Abstract
MTHFR is an important enzyme in the metabolism of folic acid and is crucial for reproductive function. Variation in the sequence of MTHFR has been implicated in subfertility, but definitive data are lacking. In the present study, a detailed analysis of two common MTHFR polymorphisms (c.677C>T and c.1298A>C) was performed. Additionally, for the first time, the frequencies of different MTHFR alleles were assessed in preimplantation embryos. Several striking discoveries were made. Firstly, results demonstrated that maternal MTHFR c.1298A>C genotype strongly influences the likelihood of a pregnancy occurring, with the 1298C allele being significantly overrepresented amongst women who have undergone several unsuccessful assisted reproductive treatments. Secondly, parental MTHFR genotypes were shown to affect the production of aneuploid embryos, indicating that MTHFR is one of the few known human genes with the capacity to modulate rates of chromosome abnormality. Thirdly, an unusual deviation from Hardy–Weinberg equilibrium was noted for the c.677C>T polymorphism in subfertile patients, especially those who had experienced recurrent failure of embryo implantation or miscarriage, potentially explained by a rare case of heterozygote disadvantage. Finally, a dramatic impact of the MTHFR 677T allele on the capacity of chromosomally normal embryos to implant is described. Not only do these findings raise a series of interesting biological questions, but they also argue that testing of MTHFR could be of great clinical value, identifying patients at high risk of implantation failure and revealing the most viable embryos during in vitro fertilisation (IVF) cycles.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Folic acid is an important B vitamin essential for human reproduction (Tamura and Picciano 2006). The processing of folic acid and other dietary folates is vital for many key processes such as amino acid metabolism, purine and pyrimidine synthesis, and methylation of nucleic acids, proteins and lipids (Wagner 1995). These folate-dependent functions are required for DNA synthesis and repair, control of gene expression and many other biological processes of fundamental importance for cell division and embryo development (Kim et al. 2009; Obican et al. 2010).
Folate deficiency (genetically determined or due to dietary restriction) results in a higher frequency of uracil misincorporation into DNA, disruption of nucleic acid integrity, slower DNA replication and an increased risk of chromosome breakage. Affected cells experience elevated rates of apoptosis and necrosis (Blount et al. 1997; Huang et al. 1999; Kimura et al. 2004). Insufficient folate or folic acid intake has also been shown to negatively affect specific reproductive functions; it has a detrimental effect on many processes involved in oocyte development, acquisition of endometrial receptivity, embryo implantation and also in the maintenance of pregnancy (Forges et al. 2007; Gmyrek et al. 2005; Hussein 2005; Thaler and Epel 2003). Several animal model studies have demonstrated that maternal folate deficiency prior to conception and during gestation has a negative effect on female fertility and foetal viability, emphasising the important role of folate during mammalian folliculogenesis and foetal development (Gueant et al. 2013; Kumar et al. 2013; Maloney et al. 2007).
Numerous variations in genes involved in folate metabolism have been identified. In some cases, these mutations and polymorphisms alter the efficiency of pathways involved in folate generation and processing. In terms of prevalence and impact, genetic variations affecting the 5,10-methylenetetrahydrofolate reductase gene (MTHFR) are among the most biologically important. MTHFR is a key enzyme that plays an important role in catalysing the conversion of 5,10-methylenetetrahydrofolate into 5-methylenetetrahydrofolate, the predominant circulating form of folate. This molecule provides the single carbon needed for the synthesis of nucleotides, the remethylation of homocysteine to methionine, the synthesis of S-adenosylmethionine and the methylation of DNA, proteins, neurotransmitters and phospholipids (Frosst et al. 1995; Yamada et al. 2005).
More than 20 DNA sequence variants and polymorphisms within the MTHFR gene have been described (Sibani et al. 2000). Two of the most investigated are single nucleotide polymorphisms (SNPs) at the mRNA positions 677 (rs1801133) and 1298 (rs1801131) (Frosst et al. 1995; van der Put et al. 1998). The well-characterised MTHFR c.677C>T transition, which results in an alanine to valine substitution (p.Ala222Val) in the predicted catalytic domain of MTHFR, renders the enzyme thermolabile and leads to a reduction in MTHFR activity. Homozygous and heterozygous individuals have in vitro MTHFR activity reduced by about 70 and 35 %, respectively (Frosst et al. 1995). Homozygosity for the 677T allele is associated with elevated circulating homocysteine in some individuals, predominantly those who have a low plasma folate level (Jacques et al. 1996). In these individuals, the level of plasma homocysteine can be lowered by folic acid supplementation (Malinow et al. 1997).
The other common polymorphism in the MTHFR gene, c.1298A>C transversion, results in a glutamate to alanine substitution (p.Glu429Ala) within a presumed regulatory domain of MTHFR (van der Put et al. 1998; Weisberg et al. 1998). The 1298C allele leads to decreased enzyme activity, although to a lesser extent than the 677T allele. Individuals who are homozygous for the 1298C allele have about a 40 % reduction in enzyme activity in vitro, but do not appear to have higher plasma homocysteine levels than controls (Malinow et al. 1997; van der Put et al. 1998; Weisberg et al. 1998). However, individuals who are compound heterozygous for the 677T and the 1298C alleles (MTHFR c.677C/T plus c.1298A/C genotype) have a 40–50 % reduction in enzyme activity in vitro and a biochemical profile similar to that seen among 677T homozygotes, with increased homocysteine and decreased folate levels. The c.1298A>C polymorphism by itself may have clinically important effects under conditions of low folate intake or during times of high folate requirements, such as pregnancy and embryogenesis (van der Put et al. 1998).
Despite the fact that many studies have explored the relationship between MTHFR polymorphisms and aspects of human reproduction, the biochemical influence and clinical relevance of these polymorphisms are still debated. Some authors have reported an association of certain genotypes with an increased risk of miscarriage, a potential consequence of poor vascularization of the placental area of individuals carrying minor alleles (Cao et al. 2013; Kim et al. 2011; Nair et al. 2012; Nelen et al. 2000). Others have described a link between c.677C>T and c.1298A>C polymorphisms and the likelihood of aneuploid conceptions, pointing out the possible influence of MTHFR on chromosome non-disjunction and other processes involved in chromosome segregation (Hassold et al. 2001; Hobbs et al. 2000; James et al. 1999; Kim et al. 2011). More recent reports have explored the impact of these polymorphisms in patients undergoing IVF treatment, suggesting an influence of some MTHFR variants on embryo quality (assessed morphologically) and upon implantation potential (Laanpere et al. 2011; Soldo et al. 2012). However, these findings remain controversial, with other studies failing to detect evidence of the proposed associations (D’Elia et al. 2014; Marci et al. 2012; Poursadegh Zonouzi et al. 2012). To clarify the true situation, especially in the context of assisted reproductive cycles, a detailed evaluation of the relationship between MTHFR polymorphism and different reproductive outcomes, using highly accurate genotyping methods, is urgently required. In particular, there remains a paucity of data from the critical preimplantation stage of development, a time when a deficiency in folate biosynthetic pathways might be expected to have a range of negative effects, some with profound implications for embryo viability. The present study involved an exhaustive examination of the incidence of two of the most common MTHFR polymorphisms in a large population of infertile patients and, for the first time, in embryos during the first few days of life (prior to implantation). Results from this study may help to settle some of the present controversies about the influence of MTHFR on the first stages of pregnancy and provide answers to the following questions: firstly, does MTHFR have a significant influence on the fertility of groups of patients with specific reproductive histories (e.g. patients with a history of recurrent miscarriages or patients suffering repetitive implantation failure after IVF treatment)? Secondly, do maternal and paternal MTHFR genotypes influence the frequency at which aneuploid embryos are produced? Finally, does the MTHFR genotype have an impact on the developmental competence of an embryo? It was hoped that this detailed analysis, including genotyping of embryos as well as adults, would help to define the aspects of human reproduction affected by inherited defects of the MTHFR gene.
