Abstract
The rugged topography of the Himalayan region has hindered large-scale human migrations, population admixture and assimilation. Such complexity in geographical structure might have facilitated the existence of several small isolated communities in this region. We have genotyped about 850,000 autosomal markers among 35 individuals belonging to the four major populations inhabiting the Himalaya and adjoining regions. In addition, we have genotyped 794 individuals belonging to 16 ethnic groups from the same region, for uniparental (mitochondrial and Y chromosomal DNA) markers. Our results in the light of various statistical analyses suggest a closer link of the Himalayan and adjoining populations to East Asia than their immediate geographical neighbours in South Asia. Allele frequency-based analyses likely support the existence of a specific ancestry component in the Himalayan and adjoining populations. The admixture time estimate suggests a recent westward migration of populations living to the East of the Himalaya. Furthermore, the uniparental marker analysis among the Himalayan and adjoining populations reveal the presence of East, Southeast and South Asian genetic signatures. Interestingly, we observed an antagonistic association of Y chromosomal haplogroups O3 and D clines with the longitudinal distance. Thus, we summarise that studying the Himalayan and adjoining populations is essential for a comprehensive reconstruction of the human evolutionary and ethnolinguistic history of eastern Eurasia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Himalayan mountain ranges extend from Pakistan in the West to the northeastern Indian states and northern Burma in the East. More than three hundred distinct ethnic groups and language communities have been documented in this region (van Driem 2001). The number of individuals living in the whole area is small due to the cold climate and some inhospitable alpine habitats. It has been estimated that the population size of the Himalayas tripled only in recent times, i.e., from 1901 to 1980 (Nandy and Rao 2001). Trans-Himayalan is the second most populous language family in the world, comprising more than 300 different languages, which can be divided into 42 subgroups (van Driem 2001). Out of the 42 major linguistic subgroups, 30 live to the south of the Himalayas, 7 to the North and East, and the remaining 5 are distributed over both the southern and northern flanks (van Driem 2001).
The prehistoric human settlement in this region remains poorly studied. It is still unclear when the first human activities occurred in this region. The archaeological record in Nepal suggests a long prehistory of human habitation, with abundant lithic material from the Early, Middle and Late Palaeolithic periods as well as through the Mesolithic and Neolithic periods (Corvinus and Erlangen Institut für Ur-und Frühgeschichte 2007). An Early Palaeolithic presence of the Acheulian tradition in the Early Middle Pleistocene is indicated by Acheulian bifacial sand flake tools industry, with Levallois technology appearing in the Middle Palaeolithic. Further, archaeological record suggest an increase in population density at the end of the last glacial maximum (Field et al. 2007). The foothills of the central Himalayas straddle the so-called Movius line, represent a transition zone between two traditions of lithic expression and show affinity with coetaneous Occidental technocomplexes as well as with technocomplexes in Southeast Asia (Schick 1994). The Acheulian artefacts in the Śivālik region were made by archaic humans about 0.7 Ma (Khanduri 2002; Ganjoo and Ota 2012; Leipe et al. 2014).
The Tibetan plateau was likewise occupied by modern humans during their first arrival in Asia (Qiu 2015), and the Neolithic presence of modern humans both North and South of the Himalayas is archaeologically widely attested (Khanduri 2002; Tewari 2007; Ganjoo and Ota 2012; Wang et al. 2012; Leipe et al. 2014). During the Neolithic time, many pastoralist communities have made permanent settlements in this region. One such pastoral community, the Changpa tribe in Ladakh, were thought to have originated from the ancient tribes on the Tibetan Plateau (Ganjoo and Ota 2012; Leipe et al. 2014).
