Abstract
A genome-wide association study (GWAS) identified tumor necrosis factor superfamily member 15 (TNFSF15) as the strongest associated gene with susceptibility to primary biliary cirrhosis (PBC) outside the HLA loci in the Japanese population. However, causal functional variants of the TNFSF15 locus and the molecular mechanism underlying disease susceptibility have not been clarified. Here, to identify the functional causal variants of the TNFSF15 locus, integrated analysis comprising in silico analysis, a case–control association study and in vitro functional analysis was performed. Initially, 32 functional candidate single-nucleotide polymorphisms (SNPs) in the expression regulatory motifs, the coding region, or the untranslated regions (UTRs) of the TNFSF15 locus were selected by in silico analysis. By the case–control association studies using PBC patients (n = 1279) and healthy controls (n = 1091) in the Japanese population, rs4979462 [P = 1.85 × 10−14 (our previous study)], rs56211063 (P = 2.21 × 10−14), and rs55768522 (r 2 = 1 with rs4979462) were likely candidates for causal variants. Among these SNPs, rs4979462 was identified as the causal variant by in vitro functional analysis using luciferase assay and electrophoretic mobility shift assay (EMSA). Super-shift assay clarified that PBC-susceptible allele of rs4979462 generated a novel NF-1 binding site. Moreover, higher endogenous TNFSF15 protein and mRNA expression levels were observed in individuals with the PBC-susceptible allele of rs4979462. This study identified the causal variant for PBC susceptibility in the TNFSF15 locus and clarified its underlying molecular mechanism. TNFSF15 and NF-1 are considered to be potential targets for the treatment of PBC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Primary biliary cirrhosis (PBC) is a chronic and cholestatic liver disease that is presumed to be caused by an autoimmune reaction against biliary epithelial cells leading to the destruction of intrahepatic bile ducts, portal inflammation, liver cirrhosis, and hepatic failure (Kaplan and Gershwin 2005). The concordance rate of PBC in identical twins is higher than that in other autoimmune diseases (Selmi et al. 2004), and the relative sibling risk (λ S) in PBC is estimated to be 10.5 (Jones et al. 1999). These findings may indicate the contribution of strong genetic factors to the development of PBC. Genome-wide association studies (GWAS) and subsequent meta-analyses in European descent have identified HLA and over 20 non-HLA susceptibility loci (IL12A, IL12RB2, STAT4, IRF5, IKZF3, MMEL1, SPIB, DENND1B, CD80, IL7R, CXCR5, TNFRSF1A, CLEC16A, NFKB, RAD51L1, MAP3K7IP1, PLCL2, RPS6KA4, TNFAIP2, 7p14, and 16q24) as the genes associated with susceptibility to PBC (Hirschfield et al. 2009, 2010; Liu et al. 2010; Mells et al. 2011). We recently performed a GWAS for PBC in the Japanese population and identified tumor necrosis factor superfamily member 15 (TNFSF15) and POU domain class 2 associating factor 1 (POU2AF1) as new genes contributing to susceptibility to PBC in the Japanese population (Nakamura et al. 2012). However, causal functional variants in the TNFSF15 locus and their molecular mechanisms that underlie the disease susceptibility to PBC have not been clarified.
TNFSF15 is a member of the tumor necrosis factor (TNF) superfamily that interacts with death receptor 3 (DR3, known as TNFRSF25) (Migone et al. 2002). TNFSF15 is produced by inflammatory cytokine (TNFα or IL-1)-stimulated endothelial cells, or by microbial antigen [Toll-like receptor (TLR) ligands]-stimulated monocyte and dendritic cells (Cassatella et al. 2007; Shih et al. 2009; Yue et al. 1999). TNFSF15 has been reported to be involved in apoptosis, cell proliferation, and polarization to Th1 and Th17 cells (Haridas et al. 1999; Bayry 2010; Bamias et al. 2010; Zhou et al. 2014). Furthermore, serum TNFSF15 levels are significantly increased in PBC as well as in other inflammatory diseases that are caused by altered immunological reactions (Aiba et al. 2014; Bamias et al. 2003, 2008, 2011; Bull et al. 2008). Genetic polymorphisms of human TNFSF15 have also been reported to be associated with susceptibility to Crohn’s disease (CD), ulcerative colitis (UC), ankylosing spondylitis (AS), and leprosy (Barrett et al. 2008; Zhang et al. 2009; Zinovieva et al. 2009; Latiano et al. 2011; Yamazaki et al. 2013). However, SNPs in the TNFSF15 locus that mediated susceptibility to each disease were not in strong linkage disequilibrium with rs4979462, which was the SNP in the TNFSF15-TNFSF8 region that showed the strongest association with susceptibility to PBC in the Japanese population in our previous GWAS (Nakamura et al. 2012).
In the present study, SNPs in the TNFSF15-TNFSF8 region that are located in expression regulatory motifs, the coding region, or in untranslated regions (UTRs) were selected as candidate functional SNPs. The causal variant for susceptibility to PBC was identified by integrated analysis which comprised in silico analysis, a case–control association study in the Japanese population, and in vitro functional analysis. Moreover, further in vitro functional analyses and expression analyses were performed to clarify the molecular mechanism of the disease susceptibility to PBC that was caused by the TNFSF15 causal variant rs4979462.
Materials and methods
Subjects
The information regarding PBC patients and healthy controls who participated in this study has been provided in Nakamura et al. 2012. This study was approved by the Research Ethics Committee of the Graduate School of Medicine, The University of Tokyo.
