Abstract
Hereditary myopathy with lactic acidosis (HML) is caused by an intron mutation in the iron-sulphur cluster assembly gene (ISCU) leading to incorporation of intron sequence into the mRNA. This results in a deficiency of Fe–S cluster proteins, affecting the TCA cycle and the respiratory chain. The proteins involved in the Fe–S machinery are evolutionary conserved and shown to be fundamental in all organisms examined. ISCU is expressed at high levels in numerous tissues in mammals, including high metabolic tissues like the heart, suggesting that a drastic mutation in the ISCU gene would be damaging to all energy-demanding organs. In spite of this, the symptoms in patients with HML are restricted to skeletal muscle, and it has been proposed that splicing events may contribute to the muscle specificity. In this study we confirm that a striking difference in the splicing pattern of mutant ISCU exists between different tissues. The highest level of incorrectly spliced ISCU mRNA was found in skeletal muscle, while the normal splice form predominated in patient heart. The splicing differences were also reflected at a functional level, where loss of Fe–S cluster carrying enzymes and accumulation of iron were present in muscle, but absent in other tissues. We also show that complete loss of ISCU in mice results in early embryonic death. The mice data confirm a fundamental role for ISCU in mammals and further support tissue-specific splicing as the major mechanism limiting the phenotype to skeletal muscle in HML.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hereditary myopathy with lactic acidosis (HML, OMIM #255125) was first described in 1964 when Larsson et al. (1964) reported 14 patients, from five families, with poor physical abilities since childhood. The patients suffered from low exercise tolerance associated with muscle cramps, tachycardia and shortness of breath, and increased release of lactate and pyruvate even when subjected to a low work load. In severe episodes of the disease, patients experienced widespread fatigue, myoglobinuria and severe acidosis (Larsson et al. 1964; Linderholm et al. 1969). Biochemical analysis showed defects in complexes I, II and III of the respiratory chain and low levels of mitochondrial aconitase, all of which contain iron–sulphur (Fe–S) centres (Linderholm et al. 1990; Hall et al. 1993; Haller et al. 1991). It was therefore suggested that the patients suffered from dysfunction in synthesis, import, processing or assembly of Fe–S clusters (Hall et al. 1993). This was confirmed when a disease-specific mutation (IVS5 + 382G>C) was found in the gene coding for the iron–sulphur cluster assembly protein (ISCU) (Mochel et al. 2008; Olsson et al. 2008). The mutation consists of a single base pair change in the last intron of the gene, leading to activation of cryptic splice sites and insertion of 100 bp of intron sequence into the mRNA (Mochel et al. 2008; Olsson et al. 2008). Recently, Kollberg et al (2009) showed that the mutation in fact gives rise to two differently sized inserts, 100 and 86 bp, respectively, with the same acceptor splice site but different donor splice sites.
The ISCU protein has a central role in the formation of Fe–S clusters where it functions as a scaffold protein on which the clusters are formed. After assembly, the Fe–S clusters are delivered to different target proteins important for various processes, including the TCA cycle and the electron transport chain (Agar et al. 2000; Yang et al. 2006). Fe–S cluster assembly is a vital process that has been maintained throughout evolution, with the major proteins well conserved from prokaryotes to mammals (Lill and Muhlenhoff 2008). The fundamental role of the Fe–S cluster assembly has also been demonstrated by the knock-down of several of the components involved in the assembly process in different species. In yeast, double deletion of the S.cerevisiae ISCU homolog’s ISU1 and ISU2 has been shown to be lethal (Tong and Rouault 2000; Schilke et al. 1999) and in both zebrafish and yeast the deletion of Glutaredoxin 5, a gene shown to be required for Fe–S cluster assembly, leads to serious defects (Wingert et al. 2005; Rodriguez-Manzaneque et al. 2002). Furthermore, knock-out of Frataxin (FXN), the believed iron-donor in the Fe–S cluster assembly machinery, results in early embryonic death in mice (Cossee et al. 2000). These and other studies show that removal of any of the key factors in the Fe–S cluster assembly is incompatible with life in many species. This would also suggest that a loss of ISCU in humans, as well as other mammals, would be detrimental. Still, the drastic mutation observed in HML patients does not result in a drastic systemic phenotype. The patients are not severely affected unless the metabolic system is put under stress by exercise or extreme diets such as fasting. One reason for the relatively mild symptoms is that the phenotype seems to be restricted to skeletal muscle leaving other energy-demanding organs, such as the heart and central nervous system, unaffected. In contrast to this, individuals homozygous for mutations in FXN suffer from Friedreich’s ataxia, a disease associated with both neurodegeneration and severe cardiomyopathy (Muhlenhoff et al. 2002). It has, however been shown that patients who are compound heterozygous for the HML mutation and a missense mutation in exon 3 (c.149G>A) of ISCU show a more severe disease phenotype, including cardiomyopathy (Kollberg et al. 2009). This indicates that ISCU is important for the Fe–S assembly also in the heart, which puts further emphasis on the question of why there is a selective involvement of skeletal muscles in HML. Recently, Sanaker et al. (2010) reported differences in splicing of ISCU mRNA between muscle, myoblasts, fibroblasts and blood, which suggests that tissue-specific splicing events may be responsible for the lack of a systemic phenotype.