Genetic studies so far, have suggested two different schools of thoughts (Torroni et al. 1994; Su et al. 2000; Gayden et al. 2007; Qin et al. 2010; Wang et al. 2012; Huerta-Sánchez et al. 2014; Bhandari et al. 2015; Zhang et al. 2017). Some studies have suggested that the Himalayan border formed a natural barrier for migratory animals including humans (Papiha et al. 1989; Aggarwal et al. 2003; Gayden et al. 2007). However, there is also evidence supporting the Himalaya as a corridor for human migrations (Debnath et al. 2011; Wang et al. 2012; Bhandari et al. 2015; Kang et al. 2016). These studies were mainly based on a limited number of samples as well as low-resolution markers. Moreover, to explore a detailed genomic ancestry of this region, one needs to cover the wider geographic region as well as populations, whereas previous studies were largely inclined towards Tibetan and Sherpa populations. Thus, to establish the temporal and spatial origin of Himalayan and adjoining populations (HAAPs) and their affinity with East and South Asians, we have undertaken an extensive study with more than 790 samples from the political boarders of India, however, their geographical spread is across the Himalayas (Fig. 1; Supplementary Table 1a; and Supplementary Text). Additionally, we have also assayed two phenotype markers, the 1540C variant in EDAR, known to be positively selected among East Asians (Bryk et al. 2008) and rs1426654 pigmentation gene SLC24A5 associated with lighter skin colour among South Asians (Jonnalagadda et al. 2011; Basu Mallick et al. 2013; Mishra et al. 2017). (Supplementary Table 8). We analysed these data to address four major questions: Which geographic region or ethnic group share the closest genetic affinity with the HAAPs? Do the HAAPs have any specific ancestry component? Since we included large number of populations from a diverse region of Himalaya, we also addressed whether we see any genetic cline of HAAPs? Is there any sex-specific admixture pattern, as we see among Indian Austroasiatic speakers?
Results and discussion
HAAP’s specific ancestry and genetic cline
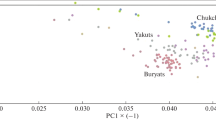
First, we merged the high-quality autosomal SNP data of 35 samples from this study with the available published data. The combined data was thereafter used in all our autosomal analyses (Supplementary Table 1b and Supplementary Text). To have an insight at the inter- and intra-population variation among the HAAPs, we computed the Fst distances (Supplementary Fig. 1 and Supplementary Table 2). The HAAPs showed closest and largely similar genetic distances with the East Asian and Southeast Asian populations, followed by the Central Asians. When we compared the intra-population distances among the HAAPs; Kusunda, Khasi and Sherpa showed larger distances, likely due to drift and, in the case of Kusunda and Khasi, perhaps due to linguistic isolation (Supplementary Table 3). Highlander Ladakh (India) pastoral population Changpa was closest to Tibetan followed by Subba and Sherpa populations. Among the South Asian populations, apart from Tibeto-Burman speakers, Munda speakers showed the closest affinity to the studied groups, probably due to the shared East/Southeast Asian ancestry (Chaubey et al. 2011). The principle component (PC) analysis of Eurasian groups placed the HAAPs closer to the East/Southeast Asians (Fig. 2a), supporting the Fst analysis. To see the placement of the HAAPs within the East/Southeast Asian cluster, we ran a separate PC analysis among the HAAPs along with the East and Southeast Asian populations (Supplementary Fig. 2). It is interesting to note that the HAAPs do not assemble in the defined East and Southeast Asian cline, instead they form their own cline. In the wider picture of a Eurasian PC plot, this cline is also visible and we see that it is stretched between East/Southeast Asia and South Asia (Fig. 2). This suggests that the HAAPs are admixed populations with higher East Asian-like and lower South Asian-like ancestries, whereas, the admixture observed in South Asian Munda (Austroasiatic) populations is characterised by the opposite pattern i.e., higher South Asian-like and lower East Asian-like ancestries (Chaubey et al. 2011).