Materials and methods
Ethics statement
Ethical approval and signed patient consent for research had been obtained for all patient samples used in this study. No embryos were biopsied specifically for the purpose of this study. The embryo DNA samples assessed consisted of surplus whole genome amplification products, surplus following routine aneuploidy screening. Ethical approval for this study was obtained from the North London REC 3 (10/H0709/26) and Western IRB (20060680 and 20131473).
Cohort selection
The initial study group consisted of 138 patients (92 females and 46 males) undergoing assisted reproductive treatment (ART) and having aneuploidy screening of their embryos for a variety of reasons including: (1) recurrent miscarriage, i.e. two or more failed clinical pregnancies (n = 51); (2) repetitive implantation failure, i.e. 3 or more IVF treatments including transfer of embryos to the uterus but no pregnancy (n = 38); (3) advanced maternal age, i.e. over 37 years of age (n = 17). Additionally a well-matched control population was assessed, composed of 161 fertile individuals (83 females, 78 males) that had previously achieved at least one successful pregnancy. Both groups were of varied ethnic origin, primarily European, but also including individuals of North African and Southeast Asian descent. A second independent data set comprising 202 individuals (101 couples: 28 fertile couples, 62 subfertile couples and 11 couples with a history of recurrent miscarriage) was available for confirmatory analysis using an alternative methodology (see below, CarrierMap, Recombine, USA). In addition to the DNA samples from adults, 193 specimens from blastocyst stage embryos, previously screened for aneuploidy using microarray comparative genomic hybridisation (aCGH), were also available for analysis.
MTHFR genotyping
Genomic DNA was extracted from blood using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). Amplification of the c.677C>T region of the MTHFR gene was performed using the primer pairs 5′-TGAAGGAGAAGGTGTCTGCGGGA-3′ (forward primer) and 5′-AGGACGGTGCGGTGAGAGTG-3′ (reverse primer) described by Frosst et al. (1995). Amplification of the c.1298A>C region of the MTHFR gene was accomplished using the forward (5′-CTTTGGGGAGCTGAAGGACTACTAC-3′) and reverse (5′-CACTTTGTGACCATTCCGGTTTG-3′) primers reported by van der Put et al. (1998). PCR amplifications (25 µl volume) consisted of the following: 5 ng of genomic DNA in the case of patients and controls or 0.5 µl of Sureplex-amplified DNA in the case of biopsied material from preimplantation embryos; 10 pmol each of the c.677C>T or c.1298A>C forward and reverse primers; 0.625 units of Perfect Taq DNA polymerase (5 prime GmbH, Hamburg, Germany); 0.2 mM of dNTPs (Thermo Scientific, Colchester, UK) in 1× PCR Buffer (5 prime GmbH, Hamburg, Germany). Amplification of the correct fragment was initially confirmed by uni-directional DNA sequencing followed by comparison and alignment to the NCBI Reference Sequence NG_013351.1.
PCR was followed by minisequencing as described by Zetterberg et al. (2002) with some modifications. Prior to the minisequencing reaction, 0.5 µl of PCR product was purified using ExoSAP-IT (Affymetrix, High Wycombe, UK) following the manufacturer’s instructions. Minisequencing was then conducted in a final volume of 5 μl, consisting of 0.5 µl of each of the purified MTHFR c.677C>T and c.1298A>C PCR products, 2.5 µl SNaPshot Multiplex Ready Reaction Mix (Applied Biosystems, UK) and 1 pmol of each of the minisequencing primers: 5′-T(20)GCGTGATGATGAAATC-G-3′ (reverse primer) for MTHFR c.677C>T; 5′-GGAGCTGACCAGTGAAG-3′ (forward primer) for c.1298A>C. The poly (T) sequence of the former was added to modify the electrophoretic mobility of the primer. The cycling protocol was 25 cycles of 96 °C/10 s, 50 °C/5 s and 60 °C/30 s. Minisequencing products were analysed by capillary electrophoresis on an ABI 3130 Genetic Analyzer (Applied Biosystems, UK). The reaction included 9.25 μl Hi-Di Formamide (Applied Biosystems, UK), 0.25 μl GeneScan-120LIZ Size standard (Applied Biosystems, UK) and 0.5 μl of minisequencing product. Incubation at 95 °C for 3 min to denature the minisequencing product was performed prior to electrophoresis. Data were subsequently visualised and analysed using the GeneMapper software (Applied Biosystems, UK).
Embryo DNA samples consisted of trophectoderm biopsy specimens (~5 cells removed at the blastocyst stage), which had been subjected to whole genome amplification (SurePlex, Rubicon, USA). The biopsies were performed for the purpose of routine preimplantation genetic screening (PGS) using microarray comparative genomic hybridisation (aCGH) (described below). To assess the polymorphisms, surplus SurePlex DNA was subjected to the same protocol as genomic DNA.
Confirmatory analysis on an independent population
An entirely independent data set was available from 202 individuals (101 couples) evaluated using an alternative, microarray-based, methodology (CarrierMap, Recombine, USA). The CarrierMap test is primarily used for preconception carrier screening, evaluating couples considering starting a family for the presence of >1700 mutations responsible for more than 250 different genetic conditions. However, a number of polymorphisms of potential relevance to reproduction are also assessed, including the MTHFR c.677C>T and c.1298A>C polymorphisms. The data were divided into three separate groups: 28 of the couples tested were considered to be fertile, having achieved at least one pregnancy without the help of any assisted reproductive treatments (average of 1.9 pregnancies per couple) and having had no more than one previous miscarriage; 62 couples had a history of infertility and had undergone treatment using IVF or intrauterine insemination (IUI); 11 couples had received a formal diagnosis of recurrent miscarriage.
Embryo aneuploidy testing
Embryo chromosome analysis was accomplished by the use of a well validated aCGH method following the protocol described by Alfarawati et al. (2011). Diagnosis of embryos was performed at the blastocyst stage and involved sampling and testing of 5 to 10 trophectoderm cells. Briefly, cells were lysed and their DNA amplified using whole genome amplification (SurePlex, Rubicon, USA). Amplified sample and reference DNAs from chromosomally normal individuals were labelled with Cy3 and Cy5 fluorochromes, respectively, and then hybridized to the probes of a bacterial artificial chromosome (BAC) microarray (24Sure, Illumina, Cambridge, UK). Chromosome losses and gains were revealed by differences in the fluorescence intensity corresponding to sample and reference DNAs for BAC probes derived from the affected chromosome or chromosomal region. Although primarily employed for the purpose of detecting losses and gains of entire chromosomes, copy number changes affecting chromosomal segments of 6 Mb or greater are also accurately detected with this approach (Colls et al. 2012). Labelling of the amplified samples, hybridization to microarray slides, post-hybridization washes and analyses were performed as described elsewhere (Alfarawati et al. 2011). Published values for the accuracy rate for aCGH are >95 % for biopsy specimens consisting of small numbers of trophectoderm cells (Fragouli et al. 2011; Wells et al. 2014).
Embryo transfers
Fresh single embryo transfers of euploid blastocysts of good morphological quality according to Gardner´s classification (Balaban et al. 2011) were performed in all cases.
Statistical analyses
MTHFR c.677C>T and c.1298A>C allele and genotype frequencies were determined for each of the three populations initially investigated (subfertile patients, fertile controls and preimplantation embryos) and compared using a Chi-squared goodness of fit test. Genotype frequencies in all groups were assessed for compliance with the Hardy–Weinberg equilibrium using GENEPOP v.4.2 software (Raymond and Rousset 1995; Rousset 2008). This software was also used to evaluate the existence of linkage disequilibrium (LD) with a goodness of fit G-test. Comparisons of patient genotypes with regards to sex, maternal age and reproductive history were also carried out using the same analysis. Similar analyses were carried out for the three populations with data obtained from Recombine (subfertile patients, fertile controls and patients with recurrent miscarriage). The proportion of aneuploid embryos (aneuploidy rate) produced by patients with specific MTHFR genotypes was assessed and compared using an ANCOVA test using maternal age as a covariant. Aneuploidy rates of different indication patient subgroups (recurrent miscarriage, repetitive implantation failure and maternal age) were compared using a T test. In the case of preimplantation embryos, comparisons of the genotype frequencies of chromosomally normal and aneuploid embryos, normal embryos with successful or failed implantation, and embryos from patients with a variety of referral reasons were also determined and compared using Chi-squared goodness of fit test.
