Abstract
Hypoxia is commonly found in human solid cancers and serves as a selective environment for the survival of aggressive cancer cells and as protection from anti-cancer therapies. In addition to a shift to anaerobic metabolism, the cellular response to hypoxia includes cessation of cell division and/or cell death. These mechanisms have still not been defined. Identification of the members of hypoxia-induced growth arrest pathways remain incomplete. We have undertaken an expression microarray analysis of the cellular response to hypoxia in diverse cell lines. An identified cohort of genes is reliably upregulated in various cells in response to hypoxia, as validated by reverse-transcriptase polymerase chain reaction (RT-PCR). One of the upregulated targets corresponds to an expressed sequence tag encoded by JMJD1A (a gene also known as JHDM2A), which has been identified as a histone demethylase that regulates the transcription of androgen receptor targets. We confirm, by RT-PCR, the upregulation of JMJD1A after hypoxia and desferroxamine treatment in multiple cell lines. We also show that JMJD1A is predominantly, but not exclusively, a nuclear protein. Immunofluorescent staining of HeLa cells shows a shift of cytoplasmic JMJD1A into the nucleus on hypoxia treatment. Immunohistochemical staining has revealed that JMJD1A is widely expressed in tissues, even in cells that are not known to express the androgen receptor, and is significantly increased in smooth muscle cells upon hypoxia treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hypoxia plays a critical role in the development, progression, and treatment of a large number of cancers. Hypoxia also activates the expression of the histone deacetylases, which alter chromatin expression and may also contribute to tumor development. The presence of hypoxic growth conditions can also have a significant impact on the effectiveness of standard cancer treatments. Hypoxic tumors are resistant to radio- and chemotherapies since these treatments rely upon the generation of oxygen free radicals to induce lethal DNA damage (Gatenby et al. 1988; Schwickert et al. 1995; Tomida and Tsuruo 1999).
Exposure to hypoxia induces the stabilization of hypoxia inducible factor 1 (HIF-1; Semenza and Wang 1992; Wang and Semenza 1993a). In hypoxia, the HIF-1 heterodimer can bind to hypoxia-responsive elements that activate a number of hypoxia-responsive genes. Some examples of HIF-1 targets include the transferrin gene required for iron metabolism (Rolfs et al. 1997), the VEGF (vascular endothelial growth factor) gene required for angiogenesis (Forsythe et al. 1996), glucose transporter genes (GLUT1, GLUT2; Ebert et al. 1995), insulin growth factor II required for proliferation (Beilharz et al. 1995), and CAIX (carbonic anhydrase) required for endothelial adhesion and pH regulation(Wykoff et al. 2000). HIF-1α is rapidly degraded in the presence of O2 by the ubquitin-mediated proteosome pathway (Huang et al. 1998; Salceda and Caro 1997). Treatment of cells with iron chelators, such as desferroxamine, which chelate iron from HIF-1α proline hydroxylase, stabilizes HIF-1α and increases the expression of hypoxia-associated genes, such the glycolytic enzymes, erythropoetin, and angiogenic proteins (Gleadle et al. 1995; Minchenko et al. 2003; Oexle et al. 1999; Wang and Semenza 1993b).
In addition to the shift to an anaerobic metabolism, the normal cellular response to hypoxia includes cessation of cell division (Amellem and Pettersen 1991; Krtolica and Ludlow 1996) and apoptosis (Graeber et al. 1996; Green and Giaccia 1998). As hypoxia is accompanied by considerably decreased ATP synthesis, this protective mechanism is logical. Cells in reduced [O2] that are in the late S, G2, and M phases will finish the cycle and arrest in G1, unless the hypoxia is severe (Green et al. 2001), whereas cells in G1 and early S phase remain in these phases (Amellem and Pettersen 1991). The mechanism by which hypoxia regulates the cell cycle is currently unknown.
One approach to studying hypoxia-regulated mediators of cell proliferation or death is to use a genome-wide approach to identify novel genes expressed in hypoxia, as has been previously been attempted, initially by using differential display (O'Rourke et al. 1996; Park et al. 2000). This method, although useful in identifying a small number of novel genes, is arduous, subject to false positives (Sung and Denman 1997), only identifies small numbers of transcripts discretely resolved in a sequencing gel, and does not allow for quantitative gene expression profiling between two cell populations. Nonetheless, one group has identified a gene, Proxy-1/NDRG1, that appears to be specifically upregulated in hypoxic breast carcinoma cells (Park et al. 2000). Several expression microarray strategies have resulted in the identification of genes that are upregulated in hypoxia (see below; Bando et al. 2003; Beitner-Johnson et al. 2001; Koong et al. 2000; Narravula and Colgan 2001; Seta and Millhorn 2004; Seta et al. 2002; Sonna et al. 2003), usually by evaluating one or a limited number of cell lines. Some of these are genes with an obvious relationship to anaerobic metabolism, whereas others are genes that have now been shown to have a pro-apoptotic function. One of these novel genes, JMJD1A (jumonji domain 1A), which is also known as jmjC-domain-containing histone demethylase 2A (JHDM2A; Yamane et al. 2006) plays a role in stem cell differentiation and spermatogenesis and as a cofactor of the androgen receptor and is upregulated by HIF-1 genes in hypoxia (Loh et al. 2007; Okada et al. 2007; Yamane et al. 2006; Wellmann et al. 2008).
We have evaluated diverse human cell lines for genes regulated by hypoxia with the aim of identifying and characterizing novel genes that may mediate hypoxia-induced cell cycle arrest or apoptosis.
