipose-derived stem cells nuclei stained by Hoechst appeared blue. (d) Localization of the hADSCs in fibrin scaffold is shown by merging panels (b) and (c). Scale bars are 100 μm in (a) and 200 μm in (b–d)
Encapsulation of hADSCs into fibrin scaffold
Fluorescent dye Hoechst staining shows the attachment and distribution of hADSCs in the fibrin scaffold. As illustrated in Fig. 3b–d, seven days after encapsulation, seeded hADSCs efficiently attached to the periphery of the pores. Staining with the DNA binding dye confirmed that hADSCs loaded into fibrin hydrogel scaffolds attached and grew on the scaffolds.
Effects of sodium valproate on hADSCs
Viability rates of hADSCs exposed to different concentrations of VPA reduced, in a linear fashion, to 92, 87, 70, 40, and 2%, respectively (Fig. 4a). Calculated 50% inhibitory concentration (IC50) of VPA on hADSCs was 3.91 mM. LDH activity of hADSCs exposed to LDH activity was measured according to the published protocol (Bagheri-Hosseinabadi et al. 2018). The same concentrations of VPA increased to 21, 25, 33, 52, and 77%, respectively (Fig. 4b). Calculated half-maximal effective concentration (EC50) of VPA on LDH release was 4.62 mM. The results of the two tests showed a reverse correlation between viability rate of cells and LDH activity of media (R: 99.9). This implies that a high proportion of the metabolically inactive cells goes into necrotic cell death.
Concentration-related responses of hADSCs exposed to sodium valproate (exposure time 24 h). (a) WST-1 cell viability assay showing viability of hADSCs exposed to sodium valproate relative to the control wells not exposed to the agent (0 conc. column). (b) LDH enzyme release from the sodium valproate exposed hADSCs into the culture medium, as a marker of cell toxicity of VPA. LDH activity in culture supernatant was depicted as a percent related to the total LDH activity (sum of the activities of cells and medium). From these tests, the concentration of 1 mM was shown to be a safe and effective concentration. *P < 0.05, **P < 0.01, ***P < 0.001 compared to 0 concentration
Cell morphology
In the 0.5 mM VPA-treated group, no significant changes were observed in the cell morphology during four weeks after induction, compared to the negative control group (results not shown). Also, cell detachment was rare in this concentration. In the 1 mM treated group, morphological changes were observed during the 4th week as shown in Fig. 5. Ball-like, multinuclear, and fork-like cells appeared on day seven (Fig. 5b). On day 14, some of the cells become elongated and they also showed some angle-like projections in the periphery that gradually changed into myotube-like structure (Fig. 5c). On day 21 (Fig. 5d), myotube-like and star-like morphologies were developed that consistently preserved until the end of the study (Fig. 5e). Treatment of hADSCs with 1.5 mM sodium valproate for 24 h resulted in the detachment of more than 50% of the cells from the bed. However, morphological changes in the remaining cells were appeared faster than the 1 mM group (results not shown). Higher doses resulted in higher rates of cell detachment and/or cell death. Therefore, 1 mM dose was selected for the rest of the study for evaluation of morphological and molecular profiles. Proliferation capacity of cells was also affected by VPA. Initially, 1 × 105 cells were added to the flasks and the cells gradually proliferated and reached 70–80% of confluence during four weeks. Some replicates also showed colony formation potential (Fig. 5f).