Statistical significance was defined as p < 0.05 and all analyses were performed using the IBM SPSS Statistics Version 20 software (IBM Corporation, USA).
Results
MTHFR in subfertile patients
To assess the relationship between MTHFR polymorphisms and subfertility, allele and genotype frequencies for a group of patients undergoing assisted reproductive treatment were determined and compared to a fertile control population. Analyses of patient subgroups with regards to sex and reproductive history (RM, RIF or AMA) were also performed.
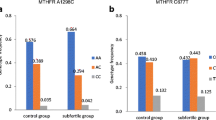
A statistically significant increase in the frequency of the less common MTHFR 1298C allele was observed in subfertile patients compared to fertile controls (p = 0.003), leading to a higher prevalence of individuals homozygous for 1298C and a lower incidence of patients homozygous for the major allele (1298A) (p = 0.01 and p = 0.02, respectively) (Table 1; Fig. 1). No differences were observed for allele or genotype frequencies for the MTHFR c.677C>T polymorphism. Analysis of the second independent population of samples tested using CarrierMap yielded similar results, with the frequency of 1298C homozygotes doubled in subfertile couples (0.137 vs 0.007, χ 2 test, p < 0.001).
Distribution of MTHFR c.677C>T and c.1298A>C genotypes in the study groups. a MTHFR c.677C>T genotype frequencies in subfertile and fertile control groups. b MTHFR c.1298A>C genotype frequencies in subfertile and fertile control groups. χ 2 test was used to estimate whether the genotypic frequencies differed significantly among the study groups. Significantly different groups (p < 0.05) are highlighted with an asterisk
When data were subdivided by sex, the difference in the MTHFR c.1298A>C genotype frequency was clearly apparent in female patients. In comparison to controls, subfertile female patients displayed a significantly higher incidence of MTHFR c.1298CC homozygotes and a lower frequency of MTHFR c.1298AA homozygotes (p < 0.01) (Table 2). In the male group a similar tendency was observed, but it was less pronounced and did not reach statistical significance. Within the fertile control group there were no differences in genotype frequencies between the two sexes. Examination of MTHFR c.677C>T polymorphism did not reveal any differences in the incidence of particular genotypes between males and females (Table 2).
Genotype distributions of both the MTHFR c.677C>T and c.1298A>C polymorphisms in the fertile control group were in Hardy–Weinberg equilibrium. However, a deviation from expected genotype frequencies was found for MTHFR c.677C>T in subfertile patients due to a deficit of heterozygotes (p < 0.01) (Fig. S1). Analysis of patient subgroups revealed that this was particularly apparent in couples with a history of failed implantation or miscarriage. Investigation of the independent set of samples tested using CarrierMap yielded concordant data, with genotype frequencies in the subfertile group and in patients with recurrent miscarriage differing significantly from those expected for a population in equilibrium (p < 0.001 and p < 0.01 respectively) (Fig. S2).
When both MTHFR polymorphisms were analysed in combination, no individuals carrying three or four mutant alleles (MTHFR c.677CT/c.1298CC, c.677TT/c.1298CA, c.677TT/c.1298CC) were detected in any of the populations studied, suggesting linkage disequilibrium between these two loci (Table S1). Based upon allele frequencies and protein function, it is likely that the ancestral MTHFR gene had a 677C/1298A haplotype. Mutation at the 677 nucleotide position later produced a 677T/1298A haplotype and, in an independent event, mutation at position 1298 produced a 677C/1298C haplotype. Consequently, 677T/1298C haplotypes can only be formed by recombination within the MTHFR gene, an extremely unlikely occurrence given the close proximity of the two polymorphisms. The existence of linkage disequilibrium was confirmed by G-test analysis (p < 0.05). The specific combination of 677T and 1298C alleles did not appear to have any additive effect in terms of impact on fertility in this patient population.
Patients were divided into groups according to their previous reproductive history, in an effort to determine whether differences in genotype frequencies with respect to the control population were related to subfertility in general or to particular reproductive problems. Analysis revealed that the significant excess of 1298C homozygotes observed in the subfertile group was primarily attributable to patients that had experienced repetitive implantation failure (RIF, i.e. three of more in vitro fertilisation treatments, including transfer of embryos to the uterus, but no pregnancy) relative to the control group (p < 0.001) (Table 3). The difference in the incidence of 1298C homozygotes amongst patients with RIF was equally apparent for both males and females. No significant differences in genotype frequency were seen for any other category of patient [recurrent miscarriage (RM) or advanced maternal age (AMA)].
To summarise, results from this section showed an increase in the frequency of MTHFR c.1298C homozygotes in the group of subfertile patients compared to controls. More detailed analysis revealed that this difference was most apparent in female patients and primarily attributable to couples who had experienced repetitive failed IVF cycles.
Parental MTHFR genotype and embryo aneuploidy
To assess the influence of parental MTHFR alleles on embryo aneuploidy, the MTHFR genotypes of patients undergoing IVF were considered in relation to the proportion of chromosomally abnormal embryos they produced (aneuploidy rate). Factors such as gender or reproductive history were also explored.
A significantly higher aneuploidy rate was found in those patients presenting with at least one MTHFR minor allele (MTHFR 677T and/or MTHFR 1298C) compared to those patients with none (ANCOVA test, p = 0.038) (Table 4). Patients homozygous for the major alleles showed a mean embryo aneuploidy rate (±SEM) of 57.9 % (±6.6), considerably lower than the 70.3 % (±2.4) average for patients carrying a minor allele for at least one of the two MTHFR polymorphisms studied. When patients were divided according to their sex, it became clear that maternal genotype was associated with embryo aneuploidy (p = 0.030), whereas the paternal genotype did not have a significant effect.
When the embryo aneuploidy rates and MTHFR genotypes for patients with different indications (AMA, RIF or RM) were compared, results showed that in the case of AMA and RM patients the presence of one minor allele in at least one of the two MTHFR polymorphisms analysed was associated with a significantly higher level of affected embryos compared to those patients homozygous for the major alleles (p = 0.045 and p = 0.048, for AMA and RM, respectively). Again, these effects were only observed in relation to maternal genotype. No differences were found in the group of RIF patients (p = 0.72) (Table 4).
Results from this section show an influence of parental MTHFR genotype on embryo aneuploidy. The impact of MTHFR genotype on the production of aneuploid embryos was most pronounced for women of advanced reproductive age (>37 years) or those with a history of recurrent miscarriage.
The effect of embryonic MTHFR genotype
To assess the existence of linkage disequilibrium between the two polymorphisms studied and analyse the relationship between embryonic MTHFR genotype and embryo competence, the frequency and distribution of MTHFR c.677C>T and c.1298A>C alleles and genotypes were explored in preimplantation embryos at the blastocyst stage (5 days post-fertilisation).
MTHFR c.677C>T and c.1298A>C allele and genotype frequencies were calculated for the 193 embryos tested. No significant differences were found when embryo allele and genotype frequencies were compared to those seen in adults (Tables S2 and S3). When both MTHFR c.677C>T and c.1298A>C polymorphisms were analysed in combination, no embryos carrying 3 or 4 mutant alleles (677CT/1298CC or 677TT/1298CA) were detected, mirroring the findings in patients. No significant differences were observed when the genotype frequencies of euploid and aneuploid embryos were compared.
Conversely, when comparing the genotype frequencies of chromosomally normal embryos that successfully implanted (n = 19) with those euploid embryos that failed to implant (n = 27), significant differences were observed for MTHFR c.677C>T (p < 0.01). The incidence of embryos homozygous for the MTHFR 677 minor allele (677T) was elevated in non-viable embryos compared to those that successfully formed on-going pregnancies (Fig. 2). One in four embryos experiencing failed implantation was homozygous for 677T, whereas only one of the 19 embryos that produced a pregnancy was homozygous for the same allele. The risk of implantation failure was twofold higher for 677T homozygotes compared with 677C homozygotes (87.5 versus 45 %) (Fig. 3). Interestingly, significant differences in MTHFR c.677C>T genotype frequencies were also found between those embryos that failed to implant and the adult population composed of fertile and subfertile subjects (p = 0.04) (Fig. S3). No such differences were detected between adults and embryos that successfully implanted (Table S3).