Materials and methods
Cells and tissue culture
Human mammary epithelial cells were grown in their recommended growth media (Clonetics/Lonza, Basel Switzerland) and cultured as recommended by Clonetics/Lonza. Hs68 human fibroblasts (passages 30-50) were grown in low bicarbonate Dulbecco’s modified Eagle’s medium (DMEM). HeLa (cervical carcinoma), MCF7 (breast carcinoma), SK-BR-3 (breast carcinoma), Hep3B (liver carcinoma), and Daoy (medulloblastoma) were seeded on 175-cm2 dishes at 1×105 cells/ml in 20 ml of their recommended growth media (ATCC, Manassas, Va., USA). Cells were incubated at 5% CO2/37°C for 48 h to 50%-60% confluency prior to exposure to hypoxic conditions. Hypoxia was established by incubating cells in a GasPak anaerobic incubator (BD Bioscience, Mississauga, ON, Canada) for various times (6, 12, and/or 24 h), which reduces O2 levels while maintaining CO2 levels at approximately 5%. O2 levels were monitored by a methylene blue indicator strip, which bleaches white at 0.1%-0.5% O2 levels, following which the timing of hypoxia was measured. Control cells were incubated together with hypoxic samples under a normal tissue culture gas mixture (20% O2/5% CO2). Untreated cells were incubated under identical conditions, with the addition of an equal (200 μl) volume of phosphate-buffered saline (PBS) as an untreated control. Desferroxamine, a hypoxia mimic, was used at a concentration of 260 μmol.
RNA isolation and purification
Total cellular RNA was isolated by using TRIzol LS (Invitrogen, Carlsbad, Calif., USA) according to manufacturer’s protocol, which was modified as follows: RNA was precipitated by using 0.5 volumes of isopropanol and 0.5 volumes of 1.2 M NaCl/0.8 M sodium citrate to prevent proteoglycan contamination. RNA was resuspended in diethyl pyrocarbonate (DEPC)-treated H20, quantitated, and used immediately or precipitated and then stored in 100% ethanol at -80°C for later use.
Expression microarray analysis
Our methodology for expression microarray analysis was as described elsewhere (Box et al. 2008). Briefly, 50 μg total RNA was used to synthesize cDNA in the presence of oligo-dT primers and Cy3- or Cy5-labeled dCTP. After purification, the labeled cDNA was mixed and allowed to hybridize to Calgary Microarray Facility Operon oligonucleotide 14 K human DNA microarray chips by standard Facility procedures. Following overnight incubation, the chip slides were washed at Tm-20 and evaluated by using a Perkin Elmer ScanArray 5000 array scanner and QuantArray software. Two independent sets of "fluor flips" were performed, for a total of four experiments.
Expression microarray data analysis
ScanArray data from all experiments and time points, including "fluor flips", were pooled into a single data set for each cell line treatment. By using tools from Gene Traffic (v3.0), weak spots were eliminated, ratios of treated to untreated were calculated, and the ratios were averaged for each time point and gene for the four replicates. The averages were transformed to log2 values, and the data were normalized by using LOWESS. Average log2 values from equivalent time points were plotted by using Excel 2002.
Semi-quantitative reverse-transcriptase polymerase chain reaction
RNA (5 μg) resuspended in DEPC-treated water was incubated with 1 U random hexamer DNA primers, 10 mM dithiothreitol, 200 μM each dNTP with reaction buffer as supplied by the manufacturer and 200 U Superscript II and 40 U RNAse OUT RNAse inhibitor (Invitrogen) at 42°C for 1 h. Before use, the enzyme component was de-activated by heat at 70°C for 10 minutes.
Semi-quantitative polymerase chain reaction and gel electrophoresis
The components of the polymerase chain reaction (PCR) were as follows: 2.5 μl 10× MgCl2-PCR buffer, 1.5 Mg2+, 1 μl JMJD1A forward primer (5’-AGC-CAG-TGA-TGG-TGT-CTG-GAG-TG-3’), 1 µl JMJD1A reverse primer (5’-GGG-CCC-AGA-TCT-GGC-CGA-AC-3’), 1 µl 10 mM dNTPs, 16.8 µl double-distilled water, 0.2 µl Platinum Taq polymerase, 1 µl cDNA. All PCRs were performed in the T Gradient thermal cycler (Biometra, Montreal, PQ, Canada). The PCR program used was as follows: 95°C for 2 min, followed by 20, 30, or 40 cycles of 95°C for 1 min, 57°C or 66°C for 30 s, 72°C for 30 s, a final annealing step of 72°C for 10 min, and holding at 4°C. At the end of 20, 30, and 40 cycles, a pair of untreated and hypoxia-treated samples was removed from the T Gradient and held on ice until the last of the samples finished 40 cycles of amplification. An annealing temperature of 57°C or 66°C was determined by gradient PCR in order to ensure that an optimal annealing temperature was used.
Antibodies to JMJD1A
Production of rabbit antibody to JMJD1A was contracted to Open Biosystems (Huntsville, Ala., USA), and the antisera were generated and purified according to their established procedures. Briefly, two female rabbits were primed with a mixture of 250 μg each of two different peptides for JMJD1A (JMJ-N: CVEFDGESWRKRRWIEVYS; JMJ-C: CHAVKDAVAMLKASESSFGKP conjugated through the amino-terminal cysteine to keyhole limpet hemocyanin) administered subcutaneously over four sites. Animals were boosted at day 14 with a mixture of 100 μg of each peptide, over four subcutaneous sites. The rabbits were boosted again three more times at 28-day intervals, and bleeds after day 56 were monitored monthly by enzyme-linked immunosorbent assay (ELISA) to unconjugated peptide. Post 56-day bleeds were pooled from the appropriate rabbit and purified by affinity chromotography over a column with the relevant peptide antigen. Immunopurified antisera was eluted by a stepwise pH gradient, neutralized with borate buffer, quantitated, and retitrated by ELISA. Immunopurified antisera from two different rabbits (anti-JMJ-7505, anti-JMJ-7506) were prepared against the JMJ-C peptide. Unfortunately, the JMJ-N peptide elicited no detectable immune response in either rabbit and was used as a control peptide in the peptide block experiments. Immunopurified JMJ antibody was diluted 1:1 with 60% glycerol, aliquoted, and stored frozen at -20°C until use.