Genotyping
The genotyping of 32 functional tag SNPs in the TNFSF15 locus was performed using the DigiTag2 assay (Nishida et al. 2012). The primer sequences are listed in Supplementary Table 1.
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts were obtained from Jurkat T cells that were stimulated with or without 50 ng/ml of 12-O-tetradecanoylphorbol-13-acetate (TPA) and 0.5 µM of calcium ionophore (CI), or from HepG2 cells that were stimulated with or without 100 ng/ml of Interleukin-6 (Active Motif, Carlsbad, CA). The EMSA was performed using a LightShift Chemiluminescent EMSA Kit (Thermo-Fisher Scientific, Waltham, MA), according to the manufacturer’s instructions, using biotin-labeled double-stranded oligonucleotide probes corresponding to each major and minor allele. The sequences of the oligonucleotide probes are shown in Supplementary Table 2. Nuclear extract (2.5 µg/ml) and the biotin-labeled probes (10 fmol/µl) were incubated for 30 min at 4 °C. For the supershift experiments, 2 µg/µl of a rabbit anti-human NF-1 antibody (N-20 X; Santa Cruz Biotechnologies, Santa Cruz, CA) and 2.5 µg/ml of nuclear extract were incubated for 1 h at 4 °C, and subsequently 10 fmol/µl of oligonucleotide probes were added and incubated for 30 min at 4 °C. Three independent experiments were performed in each assay.
Luciferase assay
The 5′ regulatory region and intron 1 of TNFSF15 were amplified from human genomic DNA using PCR with specific primers (Supplementary Table 3). The PCR products were subcloned into the reporter gene pGL4.23 (luc2/minP) vector (Promega, Madison, Wis). Minor alleles of each SNP were constructed by PCR-directed mutagenesis of these plasmids using allele-specific primer sets (Supplementary Table 3). The pGL4.74 [hRluc/TK] vector was used to normalize for variations in transfection efficiency. These plasmids (pGL4.23: 500 ng; pGL4.74: 50 ng) were transfected into HuCCT1 or Jurkat T cells using Lipofectamine LTX & plus reagents (Thermo-Fisher Scientific). Luciferase activities were determined using the Dual-Luciferase Reporter Assay system (Promega). Three independent experiments were performed in each assay.
ELISA
TNFSF15 levels in tenfold diluted serum were measured using the Human TNFSF15 ELISA Development Kit (PeproTech, Rocky Hill, CT, USA) as previously described (Aiba et al. 2014). When the TNFSF15 levels were above the range of detection (>40 ng/ml), 20-fold diluted serum was used in this ELISA. The absorbance was measured at 450 nm. All samples were run in duplicate.
GENEVAR
The correlation between the TNFSF15 rs4979462 genotype and TNFSF15 mRNA expression was examined using the mRNA data of lymphoblastoid cell lines derived from 44 HapMap JPT (Japanese in Tokyo, Japan) individuals that were available from the database of the Gene Expression Variation (GENEVAR) project at the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/humgen/genevar/) (Stranger et al. 2012).
Statistical analysis
Allele frequencies and agreement with Hardy–Weinberg Equilibrium were calculated using a χ 2 test at each locus. The relative luciferase activity and mRNA expression level of TNFSF15 were compared between major and minor alleles of each SNP using Student’s t test. Serum TNFSF15 protein expression levels were compared between major and minor alleles of rs4979462 using Mann–Whitney U test. P values < 0.05 were regarded as statistically significant.
Results
Selection of candidate functional variants in the TNFSF15-TNFSF8 region
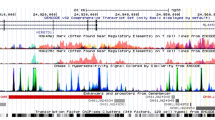
In our previous GWAS, 3,602 SNPs in the 200 kb of the TNFSF15-TNFSF8 genomic region, which contains the gene with the strongest association with PBC susceptibility outside of HLA loci in the Japanese population (Nakamura et al. 2012), were registered in the dbSNP database (http://www.ncbi.nlm.nih.gov/snp/) (Fig. 1a, b). To identify the causal variant for susceptibility to PBC in this TNFSF15-TNFSF8 region, candidate functional SNPs were selected by the following process:
Selection of candidate functional variants in the TNFSF15-TNFSF8 genomic region. a The linkage disequilibrium (LD) structure of the TNFSF15-TNFSF8 region. LD was calculated from HapMap Japanese in Tokyo (JPT) genotyping data. Pairwise D′ values for all combinations of single nucleotide polymorphisms (SNPs) are shown in the red scale. b Overview of SNP selection using in silico analysis. There were 781 SNPs whose minor allele frequency (MAF) in the TNFSF15-TNFSF8 region was greater than 0.01. Of these SNPs, 73 SNPs that are located in a transcription regulatory element (DNase I hyper-sensitivity cluster or H3K27Ac marks) and that change the binding of transcription factors, coding regions, or untranslated regions (UTRs), were selected. Of the selected 73 SNPs, 32 SNPs were selected as tag SNPs. c, d Result of a case–control association study using data of PBC patients (n = 1274) and healthy individuals (n = 1091) in the Japanese population. Each dot shows the p value (c) or odds ratio (OR) (d) for tag SNPs or for the SNPs that show complete linkage disequilibrium (r 2 = 1) with genotyped tag SNPs. The three SNPs shown in the red ellipse (rs4979462, rs56211063, and rs55768522) are the candidate functional variants. e Position of the three candidate functional variants in the TNFSF15-TNFSF8 region. The three candidate SNPs are shown in the red rectangles
First, rare variants whose minor allele frequencies (MAFs) are less than 0.01 in populations including African Ancestry (AFR), American Ancestry (AMR), East Asian Ancestry (ASN), and European Ancestry (EUR) in the 1000 Genomes Project (http://www.1000genomes.org/) were removed. Of the remaining 781 SNPs, SNPs that are located in transcription regulatory elements, coding regions or UTRs were selected as candidate functional SNPs. Transcription regulatory elements were characterized by DNase I hyper-sensitivity cluster analysis of any of 125 cell types from the Encyclopedia of DNA Elements (ENCODE; http://genome.ucsc.edu/ENCODE/index.html), or by the presence of H3K27Ac markers in any of seven cell lines (GM12878, H1-hESC, HSMM, HUVEC, K562, NHEK, or NHLF cells) from ENCODE. Additionally, SNPs that were not predicted to change the binding of transcription factors between the major allele and the minor allele by HaploReg v2 (http://www.broadinstitute.org/mammals/haploreg/haploreg.php) were removed (Fig. 1b).