Effect of 1 mM sodium valproate treatment on the morphology of cells. (a) Non-treated hADSCs indicated a fibroblast-like cell appearance. Panels (b)–(e) were taken in 2D groups after 1–4 weeks of stimulation by sodium valproate. Representative morphology of the induced cells is marked with arrows. 1, ball-like; 2, multinuclear; 3, fork-like; 4, elongated; 5, myotube-like structure; 6, star-like projections. Panels (g) and (h) show higher magnification of the related cells at arrow points in panels (b)–(e). Panel (f) shows a colony formed by proliferating cells. Scale bars are 100 μm in all images
QRT-PCR of cardiogenic genes
The effect of sodium valproate exposure (1 mM, for 24 h) on the cardiac differentiation of 2D and 3D cultured hADSCs after four weeks was determined through assessment of relative mRNA expression levels of eight cardiomyocyte-specific genes using qRT-PCR (Fig. 6a–h). VPA-treated hydrogel-encapsulated hADSCs had significantly higher mRNA expression levels (3D vs 2D) of HAND1 (5.62 vs 3.65, ~ 1.54-fold), HAND2 (4.67 vs 2.94, ~ 1.59-fold), cTnI (3.69 vs 2.1, ~ 1.76-fold), Cx43 (1.38 vs 1, ~ 1.38-fold), βMHC (1.81 vs 1.35, ~ 1.34-fold), GATA4 (4.17 vs 2.81, ~ 1.48-fold), NKX2.5 (3.16 vs 1.9, ~ 1.66-fold), and MLC2v (5.99 vs 4.28, ~ 1.4-fold) compared to the treated hADSCs in 2D cultures.
Analyses of mRNA expression levels of eight genes by qRT-PCR. Four weeks after 24 h sodium valproate 1 mM treatment, mRNA expression levels of eight genes (a) HAND1, (b) HAND2, (c) GATA4, (d) NKX2.5, (e) cTnI, (f) βMHC, (g) MLC2v, and (h) Cx43 in the hADSCs. GAPDH mRNA levels were used for normalization of the target genes. Most of the genes had significantly higher expression in 3D group than 2D group. Each of the 3D and 2D groups had its own negative control, but only data for 2D controls (−CTL) is presented here because the differences between the two negative controls (hADSCs, cultured in the absence of VPA) were at the range of ± 2%. The positive control (+CTL) was embryonic human heart tissue. *Comparison with negative control (−CTL). # Comparison between 3D and 2D groups. # and * = P < 0.05, ## and ** = P < 0.01, ### and *** = P < 0.001
Western blotting of cardiogenic proteins
Protein expression levels of NKX2.5, as an early cardiac gene marker, cTnI, as a cardiac-specific contractile protein, and, Cx43, as the most common cardiac gap junction channel protein, were examined in the hADSCs treated with 1 mM sodium valproate, compared to the negative control group. Western blot analyses of proteins at the 4th week showed that the expression levels (3D vs 2D) of NKX2.5 (0.78 vs 0.65) and Cx43 (1.11 vs 1.08) proteins were not significantly different (Fig. 7). However, the expression of cTnI (1.04 vs 0.81) was significantly higher in the 3D group (P < 0.05). Protein expression of all 3 genes in the treated groups (both 2D and 3D) was significantly higher compared to that of the untreated cultures (not detectable).
Western blot bands (a–c) and the quantified data (a′–c′) for expression of NKX2.5 (35 KDa), cTnI (26 KDa), and Cx43 (43 KDa) cardiac-specific proteins in hADSCs four weeks after 24 h sodium valproate 1 mM exposure. Untreated 2D control cells (Neg) did not express any of the three proteins. Only 2D negative control was presented in the figure because of very low differences between the two controls. Positive control (Pos) was embryonic human heart tissue.* = P < 0.05, ns, not significant
Immunocytochemistry of cardiac troponin I and connexin 43
Expression of three cardiac proteins was tested through Western blotting analysis (Fig. 7). cTnI as a mature cardiomyocyte marker and connexin 43 as a cell junction protein were assessed by immunostaining of the cardiac differentiated cells. As can be inferred from Figs. 8 and 9, 1 mM VPA exposure effectively induced the expression of cTnI and connexin 43 proteins in the hADSCs. For quantification, two hundred cells were observed under a light microscope and the percentage of cTnI and connexin 43-positive cells was determined. Frequency of cTnI-positive cells in fibrin scaffolds (3D) (24.2 ± 15%) was significantly higher than in cell culture flask (2D) (19.8 ± 3%) (P < 0.05) (Fig. 8). Frequency of Cx43-positive cells in fibrin scaffolds (3D) (12 ± 3%) was slightly higher than in 2D culture conditions (10 ± 2%). The two proteins were absent in the control cells. In support of observations in morphologic test (Fig. 5), multinuclei and star-like differentiated cells were obvious in the panel a″.