Principal component analysis of the Eurasian populations showing the placement of the HAAPs (Upper pan). The barplot of global population ancestry pattern at K13, showing the unique and shared ancestry components of the HAAPs (Lower pan). The dark brown HAAPs specific ancestry can be seen among the populations from East, Southeast and Central Asians
To gain a better understanding of ancestral components as well as to test the scenario that has emerged from PC analysis, we inferred the ancestry components of HAAPs by using the model-based algorithm, ADMIXTURE (Alexander et al. 2009) (Fig. 2b and Supplementary Figs. 3 and 4). We have used our previously described method (Metspalu et al. 2011; Chaubey et al. 2015) to find the best K value (Supplementary Fig. 4). At K = 11, a unique ancestral component distinguishing itself from the widely known ancestries, was visible. This unique ancestry was observed to be highest among the HAAPs and was also found to be present in a majority of East/Southeast Asian and Central Asian populations (Fig. 2b). Intriguingly, the pattern of this component among East/Southeast Asian and Central Asian populations is fairly similar to the Siberian component reported earlier (Fedorova et al. 2013). To investigate whether the lack of Siberian samples in our data might have given rise to this pseudo-ancestry component, we included Siberian populations and repeated the analysis (Supplementary Fig. 5). The best K (K13) was consistent with our previous observation and supported the existence of the distinct HAAP-specific ancestry component. We also note here that the clinal pattern of the HAAPs in PC analysis is not only governed by East Asian and South Asian ancestry components, but a third local component also exists which profoundly moderates this pattern (Fig. 2a, b, and Supplementary Figs. 3 and 5). The spatial distribution of this component showed its peak in the Himalayan region with a moderate distribution across regions to the North of the Himalayas, i.e., in East Asia, Siberia and Central Asia (Supplementary Fig. 6). This component was virtually absent among South Asians (except for the HAAPs living in this region, including Tibeto-Burmans, Burusho and Hazara populations). The emergence of this component in this analysis also differentiated the HAAPs with some of the South Asian populations who received gene flow from East/Southeast Asia (Tharu and Munda). To confirm the high level of affinity of HAAPs with the populations living to the North of the Himalayas, we performed outgroup-shared drift f3 statistics (Patterson et al. 2012) (Supplementary Fig. 7). All the HAAPs largely showed a consistent pattern with a high number of alleles shared with one another as well as with East Asian populations.
The spatial distribution of the HAAP-specific ancestry up into Siberia was intriguing, considering its limited spread towards South Asia (Supplementary Fig. 6). However, one may also argue that the shared East Asian ancestry might be the reason for Siberians to show such elevated levels of this ancestry. To rule out this possibility, we masked East Asian-specific ancestry and counted the derived alleles shared between the Tibetan and the targeted populations (Supplementary Table 4). Here, we used the Dai population, who showed least HAAP-specific ancestry. The significantly higher level of alleles shared with Siberian populations opposed the above assumption and supported the independent nature of the HAAP-specific ancestry reported in Siberian populations (Supplementary Fig. 6 and Supplementary Table 4).
East Asian-specific ancestry
Since both of the ancestry components (HAAP and East Asian specific) were present among East Asian populations, we calculated aggregate East Asian ancestry (Han related) among studied populations using f4 ancestry ratio estimate implemented in AdmixTools (Patterson et al. 2012) (Table 1 and Supplementary Table 7). Among the HAAPs, the Austroasiatic Khasi showed the lowest East Asian ancestry, likely because of their distinct population history (Reddy et al. 2007; Chaubey et al. 2011). We estimated the level of gene flow for the HAAPs with the East Asian, Southeast Asian and South Asian groups using D statistics (Patterson et al. 2012). We used the Han as a proxy for East Asian and the Harijan as a representative of North Indian populations (Table 2). All the HAAPs showed a significant level of gene flow with the East Asian when compared to North Indian. However, Tharu, who live in the Tarai area, which forms a border between the Indo-Gangetic plain and the lower foothills of the Himalayas, showed a higher level of affinity with North Indian populations. We also investigated whether the Tibetan or Han share more ancestry with the HAAPs (Table 2). Not surprisingly, the Sherpa and Changpa shared a significant level of ancestry with the Tibetans, who are their close linguistic relatives, in comparison with the Han, whereas, Northeast Indians and Southeast Asians shared more ancestry with the Han. Subba (i.e., Limbu), Nyishi, Kusunda and Tharu too shared a higher number of alleles with the Han, though the difference was insignificant. All the HAAPs (except Khasi) shared a higher affinity with Tibetans when compared with the Cambodians. Notably, the HAAPs share a higher number of alleles with the Tibetans than with the Sherpa, likely due to high level of genetic drift and isolation of the Sherpa population (Table 2).
