Abstract
Congenital myopathies are a heterogeneous group of muscle disorders that are often genetically determined. Here, we investigated a boy with congenital myopathy, deafness, and neuropathy from a consanguineous Kurdish family by autozygosity mapping and whole exome sequencing. We found a homozygous nonsense mutation in SPTBN4 [c.1597C>T, NM_020971.2; p.(Q533*), NP_066022.2; ClinVar SUB2292235] encoding βIV-spectrin, a non-erythrocytic member of the β-spectrin family. Western blot confirmed the absence of the full-length 288 kDa isoform in muscle and of a specific 72 kDa isoform in fibroblasts. Clinical symptoms of the patient largely corresponded to those described for the quivering mouse, a loss-of-function animal model. Since the human phenotype of βIV-spectrin deficiency included a myopathy with incomplete congenital fiber-type disproportion, we investigated muscle of the quivering (qv4J) mouse and found complete absence of type 1 fibers (fiber-type 2 uniformity). Immunohistology confirmed expression of βIV-spectrin in normal human and mouse muscle at the sarcolemma and its absence in patient and quivering (qv4J) mouse. SPTBN4 mRNA-expression levels in healthy skeletal muscle were found in the range of other regulatory proteins. More patients have to be described to confirm the triad of congenital myopathy, neuropathy and deafness as the defining symptom complex for βIV-spectrin deficiency.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Congenital myopathies are a heterogeneous group of genetic muscle disorders with hypotonia and muscle weakness from birth. About 20 genes are associated with congenital myopathies, but often the molecular pathology is unknown (Maggi et al. 2013). Histological abnormalities comprise rods, cores, caps, central nuclei, and fiber-type predominance or uniformity (North et al. 2014). Some of the congenital myopathies are characterized by congenital fiber-type disproportion (CFTD), which is an abnormality of muscle fiber size and proportion with slow-twitch type 1 fibers being consistently smaller and more numerous than fast-twitch type 2 fibers. Sometimes one may find fiber-type 1 uniformity, in unusual cases or biopsy sites only fiber-type 1 atrophy may be seen. CFTD can be inherited as an autosomal dominant (ACTA1 MIM #255310, MYH7 MIM #160500, TPM3 MIM #255310, TPM2 MIM #609285) or recessive (RYR1 MIM #117000, SEPN1 MIM #255310) trait (Clarke 2011; North et al. 2014). Proteins involved in the pathophysiology of congenital myopathy play an important role in skeletal muscle contraction via their interaction with myosin. Others are involved in skeletal muscle calcium homeostasis or are located at the sarcomere of striated muscle where they associate with thin α-actinin filaments (North et al. 2014).
βIV-spectrin is a non-erythrocytic member of the β-spectrin family. Mutations in the Sptbn4 gene (syn. Spnb4) of the quivering mouse disturb the axon initial segments and nodes of Ranvier, which are important domains for initiation, propagation, and modulation of action potentials (Yang et al. 2004). At the nodes of Ranvier, βIV-spectrin is required for ion channel clustering (Hedstrom and Rasband 2006; Devaux 2010). Sptbn4 loss-of-function mutations in mice cause ataxia, neuromyotonia, myokymia, tremor, motor neuropathy, and central deafness (Parkinson et al. 2001; Devaux 2010). Humans with mutations in SPTBN4 (MIM *606214) have not been described. Here, we report the discovery of SPTBN4 as a novel candidate disease gene for congenital myopathy in a consanguineous Kurdish family.
Materials and methods
Human subjects
The parents of the patient provided written informed consent for all aspects of the study (Charité IRB approval EA2/107/14) and publication of the patient’s photographs according to the Declaration of Helsinki. Previous gene testing had excluded spinal muscular atrophy, Prader–Willi-Syndrome, myotonic dystrophy, myotubular and centronuclear myopathy, and mutations in COLQ, IGHMBP2, POMGNT1, and FKRP. Genomic DNA was extracted from white blood cells of all investigated family members and from a chorionic villus biopsy specimen that had been obtained for prenatal testing.
Animals
Quivering mice are characterized by progressive ataxia with hind limb paralysis, deafness and tremor. They carry mutations in the murine βIV-spectrin gene (Spnb4) and several alleles have been described which cause phenotypes of varying severity. The study here investigated the qv4J mouse, which carries a nonsense mutation in Spnb4 (p.Q1358*) that truncates the protein by 47% (Parkinson et al. 2001).