Sample images of immunofluorescence staining four weeks after induction of hADSCs by 1 mM VPA in the plate (2D) and fibrin scaffold (3D) using cTnI antibody (green) and Hoechst staining for nuclei (blue). Untreated panels are negative controls for the treated counterparts. Multinuclei and star-like cells are marked in the figure (panel a″) using red and yellow arrows, respectively. Scale bars are 100 μm in all images
Sample images of immunofluorescence staining for connexin 43 gap junction protein, four weeks after induction of hADSCs by 1 mM VPA in the plate (2D) and fibrin scaffold (3D) using connexin 43 antibody (appeared as red partially diffused dots) and Hoechst staining for nuclei (blue). Untreated panels are negative controls for treated counterparts. Scale bars are 100 μm in all images
Discussion
There are certain evidences that adipose-derived stem cells (ADSCs) have considerable similarities to cardiac cells and may easily differentiate into cardiomyocytes, if especial culture conditions prevailed (Bacakova et al. 2018). However, culture conditions necessary for inducing differentiation of ADSCs towards functional cardiomyocytes have not been fully understood. In the present study, sodium valproate (VPA) (1 mM, 24 h) was used as a differentiation-inducing agent to enforce differentiation of hADSCs towards cardiomyocyte-like cells. Sodium valproate-exposed hADSCs were cultured in fibrin scaffold, in parallel with control cultures, to undergo differentiation. After four weeks of incubation under optimal culture conditions, valproate-exposed hydrogel-encapsulated hADSCs expressed a high rate of cardiac-specific markers (including, HAND1, HAND2, cTnI, MLC2v, NKX2.5, and GATA4).
Acetylation of histones is one of the most important mechanisms that mediates regulation of gene expression, cell function, differentiation, and proliferation (Göttlicher et al. 2001). It has been already revealed that histone deacetylase (HDAC) activity is regulated by HDAC inhibitors (HDACIs) (Brookes et al. 2018; Vrba et al. 2011). Structurally, HDACIs with a molecular weight of less than 5000 may bind to the catalytic site of HDACs enzymes (Göttlicher et al. 2001) and promote cell apoptosis and inhibit cell growth and differentiation (Raut and Khanna 2016). Also, it has been reported that during various developmental stages, HDACIs contribute to and direct differentiation of stem cells into different types of cell lineages, such as hepatic, neuronal, and cardiomyocytic cells (Oyama et al. 2007; Raut and Khanna 2016; Yu et al. 2009). As an example, during early stages of cardiomyogenesis in P19 cells, HDACIs exert a regulatory role for transition of mesoderm cells into cardiomyocytes (Karamboulas et al. 2006); thereby, they play a key role in the cardiac development. Recently, it was revealed that TSA, an HDACI agent, promotes differentiation of hADSCs into cardiomyocyte-like cells and these cells express specific cardiac proteins and genes (Bagheri-Hosseinabadi et al. 2017). VPA administration, a commonly used HDACI agent, which inhibits different classes of HDAC enzymes, is associated with stroke risk level of patients with prior stroke or transient ischemic attack (Brookes et al. 2018).