Validation from haplotype-based method
To gain a better understanding of genome sharing between the HAAPs, we used the haplotype-based method, fineStructure (Lawson et al. 2012). Consistent with allele frequency-based analyses, the inter-population haplotype-based analysis also showed East and Southeast Asian as major haplotype (chunk) donors to the HAAPs (Fig. 3). The mean intra-population chunk sharing for the HAAPs was significantly higher (two tailed p value < 0.0001), than the mean chunk donated by the East Asian populations (Supplementary Fig. 8), suggesting a high level of geneflow among the HAAPs. Among the HAAPs, the Tibeto-Burman language communities represented by the Tibetan, Changpa, Sherpa, Subba, Ao Naga, Naga and Nyishi populations showed high level of chunk allotment, however, linguistically Tibeto-Burman Garo, exhibited a lower level of chunk distribution. The non-Tibeto-Burman, Khasi and Kusunda remain fairly distant from the other HAAP populations (Supplementary Fig. 8).
Runs of homozygosity and admixture event(s)
The Runs of homozygosity (RoH)(Kirin et al. 2010) analysis was in consonance with the findings of previous studies, suggesting the Sherpa to be a unique isolated population due to low-effective population size (Ne), likely due to limited number of founders (Supplementary Fig. 9). After Sherpa, Ao Naga, Kusunda and Khasi also exhibited a large number of longer RoH segments in their genome. Among the HAAPs, Tibetan and Subba showed the lowest RoH segments. It is interesting to note that none of the HAAPs showed a ‘right shift’ pattern, which is an indicator of recent consanguinity.
We used the LD-based method Alder (Loh et al. 2013) to estimate the time of admixture for the HAAP-specific ancestry and South Asian ancestry. As a HAAP proxy, we used the Sherpa, whereas as a surrogate for North Indian populations we included the Harijan population. Many tests either failed and/or yielded low p value or Z scores. However, we were able to obtain significant test results for a few of the HAAPs, which clearly suggests that the expansion of the HAAP-specific component in South and Southeast Asia happened in the last 1000 years (Table 3). The recent entry of the HAAPs towards South Asia is also evident in its limited geographical spread (Supplementary Fig. 6).
Archaic ancestry among HAAPs
In the prior knowledge that the ancient hominini genome lingers on in extant modern humans, we assessed the genetic relatedness of the HAAPs with the ancient genomes published elsewhere (Green et al. 2010; Meyer et al. 2012; Prufer et al. 2014). We first investigated the outgroup f3 statistics for the 45 Kya older Ust’-Ishim genome (Fu et al. 2014) (Supplementary Fig. 10). The East Asia, Southeast Asian and the HAAPs showed an excess of derived alleles shared with the Ust’-Ishim man, supporting the previous observations (Lu et al. 2016; Wong et al. 2017). Notably, Indian Austroasiatic populations also exhibited largely a similar pattern, likely due to East/Southeast Asian ancestry or ancient shared ancestry with Ust’-Ishim. We applied a f4 ratio test in order to estimate the Denisovan ancestry among the HAAPs (Supplementary Table 8). We also included the Philippines negrito population, Aeta, and Papuan and Melanesian populations as positive controls. Consistent with the previous work, Tibetan and Sherpa showed traces of Denisovan ancestry (Yi et al. 2010; Huerta-Sánchez et al. 2014; Sankararaman et al. 2016; Hackinger et al. 2016). Moreover, we also see the same pattern among Khasi, Ao Naga, Naga and Nyishi populations. To confirm the Denisovan introgression, we applied RD(x) statistics (Qin and Stoneking 2015). Values > 1 was considered as an indicator of Denisovan introgression. Many of the HAAPs showed values close to 1 (Supplementary Table 7). Since our data contain a lower number of SNPs, we considered Z score > 2 as an indicator of Denisovan introgression.