Among the remaining 73 SNPs, 13 SNPs had already been genotyped in our previous GWAS of PBC in the Japanese population, and 11 SNPs showed nearly perfect linkage disequilibrium with these SNPs in the 1000 Genomes Project. Additionally, 17 SNPs also showed nearly perfect linkage disequilibrium with each other in the present SNP sets. Ultimately, 32 tag SNPs were selected from the 73 SNPs as candidate functional SNPs and a case–control association study was performed using the data of 963 Japanese individuals as the initial discovery panel (487 PBC cases and 476 healthy controls) and 1402 other Japanese individuals as the replication panel (787 PBC cases and 615 healthy controls). Genotyping of the 32 tag SNPs was carried out using the DigiTag2 assay, which is a multiplex SNP typing method (Nishida et al. 2012). Principal component analysis in our previous GWAS, which used the same sample set as that of the present study, showed that all PBC cases and healthy controls in the Japanese population formed a single cluster with the HapMap-Japanese people living in Tokyo (JPT) samples, but not with the HapMap-Han Chinese living in Beijing (CHB) samples (Nakamura et al. 2012). These results indicated that the effect of population stratification was negligible. Although the genotyping of one tag SNP failed, the call rates for all of the other tag SNPs were >95 %, and no deviation from Hardy–Weinberg Equilibrium (HWE; P > 0.05) was observed in the healthy controls. Figure 1c, d shows the single-point association data (based on allele frequencies) of each genotyped tag SNP using both discovery and replication panels, of SNPs that showed r 2 = 1 with a tag SNP, or of SNPs that were genotyped in our previous GWAS for PBC in the Japanese population (Nakamura et al. 2012). We previously found that rs4979462 in the TNFSF15-TNFSF8 region showed the strongest association with susceptibility to PBC (P = 1.85 × 10−14; OR = 1.57) (Nakamura et al. 2012). The genotyping of the tag SNPs indicated that there were no SNPs in the present study that showed stronger association with susceptibility to PBC than rs4979462 (Table 1, Supplementary Table 4, and Supplementary Table 5). Of the genotyped SNPs in the present study, rs56211063, which showed r 2 = 0.98 with rs4979462 in the 1000 Genomes Project data, had the strongest association with susceptibility to PBC (P = 2.21 × 10−14; OR = 0.64). In addition, rs55768522 showed perfect linkage disequilibrium (r 2 = 1) with rs4979462 in the 1000 Genomes Project data. These three SNPs were, therefore, selected as candidate functional variants in the TNFSF15-TNFSF8 region (Fig. 1e).
Identification of the causal variant by in vitro functional analysis
Rs4979462 (located in the first intron of TNFSF15), rs55768522 (at a distance of 5.8 kb from the 5′ end of TNFSF15), and rs56211063 (at a distance of 17 kb from the 5′ end of TNFSF15) are located in transcription regulatory elements and thus binding affinities of transcription factors might differ between the major allele and the minor allele. To identify the PBC causal variant among the three SNPs, an electrophoretic mobility shift assay (EMSA) was performed using a nuclear extract of the human liver cell line HepG2 and biotin-labeled probes corresponding to the different alleles of each SNP. A difference in mobility shift between the major and the minor alleles of rs4979462 and rs56211063, but not of rs55768522, was detected (Fig. 2).
Electrophoretic mobility shift assays (EMSA) of each candidate causal variant. An EMSA of candidate causal variants using a nuclear extract of HepG2 cells and biotin-labeled probes corresponding to each allele. Rs4979462 and rs56211063 showed a difference in band shift between the major allele and the minor allele. An unlabeled probe was used as a competitor. G, A, T, and C indicate each allele
To further analyze differences in transcription efficiency between the major and the minor alleles of these SNPs, luciferase assays of rs4979462 and rs56211063 were performed using the human T cell line Jurkat and the human bile duct cell line HuCCT1 (Fig. 3a, b and Supplementary Fig. 1). Luciferase activity was significantly increased by the PBC-susceptible allele of rs4979462 compared to the major allele in both cell lines 24 h after transfection of the pGL4.23 vector (Fig. 3c, d). However, no difference in induced luciferase activity was observed between the major allele and the minor allele of rs56211063 (Fig. 3e, f). These results indicated that rs4979462 at the TNFSF15 locus is the functional variant for disease-susceptibility to PBC in the Japanese population.