In the present study, based on the molecular and morphological characteristics, it was demonstrated that VPA is an effective cardiac inducer in vitro and strongly enhances expression of cardiac-related markers (Figs. 6 and 7), and its accompaniment with fibrin scaffold synergistically promoted cardiac differentiation of hADSCs. Four important early cardiac transcription factors (NKX2.5, GATA4, HAND1, and HAND2) and four structural cardiac genes (cTnI, Cx43, MLC2v, and βMHC) that are known to be involved in cardiac differentiation and play outstanding roles in development and function of the heart tissue were overexpressed (Figs. 6 and 7). Immunofluorescence staining for connexin 43 demonstrated that in both 2D and 3D cultures, gap junctions have formed in the differentiated cells (Fig. 9). This served as another indication that the cardiac differentiation capability was real. These findings are consistent with some other studies (Hosseinkhani et al. 2007; Lim et al. 2013; Yang et al. 2012) that the expression of early and late cardiomyocyte markers has increased by HDACI in embryonic stem cells, human-induced pluripotent stem cells, and bone marrow mesenchymal stem cells. Li et al. (2014) showed that valproic acid increases differentiation of skeletal myocytes from pluripotent stem cells by enhancing histone acetylation (Li et al. 2014). They concluded that histone acetylation is an important prerequisite for myogenic differentiation. Akavia et al. (2008) showed that early transcription factors of GATA-4 and NKX2.5 are not expressed by skeletal muscle cells and are specific for cardiac muscle cells (Akavia et al. 2008). In the present study, VPA increased the expression of all the studied genes, including NKX2.5 and GATA4, in both 2D and 3D groups (Fig. 6), and most of the genes had significantly higher expression in the 3D group compared to the 2D group. For initiating cardiac differentiation, increasing the activity of Akt/GSK3β pathway is necessary (Cho et al. 2009). De Sarno et al. have shown that HDACIs like VPA activate this pathway through increasing the level of phosphorylated Akt/GSK3β. They also found phosphorylation as a slow process that requires optimal concentration and long incubation time (approximately 12 h) with VPA (De Sarno et al. 2002). In addition, the present study revealed that a large number of cardiomyocyte-like cells were observed in 3D culture condition with VPA compared to the 2D conditions. Also, the results of Western blot and immunofluorescence staining as well as qRT-PCR indicated that simultaneous usage of fibrin scaffold and VPA reinforced the potency of differentiation of hADSCs. Similar results have been presented by Lin et al. when they used fibrin scaffold and cardiac lysates for differentiation of human amniotic mesenchymal stromal cells (hAMSCs) into cardiomyogenic cells (Lin et al. 2013).
As usually most of the transplanted cells leave the heart, in the process of repair of the heart after an injury, keeping the injected cells in the damaged area is an important issue. Therefore, using a compatible scaffold is necessary for overcoming this problem. The arginine–glycine–aspartic acid (RGD) motifs in the fibrin scaffold allow cell attachment and increase cell survival and binding of growth factors for cardiac cell differentiation. Also, the proper mechanical strength of fibrin scaffold can improve differentiation of the cells into desired cell lineage (Bagheri-Hosseinabadi et al. 2018; Barsotti et al. 2011; Shachar et al. 2011). As the pore sizes of the scaffold control diffusion of nutrients, removing wastes, facilitating proliferation and migration of cells, and determining the final mechanical properties of the scaffold, in this study, the pore sizes of the scaffold used were assessed by SEM, and it was revealed that the fibrin scaffold had an interconnected porous structure with pore sizes of 70 to 145 μm (Fig. 3), which is suitable for these purposes (Loh and Choong 2013).
According to the previous studies, if MSCs were in the G0/G1 phase of the cell cycle, this can affect the differentiation capacity of these cells into cardiomyocytes (Bagheri-Hosseinabadi et al. 2018; Zhang et al. 2005). VPA can induce gathering of stem cells in the G0/G1 phase of the cell cycle (Vrba et al. 2010). Accordingly, it was proposed that this might be another reason for the success of VPA in inducing cardiac differentiation of hADSCs. Since mesenchymal cells are a heterozygous population that contains only a small fraction of the cells that are capable to differentiate into cardiomyocytes (Li et al. 2013), the first step in regenerative medicine is selecting the effective and suitable dose of the inhibitor for stem cell differentiation. According to the findings of this study, VPA at concentration of 1 mM, despite of small increase in cell detachment rate, is able to induce cardiac phenotype within 4 weeks. In agreement with the present study, previous studies have shown that HDACIs at millimolar (mM) concentrations have inhibitory activity on HDACs and have the ability to induce differentiation (Cheng et al. 2007; Gonzales et al. 2006; Marks and Xu 2009).
Conclusion
VPA has the ability to induce the differentiation of human adipose-derived stem cells into cardiomyocyte-like cells. Fibrin scaffold enhances the inductive effect of VPA in treated hADSCs by high expression of cardiomyogenic markers at mRNA and protein levels. Results of this study may improve cell-based protocols for implementation of more successful cardiac repair strategies. The results may also persuade researchers to perform further studies on VPA in fibrin scaffold using other sources of mesenchymal stem cells.