Later in the course of our studies, an immunopurified rabbit anti-peptide antibody, JMJ-AB (Abcam, Cambridge, Mass., USA; no. AB32440) became commercially available.
Western blot analysis
For analysis of protein expression, hypoxic and normoxic tissue culture cells were prepared as described above, placed on ice, and rinsed with 10 ml ice-cold PBS. The cells were lysed on the plate by the addition of 700 μl NP-40 lysis buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 10% glycerol, 1.5 mM MgCl2, 1 mM EGTA, 100 mM NaF, 1% NP-40, 10 μM sodium vanadate, 10 μM phenylmethane sulfonylfluoride, 10 μg/ml aprotinin, 5 μg/ml leupeptin). Protein concentration was determined by using the Bichronate assay (Pierce, Rockford, Ill., USA). Cell lysate (50 μg) was loaded onto a gel for SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose (for detection of HIF-1) or 0.1 μm polyvinylidene difluoride (PVDF; Millipore, Billirica, Mass., USA; for detection of proteins less than 30 kDa) by blotting overnight at 30 V/60 mA. Membranes were blocked for 1 h at room temp in 10% (w/v) powdered nonfat milk/TRIS-buffered saline with 0.1% (v/v) Tween-20 (Sigma, St. Louis, Mo., USA). Western blotting was carried out overnight at 4°C with the following antibodies: actin Ab (Neomarkers, Fremont, Calif., USA; no. S-1295, 1:2000), JMJ-AB (Abcam; 1:500), JMJ-7505 (0.5 μg/ml), and JMJ-7506 (0.5 μg/ml).
For peptide block experiments, antibody dilutions used for Western blotting were incubated overnight at 4°C with a large molar excess (0.8 μg/ml antibody with 2.5 μg/ml peptide) of blocking peptide JMJ-C (for JMJ-7505/7506) or commercial peptide (Abcam; no. AB32439) for the JMJ-AB antibody, or an irrelevant peptide (JMJ-N).
Immunofluorescent staining
Sterile glass coverslips were placed into a culture dish, and then culture medium containing 50,000 cells/ml was added. Cells were allowed to grow for 48 h until 50%-60% confluency was reached and were then treated in hypoxia chambers as before.
For one set of experiments, HeLa cells were rinsed with PBS and then fixed with 3% formaldehyde freshly prepared from paraformaldehyde in cytoskeletal buffer for 20 min at 4°C. Cells were then rinsed with PBS three times, incubated in 3% bovine serum albumin in cytoskeletal buffer at 4°C for 2 h to block non-specific binding of antibody, and subsequently incubated with 250 μl 1:500 anti-JMJ-AB antibody solution overnight at 4°C inside a humidified chamber. The following day, cells were rinsed with 0.1% Triton X-100 in PBS and twice with PBS, incubated with a 1:200 dilution of fluorescein-labeled goat anti-rabbit or fluor-488-labeled goat anti-mouse secondary antibody for 2 h at room temperature also in a humidified chamber, washed with 0.1% Triton X-100 in PBS, and rinsed twice with PBS. Glass coverslips with cells were then wet-mounted with 10 μl Prolong Gold (Invitrogen).
For anti-JMJ-7505/7506 staining, cells on coverslips were placed on ice after removal from the hypoxia chamber, washed twice in ice-cold PBS, fixed in 2% formaldehyde freshly prepared from paraformaldehyde in PBS at room temperature for 30 min, and then stored in PBS at 4°C overnight. The next day, the coverslips were blocked with 1% monoethanolamine in PBS for 30 min at room temperature followed by 10% normal goal serum in PBS at 37°C for 30 min, incubated in primary antibody (1:40 anti-JMJ-7505, 1:20 anti-JMJ-7506, or 1:20 anti-HIF-1α) in PBS/0.15% Triton X-100/15% normal goat serum) on moist paper in a covered chamber for 60 min, washed three times in PBS/0.1% Tween, incubated with secondary antibody (either Cy3-Goat anti-rabbit IgG or Cy2-Goat anti-mouse IgG; Jackson Immunoresearch, West Grove Pa., USA; nos. 111-165-144 and 115-225-166, respectively) in PBS/0.1%Tween/1% normal goat serum as above for 60 min, and then washed twice with PBS/0.1% Tween and once with PBS. Coverslips were dipped into distilled water, shaken immediately, and then mounted onto a slide with 10 μl Prolong Gold (Invitrogen). After overnight curing of the mounting medium, the cells were viewed and imaged by using a Zeiss Axiovert 200 confocal microscope.
Immunohistochemical staining
Paraffin-embedded sections were deparaffinized with xylene twice for 5 min and then rehydrated to distilled water through an ethanol series. Antigen retrieval was carried out by steaming for 20 min in 10 mM citrate-buffer (pH 6.0) and then cooling to room temperature over 20 min. The slides were stained at 37°C for 60 min with anti-JMJ-AB rabbit polyclonal antibody (Abcam), which was pre-incubated (blocked) with either JMJ-N control peptide or with 2.24 μg JMJ-AB blocking peptide (Abcam) overnight at 4°C. A Ventana iView DAB detection kit (Ventana, Tuscon, Ariz., USA) was used for visualization and the stained sections were counterstained with hematoxylin.