Autozygosity mapping
Autozygosity mapping was performed using the variant calling files (VCF) from whole exome sequencing of the patient and his parents. In these variant files, the HomozygosityMapper2012 software [http://www.homozygositymapper.org, accessed Dec 2016] (Seelow and Schuelke 2012) searched for homozygous/autozygous stretches of 300 SNPs or longer that were only homozygous in the patient but not in his parents. Homozygous stretches of >30 SNPs in the parents were excluded as candidate regions. This method yielded a total of 380 protein-coding genes that were covering 32.9 Mbp on chromosomes 7 and 19 (Fig. 2b).
Whole exome analysis
Exonic sequences were enriched from the patient and his parents using the SureSelect® V4 Human All Exon Kit (Agilent) and sequenced on a HiSeq 2000 machine (Illumina) as 101 bp paired-end fragments. FASTQ files were aligned to the human GRCh37.p11 (hg19) reference sequence using the BWA-MEM v0.7.1 aligner (Li 2013). The quality of the alignment is provided in Supplementary Table 2. Subsequently, variant VCF-files were generated for all exons ±20 bp flanking regions using the GATK v3.3 software package (DePristo et al. 2011) and sent to the MutationTaster2 software [http://www.mutationtaster.org, accessed Dec 2016] for assessment of potential pathogenicity (Schwarz et al. 2014).
As the parents were consanguineous, variants were filtered for recessive inheritance and removed if occurring homozygously either in the ExAC database in >20 cases [http://exac.broadinstitute.org, accessed Dec 2016; http://biorxiv.org/content/early/2015/10/30/030338, accessed Dec 2016] or in the 1000 Genomes project in >10 cases [http://www.1000genomes.org, accessed Dec 2016]. All relevant variants were inspected visually using the Integrative Genomics Viewer (IGV) [http://www.broadinstitute.org/igv/, accessed Dec 2016] and their segregation was verified by Sanger sequencing using gene specific oligonucleotide primers and the BigDye® (Applied Biosystems) protocol on an ABI3500 Genetic Analyzer (ThermoFischer Scientific). For verification of the SPTBN4 c.1597C>T mutation (GenBank NM_020971) we analyzed the PCR-product generated with the oligonucleotide primer pair FW: 5-CAG GGT CAC ACA GGG TCA AG-3 and REV: 5-CCC TTC CCT CTC CAT CTC CA-3.
Beyond the homozygous variant in SPTBN4 we additionally found two potentially pathogenic variants in the autozygous region (Supplementary Table 3). The variant in the FFAR2 gene encoding the free fatty acid receptor 2 was excluded as the cause of the described disease here, because FFAR2 is only expressed in blood and immune cells and Ffar2 mutations in the mouse cause a different phenotype with exacerbating inflammations of various tissues. The homozygous variant in the RASIP1 gene was excluded because the gene is only expressed in heart and lung and Rasip1 mutations in the mouse cause cardiac malformations and intrauterine growth retardation and death.
Virtual gene panel and trio analysis
To exclude mutations in genes that are known to be associated with either congenital myopathies or other muscle diseases, we specifically screened the VCF-variant files of the patient for mutations therein. The composition of these virtual gene panels is provided in the Supplementary online material. Additionally, because only one child was affected in the family, we did a trio analysis for de novo mutations but did not find any in the coding regions of protein-coding genes.
Quantitative RT-PCR analysis
Total RNA was isolated from the patient and three control fibroblast lines using the TRIzol reagent, and reversely transcribed into cDNA with SuperScript III (Invitrogen) polymerase using oligo (dT) primers. Quantitative SYBR green qPCR (Life Technologies) was performed on an ABI PRISM 7700 cycler (Applied Biosystems) using the PCR-efficiency-corrected −ΔΔCt method (Pfaffl 2001) with the SPTBN4 (GenBank NM_020971) primer pair FW: 5-CTG GAG AAC GTG GAC AAG GC-3; REV: 5-TCA GCC GGT GAT TCC CAT C-3. Expression levels were normalized to 18S rRNA (FW: 5-CAT TCG AAC GTC TGC CCT ATC-3; REV: 5-CTC CCT CTC CGG AAT CGA AC-3, GenBank NR_003286.2) and GAPDH (FW: 5-TGC ACC ACC AAC TGC TTA GC-3; REV: 5-GGC ATG GAC TGT GGT CAT GAG-3, GenBank NM_002046) as described (Relizani et al. 2014).