References
Akavia UD, Veinblat O, Benayahu D (2008) Comparing the transcriptional profile of mesenchymal cells to cardiac and skeletal muscle cells. J Cell Physiol 216:663–672
Bacakova L, Zarubova J, Travnickova M, Musilkova J, Pajorova J, Slepicka P, Kasalkova NS, Svorcik V, Kolska Z, Motarjemi H (2018) Stem cells: their source, potency and use in regenerative therapies with focus on adipose-derived stem cells—a review. Biotechnol Adv
Bagheri-Hosseinabadi Z, Salehinejad P, Mesbah-Namin SA (2017) Differentiation of human adipose-derived stem cells into cardiomyocyte-like cells in fibrin scaffold by a histone deacetylase inhibitor. Biomed Eng Online 16:134
Bagheri-Hosseinabadi Z, Mesbah-Namin SA, Salehinejad P, Seyedi F (2018) Fibrin scaffold could promote survival of the human adipose-derived stem cells during differentiation into cardiomyocyte-like cells. Cell Tissue Res 372:571–589
Barsotti MC, Felice F, Balbarini A, Di Stefano R (2011) Fibrin as a scaffold for cardiac tissue engineering. Biotechnol Appl Biochem 58:301–310
Brookes RL, Crichton S, Wolfe CD, Yi Q, Li L, Hankey GJ, Rothwell PM, Markus HS (2018) Sodium valproate, a histone deacetylase inhibitor, is associated with reduced stroke risk after previous ischemic stroke or transient ischemic attack. Stroke 49:54–61
Chen G, Yue A, Ruan Z, Yin Y, Wang R, Ren Y, Zhu L (2015) Potential of 5-azacytidine induction decidual stromal cells from maternal human term placenta towards cardiomyocyte-like cells in serum-free medium. Cell Tissue Bank 16:477–485
Cheng Y-C, Lin H, Huang M-J, Chow J-M, Lin S, Liu HE (2007) Downregulation of c-Myc is critical for valproic acid-induced growth arrest and myeloid differentiation of acute myeloid leukemia. Leuk Res 31:1403–1411
Cho J, Rameshwar P, Sadoshima J (2009) Distinct roles of GSK-3α and GSK-3β in mediating cardiomyocyte differentiation in murine bone marrow derived mesenchymal stem cells. J Biol Chem 284(52):36647–36658
Choi YS, Dusting GJ, Stubbs S, Arunothayaraj S, Han XL, Collas P, Morrison WA, Dilley RJ (2010) Differentiation of human adipose-derived stem cells into beating cardiomyocytes. J Cell Mol Med 14:878–889
De Sarno P, Li X, Jope RS (2002) Regulation of Akt and glycogen synthase kinase-3β phosphorylation by sodium valproate and lithium. Neuropharmacology 43:1158–1164
Gaustad KG, Boquest AC, Anderson BE, Gerdes AM, Collas P (2004) Differentiation of human adipose tissue stem cells using extracts of rat cardiomyocytes. Biochem Biophys Res Commun 314:420–427
Girão-Silva T, Bassaneze V, Campos LCG, Barauna VG, Dallan LAO, Krieger JE, Miyakawa AA (2014) Short-term mechanical stretch fails to differentiate human adipose-derived stem cells into cardiovascular cell phenotypes. Biomed Eng Online 13:54
Gonzales E, Chen H, Munuve R, Mehrani T, Britten-Webb J, Nadel A, Alam HB, Wherry D, Burris D, Koustova E (2006) Valproic acid prevents hemorrhage-associated lethality and affects the acetylation pattern of cardiac histones. Shock 25:395–401
Göttlicher M, Minucci S, Zhu P, Krämer OH, Schimpf A, Giavara S, Sleeman JP, Coco FL, Nervi C, Pelicci PG (2001) Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J 20:6969–6978
Grabarska A, Dmoszyńska-Graniczka M, Jeleniewicz W, Kiełbus M, Nowosadzka E, Rivero-Muller A, Polberg K, Stepulak A (2014) Valproic acid suppresses growth and enhances cisplatin cytotoxicity to larynx cancer cells. Head Neck Oncol 6:7
Grayson WL, Chao P-HG, Marolt D, Kaplan DL, Vunjak-Novakovic G (2008) Engineering custom-designed osteochondral tissue grafts. Trends Biotechnol 26:181–189