Identification of the disease causal variant in the TNFSF15 locus by in vitro functional analysis. a, b Plasmid constructs of fragments of the genomic DNA of the TNFSF15 locus used for transfection. Part of an intron 1 fragment containing rs4979462 (a) and part of the 5′ transcription regulatory region containing rs56211063 (b) were subcloned into the pGL4.23 vector. c–f The transcriptional enhancing activity of these plasmid constructs was measured by assay of luciferase (luc) activity of the transfected human T cell line Jurkat T (c, e; rs4979462 and rs56211063, respectively) and the human bile duct cell line HuCCT1 (d, f; rs4979462 and rs56211063, respectively) 24 h after transfection. Cells transfected with the PBC-susceptible allele rs4979462 showed enhanced luciferase activities compared to the minor allele of rs4979462. Values of relative luciferase activity are shown as mean ± SD. *P < 0.05 (Student’s t test)
A novel NF-1 binding site was generated by the PBC-susceptible allele of rs4979462
The PBC-susceptible allele of rs4979462 was predicted to generate a novel nuclear factor 1 (NF-1) binding site by the TFSEARCH database (http://mbs.cbrc.jp/research/db/TFSEARCHJ.html). An EMSA using probes corresponding to the major and minor alleles of rs4979462 and nuclear extracts of the human liver cell line HepG2 or the human T cell line Jurkat with or without stimulation with interleukin-6 (IL-6) or 12-O-tetradecanoylphorbol-13-acetate (TPA) and calcium ionophore (CI), respectively, indicated that stimulated HepG2 cells displayed the most efficient transcription factor binding to the PBC susceptible allele of rs4979462 (Fig. 4a). The binding of NF-1 to the PBC-susceptible allele of TNFSF15 rs4979462 was, therefore, checked by a super-shift assay using the nuclear extract of IL-6 stimulated HepG2 cells. In accordance with the predicted result, the band shift of the PBC-susceptible allele of rs4979462 was abrogated by pre-incubation with the NF-1 specific antibody before electrophoresis (Fig. 4b). This result indicated that a novel NF-1 binding site was generated by the PBC-susceptible allele of TNFSF15 rs4979462 (Fig. 4c).
Novel NF-1 binding site was generated by the PBC-susceptible allele rs4979462. a Electrophoretic mobility shift assay (EMSA) using biotin-labeled probes corresponding to each candidate variant allele and nuclear extracts of HepG2 cells and Jurkat T cells. HepG2 cells were pre-stimulated with or without Interleukin-6 (IL-6). Jurkat T cells were pre-stimulated with or without 12-O-tetradecanoylphorbol-13-acetate (TPA) and calcium ionophore (CI) (TPA + CI). A shifted band could be seen for the probe corresponding to the PBC susceptibility allele of rs4979462 in the IL-6 stimulated HepG2 cells. b Super-shift assay for analysis of NF-1 binding. The shifted band that was observed in the IL-6 stimulated HepG2 cells in (a) was abrogated by incubation with an NF-1 specific antibody before electrophoresis. c The postulated functional effect of the TNFSF15 rs4979462-G and rs4979462-A alleles. The novel binding site in the first intron of TNFSF15 that is generated by rs4979462-A enhances the transcription efficiency of TNFSF15
Higher endogenous expression level of TNFSF15 in individuals with the PBC-susceptible genotype
To assess the influence of rs4979462 on the endogenous TNFSF15 expression level, serum was acquired from 34 healthy individuals (A/A: n = 7; G/A: n = 20; G/G: n = 7) and 60 PBC patients who had not undergone treatment at the time of enrolment (A/A: n = 24; G/A: n = 31; G/G: n = 5), and the TNFSF15 protein level in each serum was measured using an Enzyme-Linked Immuno-Sorbent Assay (ELISA). The demographics and clinical characteristics of the PBC patients are shown in Table 2. Although the serum TNFSF15 protein levels were higher in patients with the PBC-susceptible allele as compared to those without the PBC-susceptible allele, this difference was not statistically significant as assessed by the non-parametric Mann–Whitney U test (Fig. 5a, b). This is probably because less sample size in the healthy individuals and the baseline of TNFSF15 protein expression in the PBC patients is higher than those in healthy individuals (Aiba et al. 2014). However, in accordance with the results of our in vitro functional assays, analysis of the GENEVAR database (http://www.sanger.ac.uk/resources/software/genevar/) indicated that the endogenous expression level of TNFSF15 mRNA was significantly higher in lymphoblastoid cell lines derived from healthy Japanese individuals (JPT) with the PBC-susceptible genotype of rs4979462 than in those with other genotypes of rs4979462 (Fig. 5c). Endogenous expression levels of TNFSF15 mRNA in healthy Chinese individuals (CHB) also showed a similar tendency (Fig. 5d). These results indicated that a higher in vivo expression level of TNFSF15 was caused by the PBC susceptible allele of rs4979462.
Difference in the endogenous expression level of TNFSF15 among people with TNFSF15 rs4979462 genotypes. a, b Endogenous TNFSF15 protein expression level in the sera of 34 healthy individuals (G/G: n = 7; G/A: n = 20; A/A: n = 7) (a) and 60 PBC patients (G/G: n = 5; G/A: n = 31; A/A: n = 24) (b). TNFSF15 protein levels were measured using an ELISA. The plotted data represent averages ± standard error of the means of triplicate assays. c, d Endogenous TNFSF15 mRNA expression extracted from GENEVAR HapMap-JPT (c) and HapMap-CHB (d) database
Discussion
In the present study, rs4979462, which is a SNP located in the first intron of human TNFSF15, was identified as the causal variant for disease susceptibility to PBC in the Japanese population by integrated analyses. These analyses included in silico analysis using several databases, a case–control association study using the same sample set as that used in our previous GWAS for PBC in the Japanese population, and in vitro functional analysis. Further analysis using a super-shift assay indicated that the disease-susceptible allele of rs4979462 generated a novel NF-1 binding site. Additionally, higher TNFSF15 mRNA expression level was associated with the disease-susceptible allele of rs4979462. These results indicated that the disease-susceptible allele of rs4979462 enhances autoimmune responses by transcriptional up-regulation of TNFSF15.