In this study, we report that splicing of the mutant ISCU transcript indeed differs between different patient tissues. The most pronounced difference was observed between muscle and heart, where the majority of the ISCU transcript was incorrectly spliced in muscle, while the correct splice form predominated in heart. The difference in splicing, between muscle and other tissues, was also reflected on a functional level. Furthermore, we show that knock-out of Iscu in mice results in early embryonic death. This suggests that complete loss of ISCU is as drastic in mammals as in lower organisms, and further support alternative splicing as a phenotype-restricting mechanism in HML patients.
Methods
Patient and control material
For RNA and protein extraction, materials from four affected individuals were used. Heart, liver, kidney as well as three different muscle autopsy samples were from individual P1, who died from an episode of the disease at age 32 (post mortem time = 24 h). An additional four muscle biopsy samples were from three affected individuals P2–4 (age 37, 49, and 50 years). None of the patients showed clinical signs of heart involvement. Control material was obtained from a total of 11 individuals. Four heart autopsy samples were obtained from two individuals, C1–2, cause of death cardiomyopathy and sudden infant death, ages 42 years and 4 months (post mortem time <24 and <36 h) and three muscle autopsy samples from two individuals, C3–4, cause of death in both cases head trauma, age 20 and 35 years (post mortem time <24 h). An additional seven control muscle biopsies were obtained from seven healthy controls, C5–11 (mean age 33 years, range 20–47 years). All tissue samples were fresh-frozen in liquid nitrogen immediately after dissection. In addition, two commercial RNA samples isolated from heart and liver were used (MVP™ total RNA human heart, lot 0480123 and MVP™ total RNA human liver, pool of three individuals, lot1170418, Stratagene). For Perls’ iron staining, formalin-fixed, paraffin-embedded autopsy material (muscle, heart, liver and pons) were used from one additional patient and control.
RNA extraction and cDNA amplification
Tissue samples were frozen in liquid nitrogen and then disrupted using a ball mill grinder or the TissueLyser LT (Qiagen). RNA was extracted from the disrupted tissues using the RNeasy micro kit (Qiagen) according to the manufacturer’s recommendations, or purchased from Stratagene. cDNA was synthesized using the SuperScript™ First-Strand Synthesis System for RT-PCR (Invitrogen) with random hexamers according to the manufacturer’s recommendations.