Distribution of MTHFR c.677C>T genotypes in chromosomally normal embryos that successfully implanted compared to those that failed to implant. χ 2 test was used to estimate whether the genotypic frequencies differed significantly among the study groups. Significantly different groups (p < 0.05) are highlighted with an asterisk
A deviation from Hardy–Weinberg equilibrium, due to a deficit of heterozygote genotypes, was apparent for the total population of embryos (p < 0.01), resembling that seen for the samples from subfertile adults. Further division of the embryos into different categories (e.g. based upon patient indication or ability to implant) resulted in groups too small for meaningful statistical analysis. Nonetheless, it may be relevant that the embryos that successfully implanted (n = 19) had genotype frequencies in line with Hardy–Weinberg expectations, while those that failed to implant (n = 27) displayed an apparent increase in the proportion of homozygotes at the expense of heterozygotes. This was, however, not statistically significant due to the small size of the sample (Fig. S4). No significant differences in genotype frequencies were found in the case of the MTHFR c.1298A>C polymorphism.
Results from this section indicate that embryonic MTHFR genotype has a significant impact on viability. Indirect data suggest that c.677C>T heterozygotes are associated with increased risk of developmental failure occurring between fertilisation and the blastocyst stage, while 677T homozygotes succeed in reaching the final stages of preimplantation development, but have a reduced probability of producing a clinical pregnancy.
Discussion
MTHFR genotype and implantation failure
This study confirmed that the MTHFR c.1298A>C genotype has a strong influence on fertility. A prevalence of the 1298C allele and a corresponding increase in the frequency of MTHFR c.1298C homozygotes were observed for patients undergoing IVF treatment compared to fertile controls. This was later confirmed in an entirely independent population of patients, analysed using an alternative methodology. Analysis of different patient subgroups revealed that the increase in 1298C prevalence was most apparent in couples with a history of multiple unsuccessful IVF treatments. Amongst these patients the frequency of the 1298C allele was increased fourfold from 0.06 (seen in fertile controls) to 0.24. Women within this category had received transfer of embryos to the uterus on at least three occasions, but without any pregnancy, indicating that the 1298C allele has a powerful impact on the ability of embryos to implant. The increase in 1298C frequency was principally observed in female patients, suggesting that reduced implantation rates may be related to an abnormal endometrial response or other maternal factors. Given the established reduction in MTHFR activity associated with homozygosity for the 1298C allele (Chango et al. 2000; van der Put et al. 1998; Weisberg et al. 1998), it can be inferred that individuals with compromised MTHFR activity are at increased risk of experiencing recurrent implantation failure. It is noteworthy that an elevated 1298C allele frequency was also observed in subfertile males, albeit to a lesser extent than seen females, suggesting that male factors related to this polymorphism might also influence the likelihood of implantation. No increase in the prevalence of the 1298C allele was detected in embryos that failed to implant compared to those that produced a viable pregnancy, arguing against the possibility of an effect at the level of the embryo.
The implantation of the blastocyst into the endometrium is a complex process that involves multiple molecular interactions between trophoblastic and endometrial cells, including coagulation and fibrinolysis processes at the embryo-maternal interface (Aflalo et al. 2004; Feng et al. 2000). It is conceivable that alteration of the functional activity of blood coagulation factors, related to diminished MTHFR activity, could affect the likelihood of implantation. There have been a number of publications reporting associations between inherited thrombophilic mutations/polymorphisms and recurrent unsuccessful IVF cycles (Azem et al. 2004; Ivanov et al. 2012; Coulam et al. 2006). It is undoubtedly true that appropriate coagulation processes, both maternal and placental, are of vital importance for pregnancy maintenance and are therefore of great relevance to miscarriage. This may be relevant to the distorted genotype frequencies of MTHFR polymorphisms observed in patients with recurrent pregnancy loss during this study. However, a role for coagulative processes at the time of implantation seems less likely. Perhaps more relevant in this regard are studies suggesting that folic acid concentration, modulated by MTHFR, has an important effect on trophoblast invasion, one of the very first steps of embryo implantation (Williams et al. 2011). Another possibility, potentially explaining increased rates of miscarriage and failure to implant, is that MTHFR variants influence levels of embryo aneuploidy (discussed below). Aneuploidy is extremely common in human preimplantation embryos and is believed to be the principal cause of implantation failure in both natural and assisted reproductive cycles (Fragouli et al. 2013; Hassold and Hunt 2001; Munne et al. 2006; Harton et al. 2013).
Unlike the c.1298A>C polymorphism, investigation of the other common MTHFR variant (c.677C>T) revealed no change in overall allele frequency for any of subfertile patient groups. However, a striking and unexpected discovery was that the two MTHFR c.677C>T alleles were not distributed in the expected proportions, resulting in a pronounced distortion of genotype frequencies (i.e. a deviation from Hardy–Weinberg equilibrium). Closer examination of the data revealed a deficit of MTHFR c.677C>T heterozygotes amongst patients with a history of recurrent implantation failure or miscarriage. Again, these data were subsequently confirmed by analysis of an independent set of samples using a different genotyping method. Interestingly, within these patient groups, there was a fourfold overrepresentation of couples in which the male and female had opposite homozygous genotypes (e.g. male homozygous for 677C and female homozygous for 677T or vice versa). Clearly, such couples can only produce heterozygous embryos, leading us to hypothesise that the two MTHFR c.677C>T alleles might be incompatible at a molecular level, leading to the formation of defective MTHFR dimers (essentially resulting in a heterozygote disadvantage) (Jacques et al. 1996; Yamada et al. 2001). An alternative possibility is that heterozygosity for c.677C>T is beneficial in terms of fertility and therefore less likely to be observed in patients undergoing fertility treatments. However, the mechanism by which the presence of any 677T alleles could be beneficial is unclear, especially considering that such alleles are known to be associated with diminished MTHFR function.
Investigation of preimplantation embryos demonstrated that deviation from expected c.677C>T genotype frequencies, mirroring those seen in the subfertile adults, can also be observed just a few days after conception. This suggests that the impact of the c.677C>T genotype must primarily be felt between fertilisation and the blastocyst stage. During this time, a period of 5 days, approximately half of all human embryos undergo developmental arrest and perish. It would be valuable to repeat the experiments described here on embryos that ceased developing during the first few days of life, to determine whether c.677C>T heterozygotes are at increased risk of developmental failure prior to blastocyst formation.
Parental MTHFR genotype and embryo aneuploidy
A significantly higher proportion of aneuploid embryos was found for patients carrying one or more alleles associated with reduced MTHFR activity compared to those with none. Patients with 677T and/or 1298C alleles showed a mean embryo aneuploidy rate 20 % higher than that of patients homozygous for the major (fully functional) MTHFR alleles. The association of MTHFR genotype with embryo aneuploidy was more evident for female patients than males, indicating that the elevated rate of chromosome abnormality is primarily related to meiotic errors occurring during oogenesis. Results are suggestive of an influence in males as well, but an increased sample size would be needed to confirm this statistically. The impact of MTHFR genotype on the production of aneuploid embryos was most pronounced for women of advanced reproductive age (>37 years) or those with a history of recurrent miscarriage. This may be an indication that these patients are particularly sensitive to problems associated with compromised MTHFR function and that an increased incidence of embryo chromosomal abnormalities may be contributing to their difficulties achieving a viable pregnancy.