Over 20 PBC susceptibility genes have been identified by GWAS and subsequent meta-analyses in subjects of European descent. However, TNFSF15 has never been identified as a susceptibility gene (Hirschfield et al. 2009, 2010; Liu et al. 2010; Mells et al. 2011), probably because the MAF of rs4979462 is markedly different between populations of Caucasian ancestry and those of Asian ancestry (MAF of Asian ancestry, 0.32; MAF of European ancestry, 0.01; according to the 1000 genomes Project). We performed a power calculation using the “Power calculator for Genetics Analysis” provided by the University of Michigan (http://csg.sph.umich.edu/abecasis/CaTS/tour1.html). The statistical power for detecting significant association of rs4979462 with PBC susceptibility in the present Japanese sample set was close to 100 % (Prevalence: 0.002; Disease Allele Frequency: 0.32; Genotype Relative Risk: 1.57), whereas it was only 32 % in Caucasians even if more than 10,000 cases and 10,000 controls were used (Prevalence: 0.002; Disease Allele Frequency: 0.01; Genotype Relative Risk: 1.57). Therefore, a much larger sample size is needed to detect a significant association of rs4979462 with susceptibility to PBC in Caucasians. This finding may indicate that rs4979462 is an Asian-specific SNP for PBC, although rs4979462 or other rarer variants of TNFSF15 may also possibly have some effect on the PBC susceptibility in subjects of European descent.
Several SNPs that are located in the human TNFSF15 locus (rs4263839, rs10982385, rs4574921, rs10114470, rs6478108, or rs6478106) have been reported to be associated with susceptibility to Crohn’s disease (CD), ulcerative colitis (UC), ankylosing spondylitis (AS), and leprosy (Barrett et al. 2008; Zhang et al. 2009; Zinovieva et al. 2009; Latiano et al. 2011; Yamazaki et al. 2013). In addition, rs6478109, a SNP that is located in the 5′-expression regulatory region of TNFSF15, which shows strong linkage disequilibrium with both rs4263839 (r 2 = 0.99, Asian populations in the 1000 genomes database) and rs6478108 (r 2 = 0.98, Asian populations in the 1000 genomes database), showed a functional difference between the major allele and the minor allele (Kakuta et al. 2009). On the other hand, rs4979462, which showed a functional difference between the major allele and the minor allele in the present study, is not in strong linkage disequilibrium with other SNPs (rs4263839, rs10982385, rs4574921, rs10114470, rs6478108, and rs6478106) that have been reported to be associated with susceptibility to diseases such as CD, UC, AS, or leprosy (r 2 < 0.8, Asian population in the 1000 genomes database). These findings indicated that the disease susceptibility to PBC that is conferred on the Japanese population by rs4976462 can occur in a disease-specific manner that is independent of the disease specificities previously reported for TNFSF15 SNPs for diseases other than PBC. Interestingly, the Reference database for Expression Analysis (RefExA; http://157.82.78.238/refexa/main_search.jsp) indicates that liver sinusoidal endothelial (LSE) cells, which are the only liver tissue/cells in this database, show the highest expression level of TNFSF15 mRNA (Supplementary Figure 2). Additionally, our previous immunohistochemical data using liver specimens from PBC patients showed that TNFSF15 expression is specific for cells that are related to PBC (biliary epithelial cells, Kupffer cells, blood vessels, and infiltrating mononuclear cells) (Aiba et al. 2014). Additionally, NF-1, for which a novel binding site is generated by the disease-susceptible allele of rs4979462, shows a higher expression level in the human liver than in other organs (Nagase et al. 2000). The characteristic expression patterns of TNFSF15 and transcription factors such as NF-1 in the liver may enhance the influence of rs4979462 in increasing susceptibility to PBC.
More and more landmark SNPs that show significant associations with susceptibility to various diseases have been detected by recent GWAS. It is absolutely essential to focus on the causal variants that are in linkage disequilibrium with significantly associated landmark SNPs in each GWAS. In the present study, in vitro functional analysis showed that the PBC-susceptible allele, TNFSF15 rs4979462, enhanced the transcriptional efficiency of TNFSF15. In addition, it was shown that NF-1 has some effect on the pathogenesis of PBC by binding to a novel NF-1 binding site in TNFSF15 rs4979462. These evidences suggest that studies that focus on the functional effect of causal variants can clarify not only the functional alteration resulting from the causal variants but can also provide the key to dissection of the molecular mechanism of development of diseases with unknown pathogenesis. Landmark SNPs of HLA, TNFSF15, POU2AF1, IL7R, IKZF3, CD80, STAT4, and NFKB1, which display significant or suggestive associations with susceptibility to PBC have also been identified in the Japanese population (Nakamura et al. 2012). Therefore, similar functional analyses that are focused on these SNPs are now underway to dissect the pathogenesis of PBC.
In our in silico analysis, the dbSNP database was used for the selection of SNPs that are located in the TNFSF15-TNFSF8 region. Although this database is the biggest SNP database, it is still possible that some SNPs are missing even after selection of common variants (MAF > 1 %). Several research groups have recently constructed public human genome variation catalogs using data that were acquired by whole-genome sequencing (WGS) or whole-exome sequencing (WES) using the next-generation sequencer (NGS) [e.g., 1000 Genomes Project]. Sequencing of the gene region that shows susceptibility to PBC will provide more accurate information for the detection of disease causing variants. Additionally, identification of other causal variants that are located in other gene loci associated with susceptibility to PBC by such types of studies will forge a new path in the prediction of disease onset.