Various tissues (kidney, liver, endomyometrium, skeletal muscle, ovarian tube, ectocervix) were obtained from anonymous surgical pathology specimens and divided into two groups: hypoxia-treated and untreated. The hypoxia-treated group was incubated in a GasPak anaerobic incubator (BD Bioscience) for 6 h in low DMEM with 10%fetal bovine serum. After incubation, specimens were washed with PBS and then fixed with 10% buffered formaldehyde. Untreated tissues were fixed with 10% buffered formaldehyde after being harvested. Sections of the paraffin-embedded tissues were deparaffinized in xylene (twice for 5 min) and then rehydrated to distilled water through an ethanol series. Antigen retrieval was carried out by steaming for 20 min in 10 mM Na-citrate buffer (pH 6.0) and then allowing to cool to room temperature over 20 min. The slides were then stained at 37°C for 45 min with anti-HIF-1α mouse monoclonal antibody (BD Transduction Laboratories, Franklin Lakes, N.J., USA; no. 610959, 1:50) or anti-JMJ-AB antibody as previously described. A Ventana iView DAB detection kit was used for visualization, and the stained sections were counterstained with hematoxylin.
Results
Expression microarray upregulation of JMJD1A mRNA
We undertook an expression microarray screen of a variety of cell lines as part of a larger project seeking to identify uncharacterized human genes involved in the regulation of proliferation or apoptosis in response to hypoxia. Some results of this project have been presented elsewhere (Box et al. 2008). By scoring for the cumulative upregulation of genes amongst three different time points and multiple cell lines, we not unsurprisingly identified many genes previously characterized as hypoxia-regulated, most of which had also been characterized as HIF-1α targets (Box et al. 2008). One of these, N-myc-related downstream regulatory gene (NDRG1, Proxy-1), is a well-known hypoxia-upregulated gene and HIF-1α target (Cangul 2004; Park et al. 2000). As can be seen in Fig. 1a, it is upregulated in all hypoxia-treated cell lines tested. From our microarray screen, however, we noted that an expressed sequence tag (AB018285) for a previously uncharacterized gene, JMJD1A, was also upregulated in all of the normal and neoplastic cell lines that we examined (Fig. 1b). This expressed sequence tag was also upregulated in Hela cells treated with desferroxamine, a hypoxia-mimic, which prominently upregulates HIF-1α (Fig. 1c).
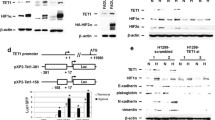
Expression microarray analysis of NDRG-1 and JMJD1A mRNA in hypoxia-treated human cell lines (Hep3B hepatocellular carcinoma, Hs68 human foreskin fibroblast, HeLa human cervical carcinoma, MCF-7 malignant breast epithelial cells, Daoy medulloblastoma, SK-BR-3 breast carcinoma, HMEC human mammary epithelial cells, HTB30 breast cancer, RD human rhabdomyosarcoma, SKNMC neuroblastoma). HeLa cells were treated with hypoxia or 260 μM desferroxamine for 6, 12, and 24 h and subjected to expression microarray analysis by using University of Calgary Operon 14 K human oligonucleotide chips. For each time point, total RNA from untreated cells seeded at the same time was used as a control. Following weak spot elimination, log transformation, and LOWESS normalization with the Gene Traffic (v3.0) tool, mean log2 treated:untreated ratios were exported into MS Excel and plotted. a, b Mean log2 treated:untreated ratios plotted over treatment time for, respectively, NDRG-1 (expressed sequence tag D87953) and JMJD1A (expressed sequence tag AB018285) with mRNA from HeLa, SK-BR-3, Hep3B, Hs68 fibroblasts, and HMEC cells (for an explanation of the colors, see insets). c Expression microarray comparison for JMJD1A between hypoxia and desferroxamine-treated HeLa cells. The x-axis denotes average log2 expression of four independent hybridizations, whereas the y-axis denotes the time in hours (with treatment times of 6, 12, and 24 h). d Expression microarray evaluation of mRNA from MCF-7, HTB30, Hep3B, Daoy, RD, SKNMC, and HeLa cells for the androgen receptor (NM_000044)
Semi-quantitative reverse-transcriptase PCR results for JMJD1A expression in various human cell lines treated with hypoxia
Total RNA was isolated from several cell lines treated with hypoxia for 24 h or from untreated controls and employed to prepare cDNA. This cDNA was used as a target for PCRs with specific primers directed to either 28 S RNA or JMJD1A intron-spanning transcript sequences. Figure 2 shows the results of PCRs with JMJD1A primers evaluated at 20, 30, or 40 cycles with cDNA from the untreated or hypoxia-treated cell lines. All of the cell lines show an increase in the amount of the JMJD1A amplicon from cDNA prepared from the hypoxia-treated cells versus the untreated cells (Fig. 2). The 28 S primer set gave similar yields between the treated and untreated cDNA sources (not shown).