Semi-qRT-PCR and qRT-PCR
For semi-qRT-PCR, short PCR (25 cycles) was performed on cDNA with addition of 32P-labelled α-dCTP to the reaction mixture. The primers used were as follows: ISCU IV-F: 5′-AGATATCGCCAAGGAGCTCT-3′ and ISCU V-R: 5′-ATTTGTAATCAGCCAGGGCG-3′. The primers will give rise to 185 or 171 bp products if the cDNA contains an intronic insertion and a product of 85 bp if it lacks an intronic insertion. The PCR products were resolved on a non-denaturing 4% polyacrylamide gel and run for 2 h at 150–200 V. Gels were fixed in 20% methanol/10% acetic acid for 15 min, then dried and visualized by phosphoimaging. Quantification of band volume was performed using Quantity One software (Bio-Rad). For qRT-PCR, cDNA was synthesized with random hexamers on 80 ng of total RNA using the SuperScript™ First-Strand Synthesis System for RT-PCR (Invitrogen). PCR experiments were performed in duplicates using FastStart Universal SYBR Green Master (ROX) mix (Roche Diagnostics), 0.45 μl of cDNA and 1.5 μl of each primer (2.5 μM) using the same cycling parameters as described before (Strom et al. 2005). The following primers were used for detection of ISCU sequences: ISCUmitoF 5′-CTACACAAGAAGGTGTTGA-3′ and ISCU IIQRTR 5′-CCCACCAGTCCAGTTCCAA-3′. GADPH was used as internal control. PCR products were detected using an ABI PRISM 7000 Sequence Detection System (Applied Biosystems). Fold change was calculated using the 2−ΔΔCt method. All statistics were calculated using Student’s t test.
SDS-PAGE and western blot analysis
For protein analysis, patient and control tissues as well as mouse (Iscu +/− and wt) tissues were used. For protein extraction, tissues were disrupted using a ball mill grinder or the TissueLyser LT (Qiagen) and dissolved in lysis buffer with phosphatase inhibitors (2% SDS, 100 mM Tris–HCl, pH 6.8, 10 mM NaF, 10 mM β-glycerol phosphate disodium and 1 mM sodium vanadate) followed by sonication. The protein extracts were separated on 8–16% Tris–HCl or 4–12% Bis–Tris Criterion™ XT precast gels (Bio-Rad) according to the manufacturer’s recommendations, and transferred to Amersham™ Hybond™ ECL nitrocellulose membranes (GE Healthcare). The membranes were blocked with 5% non-fat milk solution for 1 h at room temperature and incubated overnight at 4°C either with rabbit anti-ISCU 1/2 antibody (FL-142, 1:200), rabbit anti-SDHB (FL-280, 1:300), rabbit anti-ACO2 (A-22, 1:500) (all from Santa Cruz Biotechnology) or rabbit anti-actin (1:300, Sigma) and then with HRP-conjugated goat anti-rabbit antibody (1:50,000; Pierce) for 1 h at room temperature. Proteins were visualized by ECL advanced enhanced chemiluminescence according to the manufacturer’s recommendations (GE Healthcare).
Perls’ iron staining
Paraffin-embedded formalin-fixed muscle, heart, liver and brain samples from patient and control, and fresh-frozen muscle, heart, liver and brain from Iscu +/− and wt mice were sectioned at 8 μm and stained for iron with Perls’ Prussian Blue stain. The paraffin sections were deparaffinised and hydrated before incubation in 2% aqueous potassium ferrocyanide-HCl. Sections were counter-stained using 0.1% Nuclear Fast Red, rehydrated and mounted. For DAB enhanced Perls’staining slides were incubated with DAB solution for 20 min following the incubation in 2% aqueous potassium ferrocyanide-HCl after which the slides were rehydrated and mounted. Sections were analysed using a Nikon Eclipse E800 microscope and photographed with a Nikon Digital Camera (DXM1200).
Generation and genotyping of Iscu null mice
ES-cell lines containing Iscu deletion were obtained through the knock-out mouse project (KOMP) repository. To generate chimeras, ES-cells were injected into BALB/c host blastocysts which were transferred into pseudo-pregnant females. Male chimeras were selected by coat colour and crossed with C57BL/6J females to obtain germline transmission of the Iscu deletion. Genomic DNA was prepared from tail tips from mice, tails from E10.5 embryos, whole E7.5–9.5 embryos and whole blastocysts (E3.5). Multiplex PCR for mouse genotyping was performed using a common forward primer (SU: 5′-TCGAATCATAAACACGCCTG-3′) and two reverse primers specific for the wt (TUR: 5′-GGCTAGGTGCTTGGCTGAG-3′) and mutant (LacInZRev: 5′-GTCTGTCCTAGCTTCCTCACTG-3′) Iscu alleles. The PCR products were 801 bp for the wt allele and 353 bp for the knockout allele. PCR amplification was performed in 25 μl reactions containing 0.14 μg of genomic DNA from tails or embryos or 3 μl of blastocyst DNA preparations, 20 pmol of primer SU, 10 pmol of primer TUR, 2.5 pmol of primer LacInZRev, 1× Ampliqon Ammonium Reaction Buffer, 300 μM of each dNTP, 10% DMSO and 1.25 U of VWR taq DNA polymerase (VWR International). Cycling conditions were 35 cycles with the following steps: 95°C for 15 s, 54°C for 15 s, 72°C for 30 s in a GeneAmp PCR System 9700 (Applied Biosystems) machine. PCR fragments were separated by electrophoresis on a 1.7% agarose gel.