An influence of maternal MTHFR genotype on embryo viability was also suggested by Haggarty et al. (Haggarty et al. 2006). In their study, women homozygous for the 1298C allele were less likely to have a live birth after IVF compared to women homozygous for the more common 1298A allele. However, no chromosome analysis was carried out, so it is unclear whether the reduced viability was related to aneuploidy or other embryonic or maternal factors. Studies trying to establish an association between folic acid metabolism and preimplantation embryo development have shown that MTHFR is expressed in both human oocytes and embryos (Dobson et al. 2004, 2007). Folic acid is present in the follicular fluid and its supplementation has been associated with improved oocyte quality and the retrieval of larger numbers of mature eggs after ovarian stimulation, suggesting that MTHFR plays an important role in oocyte maturation and development (Boxmeer et al. 2008; Steegers-Theunissen et al. 1993; Szymanski and Kazdepka-Zieminska 2003). Moreover, during maturation, human oocytes express receptors involved in folic acid transport, while in Xenopus oocytes efficient movement of folate across their membrane has been demonstrated (Boxmeer et al. 2008; Dobson et al. 2004; Lo et al. 1991; Szymanski and Kazdepka-Zieminska 2003).
With regard to the specific question of chromosomal abnormality, several publications have suggested the potential for a relationship between maternal MTHFR gene polymorphisms and aneuploid pregnancy or birth. Some authors have reported an association between MTHFR mutations and the risk of conceiving a child with Down syndrome (Acacio et al. 2005; Hobbs et al. 2000; Hollis et al. 2013; James et al. 1999). The MTHFR c.1298A>C polymorphism has been linked to chromosomal aneuploidy leading to Turner Syndrome (Oliveira et al. 2008) and has also been proposed as an independent risk factor for spontaneous abortion with foetal chromosomal abnormality. Kim et al. (2011) reported a higher prevalence of heterozygosity and homozygosity involving the MTHFR 1298C allele in women with spontaneous abortions associated with foetal aneuploidy compared to those with miscarriages of normal karyotype. However, some investigators have failed to confirm such a link (Boduroglu et al. 2004; Petersen et al. 2000). Hassold and co-workers (Hassold et al. 2001) observed a significant increase in the 677T allele polymorphism in mothers of trisomy 18 conceptuses but were unable to find associations with respect to other chromosomes.
Data from the current study support the notion that MTHFR mutations are associated with an increased risk of aneuploid pregnancy and that this is mostly attributable errors occurring during female meiosis. However, the underlying mechanism has not yet been elucidated. One hypothesis is that MTHFR mutations/polymorphisms lead to aberrant DNA methylation caused by abnormal folate metabolism and that this might increase the likelihood of meiotic aneuploidy (Hassold et al. 2001; James et al. 1999). In the absence of sufficient folic acid, intracellular homocysteine accumulates, methionine resynthesis is reduced and hence methylation reactions are compromised (Balaghi and Wagner 1993; Wainfan and Poirier 1992). Both clinical and experimental studies have shown that, in lymphocytes and liver cells, genomic DNA hypomethylation can be induced by deficiency of dietary folate, as well as other factors, and that this results in a variety of features such as abnormal gene expression, DNA strand breaks, chromosomal instability and aneuploidy (Crott et al. 2001; Ji et al. 1997; Pogribny et al. 1995; Wainfan and Poirier 1992).
To our knowledge, the current study is the first to look at the influence of maternal and paternal MTHFR genotypes on the production of aneuploid preimplantation embryos. The evaluation of such an early developmental stage is particularly important when considering chromosomal abnormality because most aneuploid embryos are lost around the time of implantation. In previous studies, chromosome analyses were performed during the first trimester of pregnancy, or even later. This is already far removed from the primary aneuploidy rate at conception.
Origin of MTHFR polymorphisms
The combined analysis of both c.677C>T and c.1298A>C polymorphisms in patients, controls and day-5 preimplantation embryos revealed linkage disequilibrium between the two loci. No individuals carrying three or four low-activity MTHFR alleles were found in any of the populations studied. These results are in agreement with several previous reports (Bae et al. 2007; Chango et al. 2000; Stegmann et al. 1999; van der Put et al. 1998; Weisberg et al. 1998). The fact that certain combinations of alleles were not detected indicates that the two polymorphisms arose independently on two different wild type alleles. The close proximity of these polymorphisms to one another (2.1 kb) is such that recombination between them is extremely unlikely to occur (Friedman et al. 1999; Hanson et al. 2001; Weisberg et al. 1998).
Although concordant with most publications, our results contradict those of Isotalo et al. who found a considerable number of individuals homozygous for the MTHFR 677T allele and also heterozygous for the c.1298A>C polymorphism and proposed that the presence of 3 or 4 mutant alleles might compromise foetal viability (Isotalo and Donnelly 2000; Isotalo et al. 2000). Results obtained in the present study do not support this hypothesis. Combinations of 3–4 mutant alleles were not observed in any of the 193 preimplantation embryos analysed, suggesting that such combinations of mutant alleles are either very rare or non-existent. The most likely explanation for the conflicting results found by Isotalo and colleagues is inaccuracy of the method for examining the c.1298A>C polymorphism (Isotalo and Donnelly 2000; Zetterberg et al. 2007). The MboII RFLP method that they used has been criticised because of the potential for interference, and genotyping errors, caused by a silent polymorphism within the same exon of MTHFR. The problematic allele has a prevalence of approximately 5 % in the Canadian population that they studied (Weisberg et al. 1998).
Embryo MTHFR genotype and chromosome abnormalities
No significant differences in MTHFR c.677C>T and c.1298A>C genotype frequencies were seen when preimplantation embryos with and without chromosomal abnormalities were compared. This suggests that the increased frequency of aneuploidy observed in embryos derived from mothers carrying 677T and 1298C alleles occurs during meiosis, due to an affect in the oocyte or ovary rather than diminished MTHFR activity in the embryo itself. Complementary results were obtained by Bae et al. (2007) who explored the prevalence of MTHFR c.677C>T and c.1298A>C genotypes later in development, after a pregnancy had been established. Their analysis of miscarriages indicated that the distribution of MTHFR c.677C>T and c.1298A>C genotypes was the same regardless of whether the products of conception were euploid or aneuploid.
Embryo MTHFR genotype and implantation
A significantly higher risk of implantation failure was observed in euploid embryos homozygous for MTHFR 677T. This indicates that the embryonic MTHFR c.677C>T genotype influences the ability of an embryo to form a successful pregnancy. Indeed, data from the current study suggest that homozygosity for 677T could play a role in approximately 20 % of embryo implantation failures. Consequently, determination of the MTHFR genotype of IVF embryos, prior to choosing which embryo will be transferred to the uterus, is likely to prove highly advantageous for fertility treatments (e.g. IVF), maximising the probability that a viable embryo will be transferred.
Defective folate metabolism has been shown to have negative effects on many essential processes that could impact embryogenesis and implantation, such as synthesis of nucleotide precursors for DNA synthesis and repair and cellular methylation reactions (Balaghi and Wagner 1993; Blount et al. 1997; De Cabo et al. 1995; Duthie 1999). Early embryo development involves crucial cell division and differentiation, requiring coordinated spatial and temporal changes in gene expression (Assou et al. 2011). Disruption of DNA methylation, a fundamental feature of transcriptional modulation, is likely to have serious consequences for the developing embryo (Razin and Shemer 1995). Other than affecting gene expression, the embryo MTHFR genotype could influence viability by introducing blood coagulation problems in the placenta, increasing the risk of miscarriage. Recent studies have shown that folic acid is potentially important in a number of crucial early stages of placental development, including extravillous trophoblast (EVT) invasion and angiogenesis (Williams et al. 2011).
An important consideration, which warrants further research, is the possibility that the impact of MTHFR deficiency on embryo implantation could be mitigated by dietary folic acid supplementation (e.g. 500 µg daily as recommended for women that have had a foetus with a neural tube defect) (WHO 2007). This may be particularly valuable for patients with a history of implantation failure following IVF treatment, especially if an adverse MTHFR genotype has been detected. Furthermore, the link between MTHFR deficiency and elevated aneuploidy risk raises the question of whether increased folic acid intake in female carriers of MTHFR 677T and 1298C alleles could help to reduce the incidence of chromosomally abnormal embryos, thereby leading to improved outcomes of conception achieved naturally or using ART.