Gurvich N, Tsygankova OM, Meinkoth JL, Klein PS (2004) Histone deacetylase is a target of valproic acid-mediated cellular differentiation. Cancer Res 64:1079–1086
Hosseinkhani M, Hasegawa K, Ono K, Kawamura T, Takaya T, Morimoto T, Wada H, Shimatsu A, Prat SG, Suemori H (2007) Trichostatin A induces myocardial differentiation of monkey ES cells. Biochem Biophys Res Commun 356:386–391
Karamboulas C, Swedani A, Ward C, Al-Madhoun AS, Wilton S, Boisvenue S, Ridgeway AG, Skerjanc IS (2006) HDAC activity regulates entry of mesoderm cells into the cardiac muscle lineage. J Cell Sci 119:4305–4314
Kwiecińska P, Taubøll E, Gregoraszczuk EŁ (2012) Effects of valproic acid and levetiracetam on viability and cell cycle regulatory genes expression in the OVCAR-3 cell line. Pharmacol Rep 64:157–165
Latifpour M, Nematollahi-Mahani SN, Deilamy M, Azimzadeh BS, Eftekhar-Vaghefi SH, Nabipour F, Najafipour H, Nakhaee N, Yaghoubi M, Eftekhar-Vaghefi R (2011) Improvement in cardiac function following transplantation of human umbilical cord matrix-derived mesenchymal cells. Cardiology 120:9–18
Lemcke H, Voronina N, Steinhoff G, David R (2018) Recent progress in stem cell modification for cardiac regeneration. Stem Cells Int 2018
Li X, Yu X, Lin Q, Deng C, Shan Z, Yang M, Lin S (2007) Bone marrow mesenchymal stem cells differentiate into functional cardiac phenotypes by cardiac microenvironment. J Mol Cell Cardiol 42:295–303
Li Q, Qi L-J, Guo Z-K, Li H, Zuo H-B, Li N-N (2013) CD73+ adipose-derived mesenchymal stem cells possess higher potential to differentiate into cardiomyocytes in vitro. J Mol Histol 44:411–422
Li Q, Foote M, Chen J (2014) Effects of histone deacetylase inhibitor valproic acid on skeletal myocyte development. Sci Rep 4:7207
Lim SY, Sivakumaran P, Crombie DE, Dusting GJ, Pébay A, Dilley RJ (2013) Trichostatin A enhances differentiation of human induced pluripotent stem cells to cardiogenic cells for cardiac tissue engineering. Stem Cells Transl Med 2:715–725
Lin X, Li HY, Chen LF, Liu BJ, Yao Y, Zhu WL (2013) Enhanced differentiation potential of human amniotic mesenchymal stromal cells by using three-dimensional culturing. Cell Tissue Res 352:523–535
Loh QL, Choong C (2013) Three-dimensional scaffolds for tissue engineering applications: role of porosity and pore size. Tissue Eng B Rev 19:485–502
Marks P, Xu WS (2009) Histone deacetylase inhibitors: potential in cancer therapy. J Cell Biochem 107:600–608
Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, de Ferranti S, Després J-P, Fullerton HJ, Howard VJ (2015) American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Heart disease and stroke statistics—2015 update: a report from the American Heart Association. Circulation 131:e29–e322
Otsuki Y, Ii M, Moriwaki K, Okada M, Ueda K, Asahi M (2018) W9 peptide enhanced osteogenic differentiation of human adipose-derived stem cells. Biochem Biophys Res Commun 495:904–910
Oyama T, Nagai T, Wada H, Naito AT, Matsuura K, Iwanaga K, Takahashi T, Goto M, Mikami Y, Yasuda N (2007) Cardiac side population cells have a potential to migrate and differentiate into cardiomyocytes in vitro and in vivo. J Cell Biol 176:329–341
Planat-Benard V, Silvestre J-S, Cousin B, André M, Nibbelink M, Tamarat R, Clergue M, Manneville C, Saillan-Barreau C, Duriez M (2004) Plasticity of human adipose lineage cells toward endothelial cells: physiological and therapeutic perspectives. Circulation 109:656–663
Rangappa S, Fen C, Lee EH, Bongso A, Wei ESK (2003) Transformation of adult mesenchymal stem cells isolated from the fatty tissue into cardiomyocytes. Ann Thorac Surg 75:775–779