EDAR and SLC24A5
Several scans of positive selection on genome-wide polymorphism data from global human populations have identified the EDAR (ectodysplasin-A receptor) gene as the candidate of the strongest positive selection among East Asian populations (Sabeti et al. 2007; Bryk et al. 2008; Fujimoto et al. 2008; Kamberov et al. 2013). The non-synonymous SNP rs3827760 (370A; 1540C) has been known to be associated with a multitude of phenotypes including hair shape (Fujimoto et al. 2008), tooth (Kimura et al. 2009), sweat gland density (Kamberov et al. 2013), reduced chin protrusion and ear pinna morphology (Adhikari et al. 2016). This SNP exists in a high frequency among populations of East Asian and native American origin but essentially absent among the Indians (Dravidian and Indo-European), European and African populations (Sabeti et al. 2007; Fujimoto et al. 2008; Chaubey et al. 2011). We analysed the 1540C polymorphism in 165 individuals of HAAPs and 43 Andaman and Nicobar Islanders (Supplementary Table 8a). On average, we have observed a similar mean frequency (~ 61%) of 1540C allele among HAAPs as was reported among Indian Tibeto-Burman populations (Chaubey et al. 2011). However, this allele was highly variable among Andaman and Nicobar islanders. The Onge population, which is genetically isolated, lacks 1540C allele, whereas the Great Andamanese populations have a low frequency of this allele(6.3%), and Nicobarese have a high frequency of 63.2% (Supplementary Table 8a). Next, we genotyped 423 individuals of the HAAPs including 10 ethnic groups for rs1426654 SNP located in SLC24A5 pigmentation gene. The ancestral allele G of rs1426654 predominates in Africans and East Asians (93–97%) and the derived A allele is almost fixed among Europeans (100%) (Lamason et al. 2005). We found that the rs1426654-A allele frequencies among the Tibeto-Burman speaking groups was low, ranging from 0 to 0.13 except for Sema Naga and Bhotia that showed the frequency of 0.23 and 0.22, respectively, which otherwise does not show a deviation of maternal and paternal ancestries from their neighbouring populations (Supplementary Tables 8b and 9). The low frequency among the Tibeto-Burman speakers corroborates with our earlier study (Basu Mallick et al. 2013). The comparatively higher A allele frequency in Bhotia population can be further explained either by their intermixing with the Indo-European speakers or(and) by their habitat which constitutes the Central Himalayan region, a region known for ethnic intermixing and the fact that they also practice vertical transhumance i.e., moving to higher pastures in summer and lower valleys in winter.
Sex-specific admixture
To gain a better understanding of sex-specific admixture, we analysed the HAAPs using mtDNA and Y chromosomal markers (Table 4, Supplementary Fig. 11 and Supplementary Tables 9 and 10). For mtDNA, all the HAAPs carried frequent East/Southeast Asia-specific haplogroups supporting their overwhelming ancestry from East/Southeast Asia. However, the Sukna village population which are a heterogeneous mixture of Austroasiatic and other tribes showed a high level of South Asian maternal haplogroups. Likewise, Austroasiatic population Khasi showed a higher level of South-Asia-specific maternal ancestry. Consistent with the maternal ancestry, the paternal ancestry of the HAAPs was clearly inclined towards East/Southeast Asia. For Y chromosomal haplogroups, Sukna village tribes and Khasi did not mirror the maternal profile. Overall, the HAAPs showed a moderately male-biased admixture. However, this bias was not as severe as observed in Munda-speaking populations (Chaubey et al. 2011).
It is also interesting to note that the HAAPs showed a maternal segregation of East Asian vs Southeast-Asia-specific haplogroups. The major Southeast Asia-specific haplogroups (B4, B5, F) were rare (except for F1a which was moderate among Northeast Indian populations) in the HAAPs, whereas Tibet/East Asia-specific haplogroups (A, C and D) were frequent (Supplementary Tables 9 and 10). The Y chromosomal haplogroup (hg) D-M174 was predominant among populations living at high altitude (Tibetan, Changpa, Sherpa and Bhutia), while hg O3-M134 was dominant among populations living at lower altitude (Table 4 and Supplementary Table 9). Interestingly, we observed a decreasing trend in the frequency of hg D-M174 from Jammu and Kashmir to the northeast Indian states. However, the opposite trend was observed for hg O3-M134 (Table 4).