Muscle transcriptome analysis
Transcriptome raw data (fastq files) from four healthy male muscle samples were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/; Project GSE56787, samples SRR1398532, SRR1398533, SRR1398534, and SRR1398535) (Yao et al. 2014) and aligned to the human reference sequence (GRCh37.75) with the STAR 2.4.0.1 aligner (Dobin et al. 2013). Resulting BAM files were investigated and normalized with the CUFFLINKS 2.2.1. pipeline (Trapnell et al. 2012), which yields the relative mRNA quantities for each transcript present in the transcriptome dataset. As the quantitative measure for gene abundance we used the FPKM value (fragments per kilobase of transcript per million mapped reads), which is calculated by the CUFFQUANT module of the CUFFLINKS software. We depict the FPKM values from the above mentioned datasets of SPTBN4 along with other well known muscle genes (Fig. 3).
Results
Clinical report
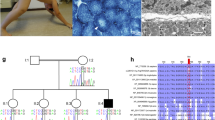
The male patient was born to healthy, first cousin parents. His elder sister is healthy. In the newborn period, he presented with general muscular hypotonia and facial weakness. Muscle tendon reflexes were absent, his motor development was delayed and he never gained head control. Feeding problems due to weak suck required gavage feeding from the age of 1 year. Cranial MRI at the age of 4 years was normal except for mildly enlarged CSF spaces. Serum CK-levels were never elevated; muscle biopsy at the age of 5 years demonstrated fiber-type 1 atrophy suggesting a rare congenital form of an unstructured congenital myopathy (Fig. 1f). At the age of 10 years the boy presented with myopathic facies, highly arched palate (Fig. 1a, b), severe distal muscle weakness, generalized muscle atrophy, scoliosis, ankle contractures (Fig. 1c–e), and severely retarded motor and mental development. Presently, he is not able to stand, sit, eat, or drink without support. He did not learn to speak. Early brainstem evoked potentials could not be elicited with 90 dB SPL while otoacoustic emissions were entirely normal. Neurography showed a combined axonal and demyelinating motor neuropathy with 30.9 m/s, 0.4 mV (peroneal nerve, normal >41.1 m/s, >2.6 mV).
a–e Clinical phenotype of the patient with a myopathic face, b highly arched palate, c distal weakness, d thenar and interdigital muscle atrophy, and e distal muscle wasting with fixed extension contractures of the feet; f The ATPase pH 4.3 stain shows fiber-type disproportion in the quadriceps muscle with reduction and atrophy of type 1 fibers (black) and predominance of type 2C (dark gray) and type 2A/B fibers (light gray); g Immunostaining of age-matched control and patient muscle with antibodies directed against β-spectrin (NCL SPEC1, clone RB C2/3D5, Novocastra UK; 1:100) and the non-erythrocytic βIV-spectrin (sc-368195, H-85, Santa-Cruz, 1:100), both expressed at the sarcolemma in normal muscle. Absence of anti-βIV-spectrin immunostaining at the sarcolemma of the patient; the remaining signals derive from interstitial autofluorescent material
Prenatal testing
Based on the molecular findings in the index patient, the parents asked us for prenatal testing in a further pregnancy, despite the fact that their child was the only patient described with a mutation in SPTBN4 so far. Chorionic villus sampling was done at 12 weeks of gestation. The fetus was heterozygous for the mutation. Maternal DNA contamination of the fetal DNA was excluded by genotyping with microsatellite markers. The boy was born at term, developed normally until now (3 months of age), and did not show any signs of muscle hypotonia and weakness.
Molecular genetics
Autozygosity mapping combined with whole exome sequencing revealed an autosomal recessive nonsense mutation in SPTBN4 (Chr19:g41,009,971C>T, GRCh37.p11 (hg19); c.1597C>T, NM_020971.2; p.(Q533*), NP_066022.2, ClinVar SUB2292235). This variant was neither listed in the 1000 Genomes, the Exome Variant Server, dbSNP, or the ExAC database. Additionally, the variant was absent from our local whole exome database (>150 exomes).