Results
The splicing pattern of ISCU differs between heart, liver and skeletal muscle
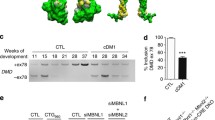
To study the splicing pattern of ISCU mRNA in different tissues, we performed semi-qRT-PCR on three energy-demanding tissues; muscle, heart and liver, from patient and controls. As expected, there was a major difference in the splicing pattern between patient and control muscle (Fig. 1a, b). In patient samples, the highest relative amount of mutant ISCU mRNA was observed in muscle, representing almost 80% of the total ISCU mRNA, with the transcript containing the 100 bp insert being the most common. Analysis of the splicing pattern in muscle biopsies from an additional three patients (P2–4) and seven controls (C5–11) confirmed the pattern observed in the autopsy samples (data not shown). In patient heart and liver, the wt ISCU mRNA predominated. In patient heart, the wt ISCU transcript made up 70% of total ISCU mRNA and the two mutant transcripts were equally represented. In liver, the wt and mutant transcripts were more equally represented with approximately 54% wt transcript (Fig. 1b). Some incorrectly spliced ISCU mRNA could also be detected in all control tissues, ranging from 3% of total ISCU mRNA in liver to 7% in muscle.
Analysis of the ISCU splicing pattern in different tissues. a Semi-qRT-PCR, using ISCU gene-specific primers, performed on cDNA from control muscle (lane 1), patient muscle (lane 2), control heart (lane 3), patient heart (lane 4), control liver (lane 5), patient liver (lane 6). The amount of PCR product loaded was adjusted to facilitate the quantification. Bands were visualized using phosphoimaging. b Quantification of the semi-qRT-PCR. The proportions of wt versus mutant transcript are shown as percentage of total ISCU in each tissue. Results are presented as mean ± SD from at least three independent experiments (*p < 0.05, **p < 0.01, ***p < 0.001; Student’s t test). All patient samples were from individual P1, control samples were from C1–4 as well as one commercial liver RNA sample (pool of three individuals)
ISCU protein is essentially absent in patient muscle but present in other tissues
To determine if the differences in splicing pattern between different tissues was reflected at the protein level, cellular extracts from patient muscle, heart, liver and kidney were analysed by western blot. In accordance with the mRNA splicing data, wt ISCU was essentially absent in patient muscle but was present in heart, liver and kidney (Fig. 2a). Since the most drastic difference in splicing was observed between muscle and heart, we compared absolute levels of ISCU protein in these tissues from a HML patient and from controls. This confirmed an absence or extremely low levels of ISCU protein in patient muscle as compared to controls (Fig. 2b, lanes 1–5). In patient heart ISCU protein was present but, also here, we observed drastically reduced levels as compared to controls. (Fig. 2b, lanes 6–9). Analysis of total mitochondrial mRNA levels of ISCU in heart of the same patient using qRT-PCR showed a decrease, with a level of only 41% as compared to controls. In both patient muscle and heart, a faint band corresponding to a faster migrating product could be seen. The size of this product corresponds to the expected size of the mutant protein (Fig. 2b).