Higher endogenous expression level of TNFSF15 has been reported in individuals with several autoimmune diseases as compared to healthy individuals, and their disease activity also showed a positive correlation with TNFSF15 expression levels (Bamias et al. 2003, 2008, 2011; Bull et al. 2008). We also recently found higher TNFSF15 expression levels in both the serum and the liver tissues of PBC patients compared to healthy controls (Aiba et al. 2014). These findings indicated that TNFSF15 has some association with the pathogenesis of PBC. In the present study, the transcription efficiency of TNFSF15 was enhanced by the disease susceptible rs4979462 allele due to NF-1 binding to the novel binding site generated by this allele. Our present findings suggested that higher expression level of TNFSF15 induced hyper-activation of several immune pathways including that of polarization to Th1 and Th17 cells, leading to specific autoimmune reactions to the small bile ducts.
In conclusion, we are the first group to identify the causal variant for susceptibility of the Japanese population to PBC in the human TNFSF15-TNFSF8 genomic region and we have clarified a molecular mechanism of disease susceptibility by the causal variant that is focused on a transcription regulatory motif in the TNFSF15 locus. These findings suggest that both TNFSF15 and NF-1 may be potential targets for the treatment of PBC.
References
Aiba Y, Harada K, Komori A, Ito M, Shimoda S, Nakamura H, Nagaoka S, Abiru S, Migita K, Ishibashi H, Nakanuma Y, Nishida N, Kawashima M, Tokunaga K, Yatsuhashi H, Nakamura M (2014) Systemic and local expression levels of TNF-like ligand 1A and its decoy receptor 3 are increased in primary biliary cirrhosis. Liver Int 34:679–688
Bamias G, Martin C 3rd, Marini M, Hoang S, Mishina M, Ross WG, Sachedina MA, Friel CM, Mize J, Bickston SJ, Pizarro TT, Wei P, Cominelli F (2003) Expression, localization, and functional activity of TL1A, a novel Th1-polarizing cytokine in inflammatory bowel disease. J Immunol 171:4868–4874
Bamias G, Siakavellas SI, Stamatelopoulos KS, Chryssochoou E, Papamichael C, Sfikakis PP (2008) Circulating levels of TNF-like cytokine 1A (TL1A) and its decoy receptor 3 (DcR3) in rheumatoid arthritis. Clin Immunol 129:249–255
Bamias G, Kaltsa G, Siakavellas SI, Papaxoinis K, Zampeli E, Michopoulos S, Zouboulis-Vafiadis I, Ladas SD (2010) High intestinal and systemic levels of decoy receptor 3 (DcR3) and its ligand TL1A in active ulcerative colitis. Clin Immunol 137:242–249
Bamias G, Evangelou K, Vergou T, Tsimaratou K, Kaltsa G, Antoniou C, Kotsinas A, Kim S, Gorgoulis V, Stratigos AJ, Sfikakis PP (2011) Upregulation and nuclear localization of TNF-like Cytokine 1A (TL1A) and its receptors DR3 and DcR3 in psoriatic skin lesions. Exp Dermatol 20:725–731
Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, Xavier RJ; NIDDK IBD Genetics Consortium, Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I, Heath S, Laukens D, Mni M, Rutgeerts P, Van Gossum A, Zelenika D, Franchimont D, Hugot JP, de Vos M, Vermeire S, Louis E; Belgian-French IBD Consortium; Wellcome Trust Case Control Consortium, Cardon LR, Anderson CA, Drummond H, Nimmo E, Ahmad T, Prescott NJ, Onnie CM, Fisher SA, Marchini J, Ghori J, Bumpstead S, Gwilliam R, Tremelling M, Deloukas P, Mansfield J, Jewell D, Satsangi J, Mathew CG, Parkes M, Georges M, Daly MJ (2008) Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet 40:955–962
Bayry J (2010) TL1A in the inflammatory network in autoimmune diseases. Nat Rev Rheumatol 6:67–68
Bull MJ, Williams AS, Mecklenburgh Z, Calder CJ, Twohig JP, Elford C, Evans BA, Rowley TF, Slebioda TJ, Taraban VY, Al-Shamkhani A, Wang EC (2008) The Death Receptor 3-TNF-like protein 1A pathway drives adverse bone pathology in inflammatory arthritis. J Exp Med 205:2457–2464
Cassatella MA, Pereira-da-Silva G, Tinazzi I, Facchetti F, Scapini P, Calzetti F, Tamassia N, Wei P, Nardelli B, Roschke V, Vecchi A, Mantovani A, Bambara LM, Edwards SW, Carletto A (2007) Soluble TNF-like cytokine (TL1A) production by immune complexes stimulated monocytes in rheumatoid arthritis. J Immunol 178:7325–7333
Haridas V, Shrivastava A, Su J, Yu GL, Ni J, Liu D, Chen SF, Ni Y, Ruben SM, Gentz R, Aggarwal BB (1999) VEGI, a new member of the TNF family activates nuclear factor-kappa B and c-Jun N-terminal kinase and modulates cell growth. Oncogene 18:6496–6504