Semi-quantitative reverse-transcriptase polymerase chain reaction (PCR) of JMJD1 from various cell lines. Hepatocellular carcinoma (Hep3B), human foreskin fibroblast (Hs68), human fibroblast (WI-38), human cervical carcinoma (HeLa), malignant breast epithelial (MCF-7), and medulloblastoma (DAOY) cells were grown for 48 h and then treated with hypoxia for 24 h (t) or left untreated (ut). Equivalent amounts of cDNA from untreated (ut) and treated (t) samples were subjected to variable numbers of PCR cycles (20 cycles, 30 cycles, 40 cycles) under conditions optimized for primer, template, and Mg++ in order to identify the amplicons prior to the plateau phase of amplification. In all samples, 28 S RNA appeared to amplify similarly with both treated and untreated samples (not shown)
JMJD1A protein is upregulated in hypoxia
Although the data from the previous experiments indicate that the JMJD1A transcript is elevated during hypoxia treatment in diverse cell lines, the regulation of the protein is of more functional significance. Figure 3 presents Western blots of cellular lysates obtained from three cell lines treated with hypoxia. All treated cell lines exhibited upregulation of HIF-1α (not shown), indicating a typical cellular response to hypoxia. In similar fashion, all three cell lines showed time-dependent upregulation of JMJD1A, indicating that the protein was also upregulated following hypoxia exposure, although all cells showed a basal level of JMJD1A expression. The effect of blocking the anti-JMJ-AB antibody with its peptide epitope was also investigated (Fig. 4). As bands were absent in the peptide-blocked blot, whereas the co-incubated anti-actin signal was similar to that of the unblocked blot, the anti-JMJ-AB antibody was considered to be specific to the JMJD1A peptide for Western blotting purposes. Likewise, the other two anti-JMJD1A antibodies (7505/7506) gave similar results when blocked with their respective peptide epitopes (not shown) indicating that all of the affinity-purified antibodies were specific. On testing the relative usefulness of the reagents for immunoprecipitation/Western blotting, the anti-JMJ-7505 antibody yielded the best signal for both immunoprecipitation and Western blotting (Fig. 5).
JMJD1A is upregulated in hypoxia-treated cell lines. Protein extracts (40 μg/lane) from cell lines HeLa, MCF-7, and Hep3B untreated (ut) or treated with hypoxia for 12 h or 24 h were loaded onto 8% acrylamide SDS-PAGE gels and transferred to PVDF. Pan-actin blots were performed for all time points to normalize protein loading amounts (not shown). a Western blots of HeLa lysates incubated with anti-JMJD1A-7505. b Western blots of MCF-7 lysates incubated with anti-JMJD1A-7505. c Western blots of Hep3B lysates incubated with anti-JMJD1A-7505. The major band corresponding to JMJD1A (~140 kDa) is indicated left
Peptide inhibition of JMJD1A immunostaining in Western blots of HeLa cell lysates. Protein extracts (40 μg/lane) from HeLa untreated (lanes a) or treated with hypoxia for 6 h (lanes b), 12 h (lanes c), or 24 h (lanes d) were loaded onto two simultaneously electrophoresed 8% acrylamide SDS-PAGE gels and transferred to PVDF. a, b Western blots incubated with a mixture of anti-pan-actin antibody and anti-JMJ-AB antibody incubated with a 50× molar excess of either an irrelevant peptide (neg Pep) or the specific blocking peptide (pos Pep), respectively. Chemiluminescent exposure was performed until the actin staining density was similar in both blots to ensure identical opportunity to generate anti-JMJ-AB signals. Actin (45 kDa) and JMJD1A (140 kDa) bands are indicated left
Immunoprecipitation of JMJD1A with three anti-JMJD1A antibodies. Total protein (500 μg) from lysate prepared from a 24-h hypoxia treatment of HeLa was used for immunoprecipitation with 5 μg/ml anti-JMJ-AB antibody (lanes b), anti-JMJ-7505 antibody (lanes c), or anti-JMJ-7506 antibody (lanes d), with control lanes (lanes a) consisting of Western blotting of 40 μg total protein lysate from HeLa. The protein A beads from immunoprecipitations were eluted by boiling in SDS loading buffer and then loaded onto a 8% acrylamide gel for PAGE. Blots were probed with equivalent amounts of anti-JMJ-AB (a), anti-JMJ-7505 antibody (b), or anti-JMJ-7506 antibody (c). The JMJD1A band at 140 kDa is indicated by arrows (left)
Immunofluorescent staining of HeLa cells with anti-JMJ antibodies
HeLa cells treated with hypoxia for 12 h were immunostained with anti-JMJ-AB antibody or anti-HIF-1α. As can be seen in Fig. 6, the anti-JMJ-AB reagent gave strong nuclear staining, as did the anti-HIF-1α antibody, in the hypoxia-treated cells. Untreated or 12-h hypoxia-treated HeLa cells were also stained with the other anti-JMJ antibodies (Fig. 7a, b: anti-JMJ-7505 antibody; Fig. 7c, d: anti-JMJ-7506 antibody). Positive staining was observed with each of these antibodies in the presence of Triton detergent for the nuclear permeablization of the Hela cells. Immunostaining with the JMJ-AB antibody was seen in the absence of Triton detergent (Fig. 7e, f). Immunostaining of all three antibodies was markedly decreased in the presence of the blocking peptide (not shown). All three antibodies showed strong nuclear staining, but the untreated cells exhibited cytoplasmic staining that was noticeably decreased in the hypoxia-treated cells.