Supplementation of embryo culture media, used during IVF treatments, should also be considered. We speculate that this might help to support homocysteine remethylation to methionine, assisting in the maintenance of the normal patterns of DNA methylation in embryos with reduced MTHFR activity. Currently, preimplantation embryos are routinely cultured in media that lack methyl donors. It would be valuable to determine whether folate-supplemented media could reduce the risk of implantation failure, miscarriage or aneuploidy. Consideration should also be given to the impact of folate supplementation on the incidence of imprinting disorders, associated with abnormal patterns of DNA methylation, which some studies suggest may be elevated in the conceptions of patients undergoing infertility treatments (Cox et al. 2002; DeBaun et al. 2003; Orstavik et al. 2003).
In conclusion, a complex picture has emerged, in which MTHFR polymorphisms modulate reproductive success on several distinct levels. The data suggest that MTHFR genotype influences implantation, aneuploidy and the viability of euploid embryos. Multiple essential processes are affected, including meiosis, embryogenesis and initiation of pregnancy. Based upon the findings of this study, MTHFR potentially joins a rare group of human genes displaying an example of heterozygote disadvantage and another select group of genes with the ability to influence aneuploidy rates. Given the relative ease with which embryonic MTHFR genotype can be determined, the two polymorphisms may represent valuable new biomarkers for the assessment of embryo competence, enhancing IVF treatments by helping to reveal the embryos most likely to produce a viable pregnancy. Another exciting possibility is that cellular pathways dependent on MTHFR could be supported through maternal dietary supplementation or, for embryos produced using ART, via the addition of folate to culture medium. While this remains highly speculative, the potential benefits, in terms of reduced risks of implantation failure, aneuploidy, abnormal imprinting and miscarriage, argue that work to examine these possibilities should be encouraged as a matter of urgency.
References
Acacio GL, Barini R, Bertuzzo CS, Couto EC, Annichino-Bizzacchi JM, Junior WP (2005) Methylenetetrahydrofolate reductase gene polymorphisms and their association with trisomy 21. Prenat Diagn 25:1196–1199
Aflalo ED, Sod-Moriah UA, Potashnik G, Har-Vardi I (2004) Differences in the implantation rates of rat embryos developed in vivo and in vitro: possible role for plasminogen activators. Fertil Steril 81(Suppl 1):780–785
Alfarawati S, Fragouli E, Colls P, Wells D (2011) First births after preimplantation genetic diagnosis of structural chromosome abnormalities using comparative genomic hybridization and microarray analysis. Hum Reprod 26:1560–1574
Assou S, Boumela I, Haouzi D, Anahory T, Dechaud H, De Vos J, Hamamah S (2011) Dynamic changes in gene expression during human early embryo development: from fundamental aspects to clinical applications. Hum Reprod Update 17:272–290
Azem F, Many A, Ben Ami I, Yovel I, Amit A, Lessing JB, Kupferminc MJ (2004) Increased rates of thrombophilia in women with repeated IVF failures. Hum Reprod 19:368–370
Bae J, Shin SJ, Cha SH, Choi DH, Lee S, Kim NK (2007) Prevalent genotypes of methylenetetrahydrofolate reductase (MTHFR C677T and A1298C) in spontaneously aborted embryos. Fertil Steril 87:351–355
Balaban B, Brison D, Calderón G, Catt J, Conaghan J, Cowan L et al (2011) Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Reprod Biomed Online 22(6):632–646
Balaghi M, Wagner C (1993) DNA methylation in folate deficiency: use of CpG methylase. Biochem Biophys Res Commun 193:1184–1190
Blount BC, Mack MM, Wehr CM, MacGregor JT, Hiatt RA, Wang G, Wickramasinghe SN, Everson RB, Ames BN (1997) Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci USA 94:3290–3295
Boduroglu K, Alanay Y, Koldan B, Tuncbilek E (2004) Methylenetetrahydrofolate reductase enzyme polymorphisms as maternal risk for Down syndrome among Turkish women. Am J Med Genet A 127A:5–10
Boxmeer JC, Brouns RM, Lindemans J, Steegers EA, Martini E, Macklon NS, Steegers-Theunissen RP (2008) Preconception folic acid treatment affects the microenvironment of the maturing oocyte in humans. Fertil Steril 89:1766–1770
Cao Y, Xu J, Zhang Z, Huang X, Zhang A, Wang J, Zheng Q, Fu L, Du J (2013) Association study between methylenetetrahydrofolate reductase polymorphisms and unexplained recurrent pregnancy loss: a meta-analysis. Gene 514:105–111
Chango A, Boisson F, Barbe F, Quilliot D, Droesch S, Pfister M, Fillon-Emery N, Lambert D, Fremont S, Rosenblatt DS, Nicolas JP (2000) The effect of 677C→T and 1298A→C mutations on plasma homocysteine and 5,10-methylenetetrahydrofolate reductase activity in healthy subjects. Br J Nutr 83:593–596
Colls P, Escudero T, Fischer J, Cekleniak NA, Ben-Ozer S, Meyer B, Damien M, Grifo JA, Hershlag A, Munné S (2012) Validation of array comparative genome hybridization for diagnosis of translocations in preimplantation human embryos. Reprod Biomed Online 24(6):621–629
Coulam C, Jeyendran RS, Fishel LA, Roussev R (2006) Multiple thrombophilic gene mutations are risk factors for implantation failure. Reprod Biomed Online 12(3):322–327
Cox GF, Burger J, Lip V, Mau UA, Sperling K, Wu BL, Horsthemke B (2002) Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am J Hum Genet 71:162–164
Crott JW, Mashiyama ST, Ames BN, Fenech M (2001) The effect of folic acid deficiency and MTHFR C677T polymorphism on chromosome damage in human lymphocytes in vitro. Cancer Epidemiol Biomark Prev 10:1089–1096
De Cabo SF, Santos J, Fernandez-Piqueras J (1995) Molecular and cytological evidence of S-adenosyl-l-homocysteine as an innocuous undermethylating agent in vivo. Cytogenet Cell Genet 71:187–192
DeBaun MR, Niemitz EL, Feinberg AP (2003) Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet 72:156–160
D’Elia PQ, dos Santos AA, Bianco B, Barbosa CP, Christofolini DM, Aoki T (2014) MTHFR polymorphisms C677T and A1298C and associations with IVF outcomes in Brazilian women. Reprod Biomed Online 28:733–738
Dobson AT, Raja R, Abeyta MJ, Taylor T, Shen S, Haqq C, Pera RA (2004) The unique transcriptome through day 3 of human preimplantation development. Hum Mol Genet 13:1461–1470
Dobson AT, Davis RM, Rosen MP, Shen S, Rinaudo PF, Chan J, Cedars MI (2007) Methylenetetrahydrofolate reductase C677T and A1298C variants do not affect ongoing pregnancy rates following IVF. Hum Reprod 22:450–456
Duthie SJ (1999) Folic acid deficiency and cancer: mechanisms of DNA instability. Br Med Bull 55:578–592
Feng Q, Liu Y, Liu K, Byrne S, Liu G, Wang X, Li Z, Ockleford CD (2000) Expression of urokinase, plasminogen activator inhibitors and urokinase receptor in pregnant rhesus monkey uterus during early placentation. Placenta 21:184–193
Forges T, Monnier-Barbarino P, Alberto JM, Gueant-Rodriguez RM, Daval JL, Gueant JL (2007) Impact of folate and homocysteine metabolism on human reproductive health. Hum Reprod Update 13:225–238
Fragouli E, Alfarawati S, Daphnis DD, Goodall NN, Mania A, Griffiths T, Gordon A, Wells D (2011) Cytogenetic analysis of human blastocysts with the use of FISH, CGH and aCGH: scientific data and technical evaluation. Hum Reprod 26:480–490