To exclude mutations in genes that are known to be associated with either congenital myopathies or with other muscle diseases, we specifically screened the VCF-variant files of the patient for mutations therein (Supplementary online material). Additionally, because only one child was affected in the family, we did a trio analysis for de novo mutations, but did not find any in the coding regions of protein-coding genes. Sanger sequencing confirmed autosomal recessive inheritance (Fig. 2a, c).
a Pedigree of the consanguineous family with segregation of the SPTBN4 mutation; b autozygous regions stretching over 300 SNPs that only occurred in the patient, but not in his healthy parents. The graph has been generated with our HomozygosityMapper2012 software. The red bars mark the two autozygous regions. c Sequence electropherograms of the c.1597C>T nonsense mutation in control, parents, and patient; d Western blot of patient and control muscle shows a reduction of the full length βIV-spectrin protein abundance in the patient. Staining with anti-α-actinin served as loading control. e Western blot of patient and control fibroblasts shows absence of the 72 kDa βIV-spectrin band in the patient, an unspecific band at ≈55 kDa and anti-β-tubulin (ab6046, Abcam, 1:10,000) serving as loading controls
To explore the effect of the mutation on the protein level, we first investigated cultured skin fibroblasts (Fig. 2e). SPTBN4 encodes a minor full-length isoform of 2564 amino acids with a predicted molecular weight of 289 kDa and a truncated major isoform of 72 kDa comprising 678 amino acids starting from amino acid position 1335 of the minor isoform (Tse et al. 2001). In two control fibroblast lines, the 72 kDa band was clearly present, while it was absent from the patient fibroblasts. As the antibody was raised against a peptide encoding AA #1661–1745 of βIV-spectrin, it would be able to bind to the truncated isoform, which does not include the premature termination codon at AA #533. The absence of the short isoform protein band in the patient indicated that nonsense-mediated mRNA decay of the full-length mRNA had occurred before or during splicing. Second, we performed Western blot analysis in human skeletal muscle from the patient and a control. We found a strong band at ≈290 kDa in control tissue corresponding to the full-length isoform of βIV-spectrin and a clear reduction of this band in the patient (Fig. 2d). Immunostaining of muscle cryosections from healthy humans located βIV-spectrin at the sarcolemma and in the muscle capillaries, while the sarcolemmal signal was clearly absent in the muscle of the patient (Fig. 1g).
qPCR with two primer pairs, one product located in the 5′-region and the other one in the 3′-region of the large mRNA that had been isolated from skin fibroblasts (Supplementary online material) demonstrated a ≈85% reduction of SPTBN4 mRNA copy numbers (patient 0.03 ± 0.01 SPTBN4/106 18S RNA, controls (n = 3) 0.18 ± 0.07). This finding would be consistent with nonsense-mediated mRNA messenger decay.
Histological investigation of mouse muscle
Anti-βIV-Spectrin staining of mouse muscle from controls revealed a strong signal at the sarcolemma and the interstitial tissue, which was absent in the muscle cryosections from the quivering mouse (qv4J) that carries a nonsense mutation in the murine Sptbn4 gene. More severe as seen in the human muscle, type 1 muscle fibers were entirely absent from the quivering mouse tibialis anterior muscle as demonstrated by the ATPase pH 4.3 staining (Supplementary Fig. 3).
SPTBN4 gene expression in skeletal muscle
Transcriptome analysis of human skeletal muscle by RNA sequencing revealed comparatively low SPTBN4 mRNA expression levels in the range of transcriptions factors (PAX7, MYOD, PPARG), proteins of the Notch pathway (NOTCH1, RBPJ, HES1), muscle growth factors (MSTN), and proteins important for the regulation of neuromuscular transmission (MUSK).
Discussion
We describe the first human patient with a loss-of-function mutation of βIV-spectrin. He presented with congenital myopathy, neuropathy, and deafness. The clinical features in a human thus largely correspond to those of the quivering mouse. Loss of βIV-spectrin in mice was associated with progressive ataxia and a hearing defect that was attributed to mislocation of voltage-gated channels at the axon initial segments and nodes of Ranvier (Parkinson et al. 2001; Komada and Soriano 2002). Alterations in the location of sodium and potassium channels in myelinated nerves slow the propagation and desynchronize action potentials. Changes in axonal membrane potential can lead to neuropathy and the alteration of membrane excitability, which might be an explanation for the severe combined axonal and demyelinating neuropathy of our patient.
As in the quivering mouse, we detected a disturbed auditory function. Otoacoustic emissions assist differentiating between pathologies located at cochlear versus higher (e.g., cochlear nerve, brainstem) levels. While the inner ear function appeared undisturbed, the brainstem conduction was severely impaired in our patient. These results exactly resemble the phenotype of the quivering mouse where cochlear morphology and function were normal, while the transmission of auditory potentials through the brainstem nuclei was abnormal thereby verifying central deafness in the quivering mouse as opposed to the most frequent causes of deafness originating from the cochlea (Bock and Steel 1983; Deol et al. 1983).