Immunofluorescent staining of untreated or 12-h hypoxia-treated HeLa cells with the three anti-JMJD1A antibodies. a, b Immunostaining with the anti-JMJ-7505 antibody in the presence of Triton detergent. c, d Immunostaining with the anti-JMJ-7506 antibody in the presence of Triton detergent. e, f Immunostaining with the anti-JMJ-AB antibody with a cytoskeletal protein staining protocol identical to that of Fig. 6. Bar 20 μm
Immunohistochemical staining of hypoxia or untreated tissues
Formalin-fixed paraffin-embedded tissue specimens were obtained from anonymous surgical pathology specimens surplus to diagnostic purposes and subjected to citrate-based antigen retrieval followed by staining with the three anti-JMJD1A antibodies. Only the anti-JMJ-AB reagent gave immunohistochemical staining. We compared smooth muscle taken from either hypoxia-treated or untreated tissues. Interestingly, JMJD1A-AB staining was more evident in hypoxia-treated smooth muscle from a variety of sources including cervix (Fig. 8a, b), arterioles, and small bowel (not shown). Smooth muscle was the only tissue that showed a clear upregulation of JMJD1A protein expression with hypoxia. HIF-1α was clearly upregulated in hypoxia-treated versus untreated tissue (Fig. 8c, d), showing that the hypoxia treatment was effective.
Anti-JMJ-AB and anti-HIF-1α immunohistochemical staining of paraffin sections of hypoxia-treated smooth muscle. Tissue was incubated in media/10% fetal calf serum for up to 2 h prior to treatment at 37°C with hypoxia or normoxia (untreated), as for the cell lines. Sections (5 μm) were stained with anti-JMJ-AB or anti-HIF-1α antibody, and immunostaining was detected by using an iView kit and a Ventana Nexes semi-automated immunostainer. a, b Anti-JMJ-AB staining of, respectively, hypoxia-treated or untreated cervical tissue. c, d Anti-HIF-1α staining of, respectively, hypoxia-treated (arrow in c) or untreated cervical vascular tissue. Bar 40 μm
Many different types of untreated epithelial tissue showed significant JMJD1A-AB staining (see examples in Fig. 9). This may have been attributable to mild hypoxia upregulating JMJD1A in both the untreated and hypoxia-treated tissues, which were derived from the same surgical specimens. Devascularization of the specimen in the surgical suite, delivery to the pathology laboratory, removal of the diagnostic specimens, and incubation of the surplus specimens for several hours may have subjected all the tissues to mild hypoxia. Indeed, we also found that HIF-2α was expressed in the untreated epithelial cells in amounts similar to those of the hypoxia-treated specimens, whereas HIF-3α was poorly expressed both in the untreated epithelial cells and hypoxia-treated specimens (not shown).
Anti-JMJ-AB immunohistochemical staining of paraffin sections of hypoxia-treated epithelial tissues. Paraffin specimens from hypoxia-treated tissues including liver (a), kidney (b), endometrium (c), and prostate adenocarcinoma (d) were stained with anti-JMJ-AB antibody (brown). Staining between hypoxia or normoxia-treated tissues (not shown) was essentially identical, and many epithelial tissues in addition to normal or malignant prostate also expressed JMJD1A (not shown). Bar 40 μm
Immunohistochemical specificity of anti-JMJD1A-AB staining
Anti-JMJD1A-AB antibody was incubated with blocking peptide, following which the immunostaining of both the germinal-center lymphocytes in the tonsil and of invasive adenocarcinoma cells was blocked, as compared with incubation with an irrelevant peptide (Fig. 10). This illustrates the specificity of the JMJD1A-AB reagent for immunohistochemical staining.
Peptide inhibition of anti-JMJD-AB antibody staining of paraffin sections (5 μm thick) of invasive carcinoma or normal tonsil tissue. Sections were incubated with anti-JMJ-AB antibody, and any immunostaining was detected with an iView kit and a Ventana Nexes semi-automated immunostainer. Left Immunostaining of invasive adenocarcinoma tissue with anti-JMJ-AB antibody pre-incubated overnight with an irrelevant peptide (a) versus the anti-JMJD1A blocking peptide (c). Right Immunostaining of tonsil tissue with anti-JMJ-AB antibody pre-incubated overnight with an irrelevant peptide (b) versus the anti-JMJD1A blocking peptide (d). Arrows indicate identical areas in the serial sections (a, c; b, d). Bar 100 μm
Discussion
The JMJ-related JMJD1A gene was originally discovered as being expressed in rat testes and was designated as testes specific gene A (Hoog et al. 1991). This gene appears not to have been studied significantly in the 15 years since its discovery, until recently, when it was identified as a binding partner for the ETS-related protein ER71 (Knebel et al. 2006). JMJD1A was purified from a Hela cell extract containing histone demethylase activity and renamed JHDM2A (Yamane et al. 2006). However, here, we continue to refer to it as JMJD1A. Both the zinc-finger and the jmjC domain, in addition to co-factors Fe(II) and α-ketoglutarate, have been shown to be essential for demethylase activity. Knockdown of JMJD1A in mouse tumor cells does not result in a change in cell morphology, although expression of a variety of developmental genes is downregulated, consistent with the role of histone demethylation in positively regulating transcription. These activities seem to be important in spermatogenesis (McGraw et al. 2007; Okada et al. 2007; Tachibana et al. 2007). In addition to interacting with the androgen receptor, JMJD1A has been shown to exhibit hormone-dependent recruitment to androgen receptor target genes, resulting in H3K9 demethylation and transcriptional activation (Yamane et al. 2006), although recent work has shown that the overexpression of the androgen receptor does not greatly alter JMJD1A transcription (Urbanucci et al. 2008). This function seems inconsistent with a role for JMJD1A in the hypoxic response, as we have shown no change in the regulation of the androgen receptor mRNA transcript in hypoxia, in diverse cell lines (Fig. 1d).