Hirschfield GM, Liu X, Xu C, Lu Y, Xie G, Lu Y, Gu X, Walker EJ, Jing K, Juran BD, Mason AL, Myers RP, Peltekian KM, Ghent CN, Coltescu C, Atkinson EJ, Heathcote EJ, Lazaridis KN, Amos CI, Siminovitch KA (2009) Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. N Engl J Med 360:2544–2555
Hirschfield GM, Liu X, Han Y, Gorlov IP, Lu Y, Xu C, Lu Y, Chen W, Juran BD, Coltescu C, Mason AL, Milkiewicz P, Myers RP, Odin JA, Luketic VA, Speiciene D, Vincent C, Levy C, Gregersen PK, Zhang J, Heathcote EJ, Lazaridis KN, Amos CI, Siminovitch KA (2010) Variants at IRF5-TNPO3, 17q12-21 and MMEL1 are associated with primary biliary cirrhosis. Nat Genet 42:655–657
Jones DE, Watt FE, Metcalf JV, Bassendine MF, James OF (1999) Familial primary biliary cirrhosis reassessed: a geographically-based population study. J Hepatol 30:402–407
Kakuta Y, Ueki N, Kinouchi Y, Negoro K, Endo K, Nomura E, Takagi S, Takahashi S, Shimosegawa T (2009) TNFSF15 transcripts from risk haplotype for Crohn’s disease are overexpressed in stimulated T cells. Hum Mol Genet 18:1089–1098
Kaplan MM, Gershwin ME (2005) Primary biliary cirrhosis. N Engl J Med 353:1261–1273
Latiano A, Palmieri O, Latiano T, Corritore G, Bossa F, Martino G, Biscaglia G, Scimeca D, Valvano MR, Pastore M, Marseglia A, D’Incà R, Andriulli A, Annese V (2011) Investigation of multiple susceptibility loci for inflammatory bowel disease in an Italian cohort of patients. PLoS One 6:e22688
Liu X, Invernizzi P, Lu Y, Kosoy R, Lu Y, Bianchi I, Podda M, Xu C, Xie G, Macciardi F, Selmi C, Lupoli S, Shigeta R, Ransom M, Lleo A, Lee AT, Mason AL, Myers RP, Peltekian KM, Ghent CN, Bernuzzi F, Zuin M, Rosina F, Borghesio E, Floreani A, Lazzari R, Niro G, Andriulli A, Muratori L, Muratori P, Almasio PL, Andreone P, Margotti M, Brunetto M, Coco B, Alvaro D, Bragazzi MC, Marra F, Pisano A, Rigamonti C, Colombo M, Marzioni M, Benedetti A, Fabris L, Strazzabosco M, Portincasa P, Palmieri VO, Tiribelli C, Croce L, Bruno S, Rossi S, Vinci M, Prisco C, Mattalia A, Toniutto P, Picciotto A, Galli A, Ferrari C, Colombo S, Casella G, Morini L, Caporaso N, Colli A, Spinzi G, Montanari R, Gregersen PK, Heathcote EJ, Hirschfield GM, Siminovitch KA, Amos CI, Gershwin ME, Seldin MF (2010) Genome-wide meta-analyses identify three loci associated with primary biliary cirrhosis. Nat Genet 42:658–660
Mells GF, Floyd JA, Morley KI, Cordell HJ, Franklin CS, Shin SY, Heneghan MA, Neuberger JM, Donaldson PT, Day DB, Ducker SJ, Muriithi AW, Wheater EF, Hammond CJ, Dawwas MF; UK PBC Consortium; Wellcome Trust Case Control Consortium 3, Jones DE, Peltonen L, Alexander GJ, Sandford RN, Anderson CA (2011) Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet 43:329–332
Migone TS, Zhang J, Luo X, Zhuang L, Chen C, Hu B, Hong JS, Perry JW, Chen SF, Zhou JX, Cho YH, Ullrich S, Kanakaraj P, Carrell J, Boyd E, Olsen HS, Hu G, Pukac L, Liu D, Ni J, Kim S, Gentz R, Feng P, Moore PA, Ruben SM, Wei P (2002) TL1A is a TNF-like ligand for DR3 and TR6/DcR3 and functions as a T cell costimulator. Immunity 6:479–492
Nagase T, Kikuno R, Ishikawa K, Hirosawa M, Ohara O (2000) Prediction of the Coding Sequences of Unidentified Human Genes. XVI. The Complete Sequences of 150 New cDNA Clones from Brain Which Code for Large Proteins in vitro. DNA Res 7:65–73
Nakamura M, Nishida N, Kawashima M, Aiba Y, Tanaka A, Yasunami M, Nakamura H, Komori A, Nakamuta M, Zeniya M, Hashimoto E, Ohira H, Yamamoto K, Onji M, Kaneko S, Honda M, Yamagiwa S, Nakao K, Ichida T, Takikawa H, Seike M, Umemura T, Ueno Y, Sakisaka S, Kikuchi K, Ebinuma H, Yamashiki N, Tamura S, Sugawara Y, Mori A, Yagi S, Shirabe K, Taketomi A, Arai K, Monoe K, Ichikawa T, Taniai M, Miyake Y, Kumagi T, Abe M, Yoshizawa K, Joshita S, Shimoda S, Honda K, Takahashi H, Hirano K, Takeyama Y, Harada K, Migita K, Ito M, Yatsuhashi H, Fukushima N, Ota H, Komatsu T, Saoshiro T, Ishida J, Kouno H, Kouno H, Yagura M, Kobayashi M, Muro T, Masaki N, Hirata K, Watanabe Y, Nakamura Y, Shimada M, Hirashima N, Komeda T, Sugi K, Koga M, Ario K, Takesaki E, Maehara Y, Uemoto S, Kokudo N, Tsubouchi H, Mizokami M, Nakanuma Y, Tokunaga K, Ishibashi H (2012) Genome-wide association study identifies TNFSF15 and POU2AF1 as susceptibility loci for primary biliary cirrhosis in the Japanese population. Am J Hum Genet 91:721–728