Fragouli E, Alfarawati S, Spath K, Jaroudi S, Sarasa J, Enciso M, Wells D (2013) The origin and impact of embryonic aneuploidy. Hum Genet 132:1001–1013. doi:10.1007/s00439-013-1309-0
Friedman G, Goldschmidt N, Friedlander Y, Ben-Yehuda A, Selhub J, Babaey S, Mendel M, Kidron M, Bar-On H (1999) A common mutation A1298C in human methylenetetrahydrofolate reductase gene: association with plasma total homocysteine and folate concentrations. J Nutr 129:1656–1661
Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, Boers GJ, den Heijer M, Kluijtmans LA, van den Heuvel LP et al (1995) A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet 10:111–113
Gmyrek GB, Sozanski R, Jerzak M, Chrobak A, Wickiewicz D, Skupnik A, Sieradzka U, Fortuna W, Gabrys M, Chelmonska-Soyta A (2005) Evaluation of monocyte chemotactic protein-1 levels in peripheral blood of infertile women with endometriosis. Eur J Obstet Gynecol Reprod Biol 122:199–205
Gueant JL, Namour F, Gueant-Rodriguez RM, Daval JL (2013) Folate and fetal programming: a play in epigenomics? Trends Endocrinol Metab 24(6):279–289
Haggarty P, McCallum H, McBain H, Andrews K, Duthie S, McNeill G, Templeton A, Haites N, Campbell D, Bhattacharya S (2006) Effect of B vitamins and genetics on success of in vitro fertilisation: prospective cohort study. Lancet 367:1513–1519
Hanson NQ, Aras O, Yang F, Tsai MY (2001) C677T and A1298C polymorphisms of the methylenetetrahydrofolate reductase gene: incidence and effect of combined genotypes on plasma fasting and post-methionine load homocysteine in vascular disease. Clin Chem 47:661–666
Harton G, Munné S, Surrey M, Grifo J, Kaplan B, McCulloh D, Griffin D, Wells D (2013) Diminished effect of maternal age on implantation after preimplantation genetic diagnosis with array comparative genomic hybridization. Fertil Steril 100(6):1695–1703
Hassold T, Hunt P (2001) To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2:280–291
Hassold TJ, Burrage LC, Chan ER, Judis LM, Schwartz S, James SJ, Jacobs PA, Thomas NS (2001) Maternal folate polymorphisms and the etiology of human nondisjunction. Am J Hum Genet 69:434–439
Hobbs CA, Sherman SL, Yi P, Hopkins SE, Torfs CP, Hine RJ, Pogribna M, Rozen R, James SJ (2000) Polymorphisms in genes involved in folate metabolism as maternal risk factors for Down syndrome. Am J Hum Genet 67:623–630
Hollis ND, Allen EG, Oliver TR, Tinker SW, Druschel C, Hobbs CA, O’Leary LA, Romitti PA, Royle MH, Torfs CP, Freeman SB, Sherman SL, Bean LJ (2013) Preconception folic acid supplementation and risk for chromosome 21 nondisjunction: a report from the National Down Syndrome Project. Am J Med Genet A 161:438–444
Huang RF, Ho YH, Lin HL, Wei JS, Liu TZ (1999) Folate deficiency induces a cell cycle-specific apoptosis in HepG2 cells. J Nutr 129:25–31
Hussein MR (2005) Apoptosis in the ovary: molecular mechanisms. Hum Reprod Update 11:162–177
Isotalo PA, Donnelly JG (2000) Prevalence of methylenetetrahydrofolate reductase mutations in patients with venous thrombosis. Mol Diagn 5:59–66
Isotalo PA, Wells GA, Donnelly JG (2000) Neonatal and fetal methylenetetrahydrofolate reductase genetic polymorphisms: an examination of C677T and A1298C mutations. Am J Hum Genet 67:986–990
Ivanov P, Tsvyatkovska T, Konova E, Komsa-Penkova R (2012) Inherited thrombophilia and IVF failure: the impact of coagulation disorders on implantation process. Am J Reprod Immunol 68:189–198
Jacques PF, Bostom AG, Williams RR, Ellison RC, Eckfeldt JH, Rosenberg IH, Selhub J, Rozen R (1996) Relation between folate status, a common mutation in methylenetetrahydrofolate reductase, and plasma homocysteine concentrations. Circulation 93:7–9
James SJ, Pogribna M, Pogribny IP, Melnyk S, Hine RJ, Gibson JB, Yi P, Tafoya DL, Swenson DH, Wilson VL, Gaylor DW (1999) Abnormal folate metabolism and mutation in the methylenetetrahydrofolate reductase gene may be maternal risk factors for Down syndrome. Am J Clin Nutr 70:495–501
Ji W, Hernandez R, Zhang XY, Qu GZ, Frady A, Varela M, Ehrlich M (1997) DNA demethylation and pericentromeric rearrangements of chromosome 1. Mutat Res 379:33–41
Kim KC, Friso S, Choi SW (2009) DNA methylation, an epigenetic mechanism connecting folate to healthy embryonic development and aging. J Nutr Biochem 20:917–926
Kim SY, Park SY, Choi JW, Kim do J, Lee SY, Lim JH, Han JY, Ryu HM, Kim MH (2011) Association between MTHFR 1298A>C polymorphism and spontaneous abortion with fetal chromosomal aneuploidy. Am J Reprod Immunol 66:252–258
Kimura M, Umegaki K, Higuchi M, Thomas P, Fenech M (2004) Methylenetetrahydrofolate reductase C677T polymorphism, folic acid and riboflavin are important determinants of genome stability in cultured human lymphocytes. J Nutr 134:48–56
Kumar KA, Lalitha A, Pavithra D, Padmavathi IJ, Ganeshan M, Rao KR, Venu L, Balakrishna N, Shanker NH, Reddy SU, Chandak GR, Sengupta S, Raghunath M (2013) Maternal dietary folate and/or vitamin B12 restrictions alter body composition (adiposity) and lipid metabolism in Wistar rat offspring. J Nutr Biochem 24:25–31
Laanpere M, Altmae S, Kaart T, Stavreus-Evers A, Nilsson TK, Salumets A (2011) Folate-metabolizing gene variants and pregnancy outcome of IVF. Reprod Biomed Online 22:603–614
Lo RS, Said HM, Unger TF, Hollander D, Miledi R (1991) An endogenous carrier-mediated uptake system for folate in oocytes of Xenopus laevis. Proc Biol Sci/R Soc 246:161–165
Malinow MR, Nieto FJ, Kruger WD, Duell PB, Hess DL, Gluckman RA, Block PC, Holzgang CR, Anderson PH, Seltzer D, Upson B, Lin QR (1997) The effects of folic acid supplementation on plasma total homocysteine are modulated by multivitamin use and methylenetetrahydrofolate reductase genotypes. Arterioscler Thromb Vasc Biol 17:1157–1162
Maloney CA, Hay SM, Rees WD (2007) Folate deficiency during pregnancy impacts on methyl metabolism without affecting global DNA methylation in the rat fetus. Br J Nutr 97:1090–1098
Marci R, Lisi F, Soave I, Lo Monte G, Patella A, Caserta D, Moscarini M (2012) Impact of 677C>T mutation of the 5,10-methylenetetrahydrofolate reductase on IVF outcome: is screening necessary for all infertile women? Genet Test Mol Biomark 16:1011–1014
Munne S, Fischer J, Warner A, Chen S, Zouves C, Cohen J, Referring Centers PGDG (2006) Preimplantation genetic diagnosis significantly reduces pregnancy loss in infertile couples: a multicenter study. Fertil Steril 85:326–332
Nair RR, Khanna A, Singh K (2012) MTHFR C677T polymorphism and recurrent early pregnancy loss risk in north Indian population. Reprod Sci 19:210–215
Nelen WL, Bulten J, Steegers EA, Blom HJ, Hanselaar AG, Eskes TK (2000) Maternal homocysteine and chorionic vascularization in recurrent early pregnancy loss. Hum Reprod 15:954–960
Obican SG, Finnell RH, Mills JL, Shaw GM, Scialli AR (2010) Folic acid in early pregnancy: a public health success story. Faseb J 24:4167–4174
Oliveira KC, Bianco B, Verreschi IT, Guedes AD, Galera BB, Galera MF, Barbosa CP, Lipay MV (2008) Prevalence of the polymorphism MTHFR A1298C and not MTHFR C677T is related to chromosomal aneuploidy in Brazilian Turner Syndrome patients. Arq Bras Endocrinol Metabol 52:1374–1381