While this work was in preparation, two other groups have reported that JMJD1A is a hypoxia-regulated protein and a transcriptional activation target of HIF-1α (Pollard et al. 2008; Wellmann et al. 2008). One study has identified JMJD1A as a potential hypoxia-inducible target because of the presence of Fe(II) and 2-oxoglutarate motifs. HIF-1α has also been shown to bind to the promoter of JMJD1A. This work has further revealed that two groups of Jumonji-containing histone demethylases respond to hypoxia, and that JMJD1A belongs to a group exhibiting the most robust response (Pollard et al. 2008). The other study has identified JMJD1A in an expression microarray comparison of the response to hypoxia in HIF-1α-competent and -deficient leukemia cell lines derived from Z-33 (Wellmann et al. 2004). Hypoxic fetal tissues express the JMJD1A transcript to a greater extent than adult tissues, and in rat tissues, the kinetics of JMJD1A transcript expression mimic those of the known HIF-1α target, VEGF. Investigation of the JMJD1A promoter has revealed eight potential HIF-1α sites. Functional studies of the promoter indicate that the upregulation of a downstream reporter is induced by the upregulation of HIF-1α, demonstrating JMJD1A to be a bona fide HIF-1 target (Wellmann et al. 2004).
Interestingly, at least five genes have been suggested as being regulated by JMJD1A (Loh et al. 2007; Okada et al. 2007). Of these five, Tcfcp2l1, Zfp67, Tnp1, and Prm1 were not present as targets in our microarray screen. TCL1a was included on our array, but its regulation did not change significantly in any cell line at any time point during the hypoxic response (not shown).
The work that we present here has thus confirmed the hypoxia-regulated expression of JMJD1A in diverse epithelial and other human cells and has characterized several immune reagents derived from JMJD1A peptides. The function of JMJD1A in hypoxia probably differs from its previously characterized roles in regulating androgen receptor or Oct 4 pathways, as the androgen receptor gene is not upregulated in hypoxia in any of the cell lines that we have studied. Perhaps this function(s) involves the translocation of JMJD1A from a cytoplasmic to a nuclear location.
References
Amellem O, Pettersen EO (1991) Cell inactivation and cell cycle inhibition as induced by extreme hypoxia: the possible role of cell cycle arrest as a protection against hypoxia-induced lethal damage. Cell Prolif 24:127–141
Bando H, Toi M, Kitada K, Koike M (2003) Genes commonly upregulated by hypoxia in human breast cancer cells MCF-7 and MDA-MB-231. Biomed Pharmacother 57:333–340
Beilharz EJ, Bassett NS, Sirimanne ES, Williams CE, Gluckman PD (1995) Insulin-like growth factor II is induced during wound repair following hypoxic-ischemic injury in the developing rat brain. Brain Res Mol Brain Res 29:81–91
Beitner-Johnson D, Seta K, Yuan Y, Kim H, Rust RT, Conrad PW, Kobayashi S, Millhorn DE (2001) Identification of hypoxia-responsive genes in a dopaminergic cell line by subtractive cDNA libraries and microarray analysis. Parkinsonism Relat Disord 7:273–281
Box AH, Yuen C, Ponjevic D, Fick GH, Demetrick DJ (2008) Signaling and apoptosis differences between severe hypoxia and desferoxamine treatment of human epithelial cells. Biochem Cell Biol 86:425-436
Cangul H (2004) Hypoxia upregulates the expression of the NDRG1 gene leading to its overexpression in various human cancers. BMC Genet 5:27
Ebert BL, Firth JD, Ratcliffe PJ (1995) Hypoxia and mitochondrial inhibitors regulate expression of glucose transporter-1 via distinct Cis-acting sequences. J Biol Chem 270:29083–29089
Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, Semenza GL (1996) Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol 16:4604–4613
Gatenby RA, Kessler HB, Rosenblum JS, Coia LR, Moldofsky PJ, Hartz WH, Broder GJ (1988) Oxygen distribution in squamous cell carcinoma metastases and its relationship to outcome of radiation therapy. Int J Radiat Oncol Biol Phys 14:831–838
Gleadle JM, Ebert BL, Firth JD, Ratcliffe PJ (1995) Regulation of angiogenic growth factor expression by hypoxia, transition metals, and chelating agents. Am J Physiol 268:C1362–C1368
Graeber TG, Osmanian C, Jacks T, Housman DE, Koch CJ, Lowe SW, Giaccia AJ (1996) Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature 379:88–91
Green SL, Giaccia AJ (1998) Tumor hypoxia and the cell cycle: implications for malignant progression and response to therapy. Cancer J Sci Am 4:218–223
Green SL, Freiberg RA, Giaccia AJ (2001) p21(Cip1) and p27(Kip1) regulate cell cycle reentry after hypoxic stress but are not necessary for hypoxia-induced arrest. Mol Cell Biol 21:1196–1206
Hoog C, Schalling M, Grunder-Brundell E, Daneholt B (1991) Analysis of a murine male germ cell-specific transcript that encodes a putative zinc finger protein. Mol Reprod Dev 30:173–181
Huang LE, Gu J, Schau M, Bunn HF (1998) Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA 95:7987–7992
Knebel J, De Haro L, Janknecht R (2006) Repression of transcription by TSGA/Jmjd1a, a novel interaction partner of the ETS protein ER71. J Cell Biochem 99:319–329
Koong AC, Denko NC, Hudson KM, Schindler C, Swiersz L, Koch C, Evans S, Ibrahim H, Le QT, Terris DJ, Giaccia AJ (2000) Candidate genes for the hypoxic tumor phenotype. Cancer Res 60:883–887
Krtolica A, Ludlow JW (1996) Hypoxia arrests ovarian carcinoma cell cycle progression, but invasion is unaffected. Cancer Res 56:1168–1173