Nishida N, Mawatari Y, Sageshima M, Tokunaga K (2012) Highly parallel and short-acting amplification with locus-specific primers to detect single nucleotide polymorphisms by the DigiTag2 assay. PLoS One 7:e29967
Selmi C, Mayo MJ, Bach N, Ishibashi H, Invernizzi P, Gish RG, Gordon SC, Wright HI, Zweiban B, Podda M, Gershwin ME (2004) Primary biliary cirrhosis in monozygotic and dizygotic twins: genetics, epigenetics, and environment. Gastroenterology 127:485–492
Shih DQ, Kwan LY, Chavez V, Cohavy O, Gonsky R, Chang EY, Chang C, Elson CO, Targan SR (2009) Microbial induction of inflammatory bowel disease associated gene TL1A (TNFSF15) in antigen presenting cells. Eur J Immunol 39:3239–3250
Stranger BE, Montgomery SB, Dimas AS, Parts L, Stegle O, Ingle CE, Sekowska M, Smith GD, Evans D, Gutierrez-Arcelus M, Price A, Raj T, Nisbett J, Nica AC, Beazley C, Durbin R, Deloukas P, Dermitzakis ET (2012) Patterns of cis regulatory variation in diverse human populations. PLoS Genet 8:e1002639
Yamazaki K, Umeno J, Takahashi A, Hirano A, Johnson TA, Kumasaka N, Morizono T, Hosono N, Kawaguchi T, Takazoe M, Yamada T, Suzuki Y, Tanaka H, Motoya S, Hosokawa M, Arimura Y, Shinomura Y, Matsui T, Matsumoto T, Iida M, Tsunoda T, Nakamura Y, Kamatani N, Kubo M (2013) A genome-wide association study identifies 2 susceptibility Loci for Crohn’s disease in a Japanese population. Gastroenterology 144:781–788
Yue TL, Ni J, Romanic AM, Gu JL, Keller P, Wang C, Kumar S, Yu GL, Hart TK, Wang X, Xia Z, DeWolf WE Jr, Feuerstein GZ (1999) TL1, a novel tumor necrosis factor-like cytokine, induces apoptosis in endothelial cells: involvement of activation of stress protein kinases (stress-activated protein kinase and p38 mitogenactivated protein kinase) and caspase-3-like protease. J Biol Chem 274:1479–1486
Zhang FR, Huang W, Chen SM, Sun LD, Liu H, Li Y, Cui Y, Yan XX, Yang HT, Yang RD, Chu TS, Zhang C, Zhang L, Han JW, Yu GQ, Quan C, Yu YX, Zhang Z, Shi BQ, Zhang LH, Cheng H, Wang CY, Lin Y, Zheng HF, Fu XA, Zuo XB, Wang Q, Long H, Sun YP, Cheng YL, Tian HQ, Zhou FS, Liu HX, Lu WS, He SM, Du WL, Shen M, Jin QY, Wang Y, Low HQ, Erwin T, Yang NH, Li JY, Zhao X, Jiao YL, Mao LG, Yin G, Jiang ZX, Wang XD, Yu JP, Hu ZH, Gong CH, Liu YQ, Liu RY, Wang DM, Wei D, Liu JX, Cao WK, Cao HZ, Li YP, Yan WG, Wei SY, Wang KJ, Hibberd ML, Yang S, Zhang XJ, Liu JJ (2009) Genome-wide association study of leprosy. N Engl J Med 361:2609–2618
Zhou M, Liu R, Su D, Feng X, Li X (2014) TL1A increased the differentiation of peripheral Th17 in rheumatoid arthritis. Cytokine 69:125–130
Zinovieva E, Bourgain C, Kadi A, Letourneur F, Izac B, Said-Nahal R, Lebrun N, Cagnard N, Vigier A, Jacques S, Miceli-Richard C, Garchon HJ, Heath S, Charon C, Bacq D, Boland A, Zelenika D, Chiocchia G, Breban M (2009) Comprehensive linkage and association analyses identify haplotype, near to the TNFSF15 gene, significantly associated with spondyloarthritis. PLoS Genet 5:e1000528
Acknowledgments
We would like to thank all the patients and volunteers who enrolled in the study. We also thank Ms. Megumi Sageshima, Ms. Yuko Hirano, Ms. Natsumi Baba, Ms. Rieko Shirahashi, Ms. Ayumi Nakayama (The University of Tokyo), and Ms. Hitomi Nakamura (National Hospital Organization Nagasaki Medical Center) for technical and administrative assistance. This work was supported by Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (#23591006, #2629318) to M.N., Grant-in-Aid for Clinical Research from the NHO to M.N., Grant from the Research Program of Intractable Disease provided by the Ministry of Health, Labor, and Welfare of Japan to M.N., Grant from Core-to-Core program (Asia-Africa Science Platforms) from the Japan Society for the Promotion of Science to K.T., Uehara Memorial Foundation to Y.H., and Takeda Foundation to Y.H.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hitomi, Y., Kawashima, M., Aiba, Y. et al. Human primary biliary cirrhosis-susceptible allele of rs4979462 enhances TNFSF15 expression by binding NF-1. Hum Genet 134, 737–747 (2015). https://doi.org/10.1007/s00439-015-1556-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-015-1556-3