Orstavik KH, Eiklid K, van der Hagen CB, Spetalen S, Kierulf K, Skjeldal O, Buiting K (2003) Another case of imprinting defect in a girl with Angelman syndrome who was conceived by intracytoplasmic semen injection. Am J Hum Genet 72:218–219
Petersen MB, Grigoriadou M, Mikkelsen M (2000) A common mutation in the methylenetetrahydrofolate reductase gene is not a risk factor for Down syndrome. Am J Hum Genet 67(Supplement 2):141
Pogribny IP, Basnakian AG, Miller BJ, Lopatina NG, Poirier LA, James SJ (1995) Breaks in genomic DNA and within the p53 gene are associated with hypomethylation in livers of folate/methyl-deficient rats. Cancer Res 55:1894–1901
Poursadegh Zonouzi A, Chaparzadeh N, Asghari Estiar M, Mehrzad Sadaghiani M, Farzadi L, Ghasemzadeh A, Sakhinia M, Sakhinia E (2012) Methylenetetrahydrofolate reductase C677T and A1298C mutations in women with recurrent spontaneous abortions in the Northwest of Iran. ISRN Obstet Gynecol 2012:945486
Raymond M, Rousset F (1995) GENEPOP (Version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Razin A, Shemer R (1995) DNA methylation in early development. Hum Mol Genet 4 Spec No:1751–1755
Rousset F (2008) genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
Sibani S, Christensen B, O’Ferrall E, Saadi I, Hiou-Tim F, Rosenblatt DS, Rozen R (2000) Characterization of six novel mutations in the methylenetetrahydrofolate reductase (MTHFR) gene in patients with homocystinuria. Hum Mutat 15:280–287
Soldo V, Cutura N, Zamurovic M (2012) Defect of methylenetetrahydrofolate reductase in a patient with ten habitual misscarriages: a case report. Clin Exp Obstet Gynecol 39:556–558
Steegers-Theunissen RP, Steegers EA, Thomas CM, Hollanders HM, Peereboom-Stegeman JH, Trijbels FJ, Eskes TK (1993) Study on the presence of homocysteine in ovarian follicular fluid. Fertil Steril 60:1006–1010
Stegmann K, Ziegler A, Ngo ET, Kohlschmidt N, Schroter B, Ermert A, Koch MC (1999) Linkage disequilibrium of MTHFR genotypes 677C/T-1298A/C in the German population and association studies in probands with neural tube defects(NTD). Am J Med Genet 87:23–29
Szymanski W, Kazdepka-Zieminska A (2003) Effect of homocysteine concentration in follicular fluid on a degree of oocyte maturity. Ginekol Pol 74:1392–1396
Tamura T, Picciano MF (2006) Folate and human reproduction. Am J Clin Nutr 83:993–1016
Thaler CD, Epel D (2003) Nitric oxide in oocyte maturation, ovulation, fertilization, cleavage and implantation: a little dab’ll do ya. Curr Pharm Des 9:399–409
van der Put NMJ, Gabreels F, Stevens EMB, Smeitink JAM, Trijbels FJM, Eskes TKAB, van den Heuvel LP, Blom HJ (1998) A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am J Hum Genet 62:1044–1051
Wagner CL (1995) Biochemical role of folate in cellular metabolism. In: Bailey LB (ed) Folate in health and disease. Marcel Dekker, New York, pp 23–42
Wainfan E, Poirier LA (1992) Methyl groups in carcinogenesis: effects on DNA methylation and gene expression. Cancer Res 52:2071s–2077s
Weisberg I, Tran P, Christensen B, Sibani S, Rozen R (1998) A second genetic polymorphism in methylenetetrahydrofolate reductase (MTHFR) associated with decreased enzyme activity. Mol Genet Metab 64:169–172
Wells D, Kaur K, Grifo J, Glassner M, Taylor JC, Fragouli E, Munne S (2014) Clinical utilisation of a rapid low-pass whole genome sequencing technique for the diagnosis of aneuploidy in human embryos prior to implantation. J Med Genet 51:553–562
Williams PJ, Bulmer JN, Innes BA, Broughton Pipkin F (2011) Possible roles for folic acid in the regulation of trophoblast invasion and placental development in normal early human pregnancy. Biol Reprod 84:1148–1153
World Health Organization (2007) Standard for maternal and neonatal care. Department of Making Pregnancy Safer and Department of Reproductive Health and Research. World Health Organization Press, Geneva
Yamada K, Chen Z, Rozen R, Matthews RG (2001) Effects of common polymorphisms on the properties of recombinant human methylenetetrahydrofolate reductase. Proc Natl Acad Sci USA 98:14853–14858
Yamada K, Strahler JR, Andrews PC, Matthews RG (2005) Regulation of human methylenetetrahydrofolate reductase by phosphorylation. Proc Natl Acad Sci USA 102:10454–10459
Zetterberg H, Regland B, Palmer M, Ricksten A, Palmqvist L, Rymo L, Arvanitis DA, Spandidos DA, Blennow K (2002) Increased frequency of combined methylenetetrahydrofolate reductase C677T and A1298C mutated alleles in spontaneously aborted embryos. Eur J Hum Genet 10:113–118
Zetterberg M, Tasa G, Palmer MS, Juronen E, Toover E, Blennow K, Zetterberg H (2007) Methylenetetrahydrofolate reductase genetic polymorphisms in patients with primary open-angle glaucoma. Ophthalmic Genet 28:47–50
Acknowledgments
Maria Enciso is funded by the Spanish Ministerio de Educacion Cultura y Deporte Fellowship. Dagan Wells is funded by the Oxford NIHR Biomedical Research Centre.
Author information
Authors and Affiliations
Corresponding authors
Additional information
M. Enciso and J. Sarasa contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
439_2016_1652_MOESM2_ESM.tif
Supplementary material 2 (TIFF 4758 kb) Fig. S1 Distribution of MTHFR c.677C>T genotype frequencies in subfertile populations and predicted frequencies based upon Hardy-Weinberg. Compliance with the Hardy-Weinberg equilibrium was assessed using GENEPOP v.4.2. All populations shown in this graph present a deviation from Hardy-Weinberg equilibrium due to a deficit of heterozygotes
439_2016_1652_MOESM3_ESM.tif
Supplementary material 3 (TIFF 7144 kb) Fig. S2 Distribution of MTHFR c.677C>T genotype frequencies in an independent set of samples investigated using CarrierMap test (Recombine) and predicted frequencies based upon Hardy-Weinberg. Compliance with the Hardy-Weinberg equilibrium was assessed using GENEPOP v.4.2. All populations shown in this graph present a deviation from Hardy-Weinberg equilibrium due to a deficit of heterozygotes
439_2016_1652_MOESM4_ESM.tif
Supplementary material 4 (TIFF 8042 kb) Fig. S3 Distribution of MTHFR c.677C>T genotype frequencies in euploid preimplantation embryos with failed implantation and the adult population composed of fertile and subfertile individuals. X2 test was used to estimate whether the genotypic frequencies differed significantly among the study groups. Significantly different groups (p<0.05) are highlighted with an asterisk
439_2016_1652_MOESM5_ESM.tif
Supplementary material 5 (TIFF 7465 kb) Fig. S4 Distribution of MTHFR c.677C>T genotype frequencies in euploid preimplantation embryos with successful and failed implantation and predicted frequencies based upon Hardy- Weinberg. Compliance with the Hardy-Weinberg equilibrium was assessed using GENEPOP v.4.2. No deviation from HWE was detected in any of the groups shown in this graph probably due to small size
Rights and permissions
About this article
Cite this article
Enciso, M., Sarasa, J., Xanthopoulou, L. et al. Polymorphisms in the MTHFR gene influence embryo viability and the incidence of aneuploidy. Hum Genet 135, 555–568 (2016). https://doi.org/10.1007/s00439-016-1652-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-016-1652-z