Loh YH, Zhang W, Chen X, George J, Ng HH (2007) Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev 21:2545–2557
McGraw S, Vigneault C, Sirard MA (2007) Temporal expression of factors involved in chromatin remodeling and in gene regulation during early bovine in vitro embryo development. Reproduction 133:597-608
Minchenko O, Opentanova I, Caro J (2003) Hypoxic regulation of the 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase gene family (PFKFB-1–4) expression in vivo. FEBS Lett 554:264–270
Narravula S, Colgan SP (2001) Hypoxia-inducible factor 1-mediated inhibition of peroxisome proliferator-activated receptor alpha expression during hypoxia. J Immunol 166:7543–7548
O'Rourke JF, Pugh CW, Bartlett SM, Ratcliffe PJ (1996) Identification of hypoxically inducible mRNAs in HeLa cells using differential-display PCR. Role of hypoxia-inducible factor-1. Eur J Biochem 241:403–410
Oexle H, Gnaiger E, Weiss G (1999) Iron-dependent changes in cellular energy metabolism: influence on citric acid cycle and oxidative phosphorylation. Biochim Biophys Acta 1413:99–107
Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y (2007) Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 450:119–123
Park H, Adams MA, Lachat P, Bosman F, Pang SC, Graham CH (2000) Hypoxia induces the expression of a 43-kDa protein (PROXY-1) in normal and malignant cells. Biochem Biophys Res Commun 276:321–328
Pollard PJ, Loenarz C, Mole D, McDonough MA, Gleadle JM, Schofield C, Ratcliffe PJ (2008) Regulation of Jumonji-domain containing histone demethylases by hypoxia inducible factor (HIF) 1-alpha. Biochem J 416:387–394
Rolfs A, Kvietikova I, Gassmann M, Wenger RH (1997) Oxygen-regulated transferrin expression is mediated by hypoxia-inducible factor-1. J Biol Chem 272:20055–20062
Salceda S, Caro J (1997) Hypoxia-inducible factor 1alpha (HIF-1alpha) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J Biol Chem 272:22642–22647
Schwickert G, Walenta S, Sundfor K, Rofstad EK, Mueller-Klieser W (1995) Correlation of high lactate levels in human cervical cancer with incidence of metastasis. Cancer Res 55:4757–4759
Semenza GL, Wang GL (1992) A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol 12:5447–5454
Seta KA, Millhorn DE (2004) Functional genomics approach to hypoxia signaling. J Appl Physiol 96:765–773
Seta KA, Spicer Z, Yuan Y, Lu G, Millhorn DE (2002) Responding to hypoxia: lessons from a model cell line. Sci STKE 2002:RE11
Sonna LA, Cullivan ML, Sheldon HK, Pratt RE, Lilly CM (2003) Effect of hypoxia on gene expression by human hepatocytes (HepG2). Physiol Genomics 12:195–207
Sung YJ, Denman RB (1997) Use of two reverse transcriptases eliminates false-positive results in differential display. Biotechniques 23:462-468
Tachibana M, Nozaki M, Takeda N, Shinkai Y (2007) Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J 26:3346–3359
Tomida A, Tsuruo T (1999) Drug resistance mediated by cellular stress response to the microenvironment of solid tumors. Anticancer Drug Des 14:169–177
Urbanucci A, Waltering KK, Suikki HE, Helenius MA, Visakorpi T (2008) Androgen regulation of the androgen receptor coregulators. BMC Cancer 8:219
Wang GL, Semenza GL (1993a) Characterization of hypoxia-inducible factor 1 and regulation of DNA binding activity by hypoxia. J Biol Chem 268:21513–21518
Wang GL, Semenza GL (1993b) Desferrioxamine induces erythropoietin gene expression and hypoxia-inducible factor 1 DNA-binding activity: implications for models of hypoxia signal transduction. Blood 82:3610–3615
Wellmann S, Buhrer C, Moderegger E, Zelmer A, Kirschner R, Koehne P, Fujita J, Seeger K (2004) Oxygen-regulated expression of the RNA-binding proteins RBM3 and CIRP by a HIF-1-independent mechanism. J Cell Sci 117:1785–1794
Wellmann S, Bettkober M, Zelmer A, Seeger K, Faigle M, Eltzschig HK, Buhrer C (2008) Hypoxia upregulates the histone demethylase JMJD1A via HIF-1. Biochem Biophys Res Commun 372:892–897
Wykoff CC, Beasley NJ, Watson PH, Turner KJ, Pastorek J, Sibtain A, Wilson GD, Turley H, Talks KL, Maxwell PH, Pugh CW, Ratcliffe PJ, Harris AL (2000) Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res 60:7075–7083
Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y (2006) JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125:483–495
Acknowledgements
We thank Erin MacRae and Claude Veillette for excellent technical assistance and Ponnampalam Thirukkumaran of the Southern Alberta Microarray Facility for performing the expression microarray hybridizations.
Author information
Authors and Affiliations
Corresponding author
Additional information
The authors acknowledge funding from the Ruth Barker Foundation and from an Alberta Cancer Board Pilot Project grant (23289). A.S. was supported by the Division of Transplant Research, University of Calgary. Salary support for M.N. was provided by the Ruth Barker Foundation. Salary support for D.J.D. was provided by Calgary Laboratory Services. A.H.B. was supported by a stipend from the CIHR-Alberta Cancer Board Strategic Training Program in Translational Cancer Research.
Rights and permissions
About this article
Cite this article
Sar, A., Ponjevic, D., Nguyen, M. et al. Identification and characterization of demethylase JMJD1A as a gene upregulated in the human cellular response to hypoxia. Cell Tissue Res 337, 223–234 (2009). https://doi.org/10.1007/s00441-009-0805-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-009-0805-y