Conclusions
In conclusion, we report a distinct genetic cline of the HAAPs, which is likely to have originated due to a novel genetic component, admixed to different degrees with East and South Asian ancestries. The HAAPs share a high level of affinity with the East Asian populations, rather than with South Asians. The spatial distribution of the HAAP-specific component is widespread over the regions to the North of the Himalayas, whereas this component experiences an abrupt fall towards the western Himalayas. In haplotype-based analysis, the HAAPs showed massive intra-population sharing of chunks than any other neighbouring groups (East Asian or South Asian), supporting a common ancestry of the populations studied here. The haplotype- and RoH-based analyses supported the isolated nature of the Sherpa population. Consistent with the previous analyses, many of the HAAPs showed a significant level of shared ancestry with Ust’-Ishim and Denisovan. Our time of admixture estimation suggested a recent dispersal of the HAAP-specific ancestry in Northeast India and northern Nepal.
Materials and methods
Detailed materials and methods have been added in the Supplementary material online. 2–5 ml of blood samples were collected from 794 individuals belonging to 16 ethnic groups. This project was carried out in accordance with the Helsinki Declaration guidelines and also permitted by the Institutional Ethical Committee of the CSIR-Centre for Cellular and Molecular Biology, Hyderabad, India. After careful removal of people related to last three generations, we landed to 792 samples which have been genotyped for mtDNA and Y chromosome markers (Supplementary Table 1a). For autosomal analysis, we genotyped 35 individuals with Affymatrix following the manufacturer specifications. After quality control, we have merged this data with the data published elsewhere (Supplementary Table 1b). We have calculated Fst distance (Weir and Cockerham 1984), performed PC (Principle component) analysis (Patterson et al. 2006), unsupervised ADMIXTURE analysis (Alexander et al. 2009), fineSTRUCTURE analysis (Lawson et al. 2012), and AdmixTool (Patterson et al. 2012), package for f3, f4 and D statistics (Supplementary Table 8). Further, we have used the RD(x) statistics defined elsewhere (Qin and Stoneking 2015), to confirm Denisovan introgression. The Runs of Homozygosity (ROH) was calculated using Plink 1.9 (Chang et al. 2015). To estimate the admixture time, we used the ALDER software (Loh et al. 2013), for HAAP populations.
References
Adhikari K, Fuentes-Guajardo M, Quinto-Sánchez M et al (2016) A genome-wide association scan implicates DCHS2, RUNX2, GLI3, PAX1 and EDAR in human facial variation. Nat Commun 7:11616
Aggarwal R, Ramadevi J, Singh L (2003) Ancient origin and evolution of the Indian wolf: evidence from mitochondrial DNA typing of wolves from Trans-Himalayan region and Pennisular India. Genome Biol 4:p6
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19:1655–1664
Basu Mallick C, Iliescu FM, Möls M et al (2013) The light skin allele of SLC24A5 in South Asians and Europeans shares identity by descent. PLoS Genet 9:e1003912
Bhandari S, Zhang X, Cui C et al (2015) Genetic evidence of a recent Tibetan ancestry to Sherpas in the Himalayan region. Sci Rep 5:16249
Bryk J, Hardouin E, Pugach I et al (2008) Positive selection in East Asians for an EDAR allele that enhances NF-kappaB activation. PLoS ONE 3:e2209
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4:7. https://doi.org/10.1186/s13742-015-0047-8
Chaubey G, Metspalu M, Choi Y et al (2011) Population genetic structure in indian austroasiatic speakers: the role of landscape barriers and sex-specific admixture. Mol Biol Evol 28:1013–1024
Chaubey G, Kadian A, Bala S, Rao VR (2015) Genetic affinity of the Bhil, Kol and Gond mentioned in epic Ramayana. PLoS One 10:e0127655
Corvinus G, Erlangen Institut für Ur-und Frühgeschichte (2007) Prehistoric cultures in Nepal: from the Early Palaeolithic to the Neolithic and the quaternary geology of the Dang-Deokhuri Dun Valleys. Otto Harrassowitz, Aryan Books International, New Delhi, p 646