In addition to the originally described quivering mouse, the human phenotype of βIV-spectrin deficiency includes congenital myopathy with incomplete congenital fiber-type disproportion. So far, the muscle of the quivering mouse had not been evaluated in detail and we were now able to demonstrate a complete absence of type 1 fibers (fiber-type 2 uniformity) in the tibialis anterior muscle of the quivering (qv4J) mouse (Supplementary Fig. 3). Western blot analysis of human skeletal muscle from the patient and a control demonstrated the presence of βIV-spectrin in muscle tissue and a clear reduction of the protein amount in the mutated muscle specimen. We found a predominantly sarcolemmal distribution of βIV-spectrin in human and in mouse skeletal muscle. Immunohistochemical analysis of the trans-sarcolemmal protein β-dystroglycan confirmed the integrity of the cell membrane (Supplementary Figure 1), and the absence of α-laminin V staining at the sarcolemma largely rules out a congenital muscular dystrophy (Supplementary Fig. 2).
Hund et al. found βIV-spectrin to associate with critical structural and regulatory proteins in excitable cells not only in the brain, but also in the heart (Hund et al. 2010). The authors postulate that βIV-spectrin may associate with key membrane domains of other tissues where it modulates local signaling pathways and alters cell excitability, e.g., the flow of sodium ions through the postsynaptic membrane at the neuromuscular junction. This might explain the early developmental defect of the muscle and the comparatively low SPTBN4 mRNA-expression levels in the range of other regulatory proteins of the skeletal muscle (Fig. 3).
Transcriptome analysis of n = 4 healthy individuals by RNA sequencing places SPTBN4 mRNA-expression levels in the low range, typically occupied by other regulatory proteins and transcription factors. Transcript abundance is depicted as FPKM values (fragments per kilobase of transcript per million mapped reads). The figure gives an overview of the relative mRNA-expression levels of genes coding for various muscle proteins. Muscle enzymes: ALDOA aldolase A, GPDH glyceraldehyde-3-phosphate dehydrogenase, PYGM glycogen phosphorylase; Components of the contractile apparatus: TNNT1 troponin T1, TPM3 tropomyosin 3; Components of the energy metabolism: ATP2A1 sarcoplasmic reticulum type Ca++-transporting ATPase, NDUFV1 NADH:ubiquinone oxidoreductase core subunit V1, PDHA1 pyruvate dehydrogenase α-subunit 1, CKMT2 mitochondrial creatine kinase 2; Proteins of the dystrophin complex: DMD dystrophin, DAG1 dystroglycan 1, DTNA α-dystrobrevin, SSPN sarcospan, SGCA/B/G/D sarcoglycan α/β/γ/δ, NOS1 nitric oxide synthase 1; Ion channels of the muscle: RYR1 ryanodine receptor 1, RAPSN receptor associated protein of the synapse, CHRNA1/B1/G/D nicotinic cholinergic receptor subunits α1/β1/γ/δ, CLCN1 voltage-gated chloride channel 1, SCN4A voltage-gated sodium channel α-subunit 4; Regulatory proteins of the muscle: PAX7 paired box factor 7, MYOD myogenic differentiation factor 1, NOTCH1 Notch 1, RBPJ recombination signaling binding protein for immunoglobulin κ J region, HES1 Hes family BHLH transcription factor 1, PPARG peroxysome proliferator-activated receptor γ, MSTN myostatin, MUSK muscle associated receptor tyrosine kinase, SPTBN4 non-erythrocytic βIV-spectrin
In conclusion, the abrogation of βIV-spectrin in muscle cells leads to a distinct form of congenital myopathy associated with neuropathy and central deafness. However, more patients have to be found to verify this triad as the defining symptom complex for βIV-spectrin deficiency. Replication will be the ultimate proof that this gene is indeed involved in congenital myopathies, and will give an idea of its frequency and presence in other cohorts.