Raut A, Khanna A (2016) Enhanced expression of hepatocyte-specific microRNAs in valproic acid mediated hepatic trans-differentiation of human umbilical cord derived mesenchymal stem cells. Exp Cell Res 343:237–247
Rice K, Hormaeche I, Licht J (2007) Epigenetic regulation of normal and malignant hematopoiesis. Oncogene 26:6697
Shachar M, Tsur-Gang O, Dvir T, Leor J, Cohen S (2011) The effect of immobilized RGD peptide in alginate scaffolds on cardiac tissue engineering. Acta Biomater 7:152–162
Sukho P, Cohen A, Hesselink JW, Kirpensteijn J, Verseijden F, Bastiaansen-Jenniskens YM (2018) Adipose tissue-derived stem cell sheet application for tissue healing in vivo: a systematic review. Tissue Eng B Rev 24:37–52
Tobita M, Orbay H, Mizuno H (2011) Adipose-derived stem cells: current findings and future perspectives. Discov Med 11:160–170
Varghese J, Griffin M, Mosahebi A, Butler P (2017) Systematic review of patient factors affecting adipose stem cell viability and function: implications for regenerative therapy. Stem Cell Res Ther 8:45
Vrba J, Dolezel P, Ulrichova J (2010) Neutrophilic differentiation modulates the apoptotic response of HL-60 cells to sodium butyrate and sodium valproate. Neoplasma 57:438
Vrba J, Trtkova K, Ulrichova J (2011) HDAC inhibitors sodium butyrate and sodium valproate do not affect human ncor1 and ncor2 gene expression in HL-60 cells. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 155:259–262
Xu W, Zhang X, Qian H, Zhu W, Sun X, Hu J, Zhou H, Chen Y (2004) Mesenchymal stem cells from adult human bone marrow differentiate into a cardiomyocyte phenotype in vitro. Exp Biol Med 229:623–631
Yang G, Tian J, Feng C, L-L Z, Liu Z, Zhu J (2012) Trichostatin a promotes cardiomyocyte differentiation of rat mesenchymal stem cells after 5-azacytidine induction or during coculture with neonatal cardiomyocytes via a mechanism independent of histone deacetylase inhibition. Cell Transplant 21:985–996
Yang J-J, Z-Q L, J-M Z, Wang H-B, Hu S-Y, J-F L, Wang C-Y (2013) Real-time tracking of adipose tissue-derived stem cells with injectable scaffolds in the infarcted heart. Heart Vessel 28:385–396
Yu IT, Park J-Y, Kim SH, Lee J-S, Kim Y-S, Son H (2009) Valproic acid promotes neuronal differentiation by induction of proneural factors in association with H4 acetylation. Neuropharmacology 56:473–480
Zeng R, Y-C C, Zeng Z, W-Q L, X-F J, Liu R, Qiang O, Li X (2011) Effect of mini-tyrosyl-tRNA synthetase/mini-tryptophanyl-tRNA synthetase on ischemic angiogenesis in rats: proliferation and migration of endothelial cells. Heart Vessel 26:69–80
Zhang F-B, Li L, Fang B, Zhu D-L, Yang H-T, Gao P-J (2005) Passage-restricted differentiation potential of mesenchymal stem cells into cardiomyocyte-like cells. Biochem Biophys Res Commun 336:784–792
Acknowledgments
This study was granted by the Physiology Research Center of Kerman University of Medical Sciences (Grant No KMU-1395/476). The authors are grateful for the help of Amirkabir University, Tehran, for electron microscopy of the samples.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Najafipour, H., Bagheri-Hosseinabadi, Z., Eslaminejad, T. et al. The effect of sodium valproate on differentiation of human adipose-derived stem cells into cardiomyocyte-like cells in two-dimensional culture and fibrin scaffold conditions. Cell Tissue Res 378, 127–141 (2019). https://doi.org/10.1007/s00441-019-03027-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-019-03027-5