Debnath M, Palanichamy MG, Mitra B et al (2011) Y-chromosome haplogroup diversity in the sub-Himalayan Terai and Duars populations of East India. J Hum Genet 56:765–771
Fedorova SA, Reidla M, Metspalu E et al (2013) Autosomal and uniparental portraits of the native populations of Sakha (Yakutia): implications for the peopling of Northeast Eurasia. BMC Evol Biol 13:127
Field J, Petraglia M, Lahr M (2007) The southern dispersal hypothesis and the South Asian archaeological record: examination of dispersal routes through GIS analysis. J Anthropol Archaeol 26:88–108
Fu Q, Li H, Moorjani P et al (2014) Genome sequence of a 45,000-year-old modern human from western Siberia. Nature 514:445–449
Fujimoto A, Kimura R, Ohashi J et al (2008) A scan for genetic determinants of human hair morphology: EDAR is associated with Asian hair thickness. Hum Mol Genet 17:835–843
Ganjoo R, Ota S (2012) Mountain environment and early human adaptation in NW Himalaya, India: a case study of Siwalik Hill Range and Leh valley. Quat Int 269:31–37
Gayden T, Cadenas AM, Regueiro M et al (2007) The Himalayas as a directional barrier to gene flow. Am J Hum Genet 80:884–894
Green RE, Krause J, Briggs AW et al (2010) A draft sequence of the Neandertal genome. Science 328:710–722
Hackinger S, Kraaijenbrink T, Xue Y et al (2016) Wide distribution and altitude correlation of an archaic high-altitude-adaptive EPAS1 haplotype in the Himalayas. Hum Genet 135:393–402
Huerta-Sánchez E, Jin X, Asan et al (2014) Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512:194–197
Jonnalagadda M, Ozarkar S, Mushrif-Tripathy V (2011) Population affinities of Parsis in the Indian subcontinent. Int J Osteoarchaeol 21:103–110
Kamberov YG, Wang S, Tan J et al (2013) Modeling recent human evolution in mice by expression of a selected EDAR variant. Cell 152:691–702
Kang L, Wang C-C, Chen F et al (2016) Northward genetic penetration across the Himalayas viewed from Sherpa people. Mitochondrial DNA 27:342–349
Khanduri B (2002) Archaeology of central Himalaya: a review. Puraratna Emerg Trends Archaeol Art Anthropol Conserv Hist Honour Shri Jagat Pati Joshi 1:213
Kimura R, Yamaguchi T, Takeda M et al (2009) A common variation in EDAR is a genetic determinant of shovel-shaped incisors. Am J Hum Genet 85:528–535
Kirin M, McQuillan R, Franklin CS et al (2010) Genomic runs of homozygosity record population history and consanguinity. PLoS One 5:e13996
Lamason RL, Mohideen M-AP, Mest JR et al (2005) SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310:1782–1786
Lawson DJ, Hellenthal G, Myers S, Falush D (2012) Inference of population structure using dense haplotype data. PLoS Genet 8:e1002453
Leipe C, Demske D, Tarasov PE et al (2014) Potential of pollen and non-pollen palynomorph records from Tso Moriri (Trans-Himalaya, NW India) for reconstructing Holocene limnology and human–environmental interactions. Quat Int 348:113–129
Loh P-R, Lipson M, Patterson N et al (2013) Inferring admixture histories of human populations using linkage disequilibrium. Genetics 193:1233–1254
Lu D, Lou H, Yuan K et al (2016) Ancestral origins and genetic history of Tibetan highlanders. Am J Hum Genet. https://doi.org/10.1016/j.ajhg.2016.07.002
Metspalu M, Romero IG, Yunusbayev B et al (2011) Shared and unique components of human population structure and genome-wide signals of positive selection in South Asia. Am J Hum Genet 89:731–744
Meyer M, Kircher M, Gansauge M-T, et al (2012) A high-coverage genome sequence from an archaic Denisovan individual
Mishra A, Nizammuddin S, Mallick CB et al (2017) Genotype-phenotype study of the middle Gangetic plain in India shows association of rs2470102 with skin pigmentation. J Invest Dermatol 137:670–677
Nandy S, Rao K (2001) Census 2001: population dynamics of Indian Himalaya. Envis Bull 9:1–6
Papiha SS, Mastana SS, Stephenson A (1989) Serogenetic investigations of Tibetans and Himachalis from Himachal Pradesh, India: genetic relationship between Tibetans and certain selected mongoloid populations. Jinrui Idengaku Zasshi 34:143–157