References
Bock GR, Steel KP (1983) Inner ear pathology in the deafness mutant mouse. Acta Otolaryngol (Stockh) 96:39–47. doi:10.3109/00016488309132873
Clarke NF (2011) Congenital fiber-type disproportion. Semin Pediatr Neurol 18:264–271. doi:10.1016/j.spen.2011.10.008
Deol MS, Frank MP, Steel KP, Bock GR (1983) Genetic deafness of central origin. Brain Res 258:177–179. doi:10.1016/0006-8993(83)91248-9
DePristo MA, Banks E, Poplin R et al (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. doi:10.1038/ng.806
Devaux JJ (2010) The C-terminal domain of βIV-spectrin is crucial for KCNQ2 aggregation and excitability at nodes of Ranvier. J Physiol 588:4719–4730. doi:10.1113/jphysiol.2010.196022
Dobin A, Davis CA, Schlesinger F et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi:10.1093/bioinformatics/bts635
Hedstrom KL, Rasband MN (2006) Intrinsic and extrinsic determinants of ion channel localization in neurons. J Neurochem 98:1345–1352. doi:10.1111/j.1471-4159.2006.04001.x
Hund TJ, Koval OM, Li J et al (2010) A βIV-spectrin/CaMKII signaling complex is essential for membrane excitability in mice. J Clin Investig 120:3508–3519. doi:10.1172/JCI43621
Komada M, Soriano P (2002) βIV-spectrin regulates sodium channel clustering through ankyrin-G at axon initial segments and nodes of Ranvier. J Cell Biol 156:337–348. doi:10.1083/jcb.200110003
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. http://arxiv.org/pdf/1303.3997.pdf. Accessed Sept 3 2015
Maggi L, Scoto M, Cirak S et al (2013) Congenital myopathies—Clinical features and frequency of individual subtypes diagnosed over a 5-year period in the United Kingdom. Neuromuscul Disord 23:195–205. doi:10.1016/j.nmd.2013.01.004
North KN, Wang CH, Clarke N et al (2014) Approach to the diagnosis of congenital myopathies. Neuromuscul Disord 24:97–116. doi:10.1016/j.nmd.2013.11.003
Parkinson NJ, Olsson CL, Hallows JL et al (2001) Mutant β-spectrin 4 causes auditory and motor neuropathies in quivering mice. Nat Genet 29:61–65. doi:10.1038/ng710
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29:e45–e45. doi:10.1093/nar/29.9.e45
Relizani K, Mouisel E, Giannesini B et al (2014) Blockade of ActRIIB signaling triggers muscle fatigability and metabolic myopathy. Mol Ther 22:1423–1433. doi:10.1038/mt.2014.90
Schwarz JM, Cooper DN, Schuelke M, Seelow D (2014) MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 11:361–362. doi:10.1038/nmeth.2890
Seelow D, Schuelke M (2012) HomozygosityMapper2012–bridging the gap between homozygosity mapping and deep sequencing. Nucleic Acids Res 40:W516–W520. doi:10.1093/nar/gks487
Trapnell C, Roberts A, Goff L et al (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. doi:10.1038/nprot.2012.016
Tse WT, Tang J, Jin O et al (2001) A new spectrin, βIV, has a major truncated isoform that associates with promyelocytic leukemia protein nuclear bodies and the nuclear matrix. J Biol Chem 276:23974–23985. doi:10.1074/jbc.M009307200
Yang Y, Lacas-Gervais S, Morest DK et al (2004) βIV Spectrins are essential for membrane stability and the molecular organization of nodes of ranvier. J Neurosci 24:7230–7240. doi:10.1523/JNEUROSCI.2125-04.2004
Yao Z, Snider L, Balog J et al (2014) DUX4-induced gene expression is the major molecular signature in FSHD skeletal muscle. Hum Mol Genet 23:5342–5352. doi:10.1093/hmg/ddu251
Acknowledgements
The authors would like to thank the patient and his parents for participation in the study.
Author information
Authors and Affiliations
Contributions
MS, EK phenotyped the patient and gathered clinical information and material; SMG performed cell culture experiments; EG, FS performed molecular genetics experiments; WS performed muscle histology; SDU, TJH provided and prepared the muscle samples of the quivering (qv4J) mouse; MS performed homozygosity mapping, sequence alignment, and bioinformatic analysis of the WES data. EK analyzed and validated the results of the WES data. EK, MS co-authored the first draft of the manuscript and contributed funding. All authors read the final version of the manuscript for intellectual content and gave their permission for publication.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Funding statement
The project was funded by the Charité-Universitätsmedizin Berlin via the “Rahel-Hirsch” Program to EK, the Deutsche Forschungsgemeinschaft (SFB 665 TP C4) to MS, and the NeuroCure Center of Excellence (Exc 257) to MS.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Knierim, E., Gill, E., Seifert, F. et al. A recessive mutation in beta-IV-spectrin (SPTBN4) associates with congenital myopathy, neuropathy, and central deafness. Hum Genet 136, 903–910 (2017). https://doi.org/10.1007/s00439-017-1814-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-017-1814-7