Patterson N, Moorjani P, Luo Y et al (2012) Ancient admixture in human history. Genetics 192:1065–1093
Patterson N, Price AL, Reich D (2006) Population Structure and Eigenanalysis. PLoS Genetics 2 (12):e190
Prufer K, Racimo F, Patterson N et al (2014) The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505:43–49
Qin P, Stoneking M (2015) Denisovan ancestry in East Eurasian and native American populations. Mol Biol Evol 32:2665–2674. https://doi.org/10.1093/molbev/msv141
Qin Z, Yang Y, Kang L et al (2010) A mitochondrial revelation of early human migrations to the Tibetan Plateau before and after the last glacial maximum. Am J Phys Anthropol 143:555–569
Qiu J (2015) Who are the Tibetans? Science 347:708–711
Reddy BM, Langstieh BT, Kumar V et al (2007) Austro-Asiatic tribes of Northeast India provide hitherto missing genetic link between South and Southeast Asia. PLoS ONE 2:e1141
Sabeti PC, Varilly P, Fry B et al (2007) Genome-wide detection and characterization of positive selection in human populations. Nature 449:913–918
Sankararaman S, Mallick S, Patterson N, Reich D (2016) The combined landscape of Denisovan and Neanderthal ancestry in present-day humans
Schick KD (1994) The Movius Line reconsidered: perspectives on the earlier Paleolithic of eastern Asia. In: Corruccini RS, Ciochon RL (eds) Integrative Paths to the Past, Prentice Hall, New jersey, pp 569–596
Su B, Xiao C, Deka R et al (2000) Y chromosome haplotypes reveal prehistorical migrations to the Himalayas. Hum Genet 107:582–590
Tewari R (2007) Some important stone images from Uttaranchal Himalaya. South Asian Stud 23:95–106
Torroni A, Miller JA, Moore LG et al (1994) Mitochondrial DNA analysis in Tibet: implications for the origin of the Tibetan population and its adaptation to high altitude. Am J Phys Anthropol 93:189–199
Van Driem G (2001) Languages of the Himalayas: an ethnolinguistic handbook. Handbuch der Orientalistik. Leiden, Brill
Wang H-W, Li Y-C, Sun F et al (2012) Revisiting the role of the Himalayas in peopling Nepal: insights from mitochondrial genomes. J Hum Genet 57:228–234
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Wong EH, Khrunin A, Nichols L et al (2017) Reconstructing genetic history of Siberian and Northeastern European populations. Genome Res 27:1–14
Yi X, Liang Y, Huerta-Sanchez E et al (2010) Sequencing of 50 human exomes reveals adaptation to high altitude. Science. https://doi.org/10.1126/science.1190371
Zhang C, Lu Y, Feng Q et al (2017) Differentiated demographic histories and local adaptations between Sherpas and Tibetans. Genome Biol 18:115. https://doi.org/10.1186/s13059-017-1242-y
Acknowledgements
During the revision of this manuscript, we have lost our senior collegue and mentor Dr. Lalji Singh, we dedicate this article to him to show our respect for his dedication to Science. We thank all the volunteers for their kind participation in this study. All the individuals assisting the sample collection are highly appreciated for their timely help and cooperation. RT was supported by CSIR_SRF and European Social Funds Doctoral Studies and Internationalisation Programme DoRa. GC is supported by Estonian team grant (PRG-213), RV was supported by Estonian Institutional Research Grants IUT24-1, KT was supported by the Council of Scientific and Industrial Research (CSIR), Government of India, LS was supported by DST, Government of India and CBM was supported by Marie Sklodowska- Curie Actions individual fellowship (Grant-70429). We acknowledge the late R Rajkumar for his contribution in sequencing some Y chromosome and mtDNA markers among the studied populations.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Deceased: Lalji Singh.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tamang, R., Chaubey, G., Nandan, A. et al. Reconstructing the demographic history of the Himalayan and adjoining populations. Hum Genet 137, 129–139 (2018). https://doi.org/10.1007/s00439-018-1867-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-018-1867-2