ISCU protein levels in different tissues, analysed by western blot. a Control muscle (lane 1) and patient muscle, kidney, heart and liver (lanes 2–5). b Control muscle (lanes 1–3), patient muscle (lanes 4–5), control heart (lanes 6–8) and patient heart (lane 9). Actin was used as a loading control. All patient samples were from individual P1, control samples were from C1–4
The defective splicing is reflected at the functional level
It has previously been shown that iron accumulates in the mitochondria of HML patient muscles as a consequence of the defect in Fe–S metabolism (Haller et al. 1991; Mochel et al. 2008). In agreement with this, iron deposits were observed in patient muscle but not in control muscle (Fig. 3). In contrast, no accumulation of iron was seen in any other patient or control tissues examined including heart (Fig. 3). Earlier analysis of muscle tissue from HML patients has shown a decrease of the Fe–S cluster-containing enzymes aconitase and SDH (Hall et al. 1993; Haller et al. 1991). We compared patient heart and muscle samples with controls to see whether the decrease of ISCU protein seen in heart also affects the levels of these Fe–S cluster-containing enzymes. As expected, the levels of aconitase and the Fe–S-containing SDH subunit (SDHB) were much lower in muscle from patients than in muscle from controls (Fig. 4, lanes 1–5). In contrast to this, the levels of both enzymes were similar in heart tissue from patient and controls (Fig. 4, lanes 6–9). There even seemed to be a slight elevation of SDHB expression in patient heart as compared to controls.
Iron accumulation in different tissues analysed by Perls’ iron staining (rows 1–2) and Perls’ DAB enhanced staining (rows 3–4) on paraffin sections (8 μm thickness) from muscle, heart, liver, and pons from a control (row 1) and a patient (rows 2–4). Iron accumulation appears as blue punctate staining (rows 1–2) or brown punctate staining (row 3)
Aconitase (ACO2) and SDHB protein levels determined by western blot. Protein extracts from control muscle (lanes 1–3), patient muscle (lanes 4–5), control heart (lanes 6–8) and patient heart (lane 9). Actin was used as a loading control. All patient samples were from individual P1, control samples were from C1–4
Complete absence of ISCU results in an embryonic lethal phenotype
If the reason for the muscle-specific phenotype seen in patients is the differential splicing observed, total loss of ISCU should result in a more severe phenotype affecting other energy-demanding tissues. To test this hypothesis, Iscu null mice were generated from ES cell lines obtained from the KOMP repository. The heterozygous mice were indistinguishable from their wt littermates in terms of physical and behavioural phenotype. However, inter-cross of heterozygous mice failed to produce homozygous Iscu null offspring (n = 100). To examine the stage of embryonic death, E7.5–10.5 embryos (n = 45) were dissected and genotyped. No Iscu −/− embryos were observed at day 7.5–10.5; however, there were remnants of resorbed embryos present. This suggests that loss of ISCU results in early embryonic death. To determine whether Iscu −/− embryos could be observed pre-implantation, stage E3.5 embryos were flushed out and genotyped. Out of nine genotyped E3.5 embryos, two were homozygous for the null allele (Fig. 5a, lanes 3–4). To see if there was any defects on enzymes important for the aerobic energy production due to the loss of one Iscu allele, we examined the heterozygous mice further. We observed reduced (20–30%) levels of ISCU protein in the heterozygous null mice in all tissues examined (Fig. 5b). This indicates that the functional Iscu allele is up-regulated, approaching wt levels. In accordance with this, we could not see any effect on the levels of the metabolic enzymes aconitase and SDHB (Fig. 5b). Furthermore, no accumulation of iron was observed in muscle or any other tissue examined (data not shown).
a PCR on DNA isolated from E3.5 blastocysts from intercross between Iscu+/− mice. Wt embryo (lane 1), heterozygous embryo (lane 2) and embryos homozygous for the null allele (lanes 3 and 4). b ISCU, aconitase (ACO2) and SDHB protein levels in muscle, heart, brain and liver of Iscu+/−mice determined by western blot. Wt mice (+/+) (lanes 1, 3, 5 and 7). Iscu+/− mice (±) (lanes 2, 4, 6 and 8). Actin was used as a loading control
Discussion
In this paper we show that mutant ISCU mRNA is differentially spliced in energy-demanding tissues of HML patients, confirming differential splicing as a likely mechanism for the muscle-specific phenotype. We show that the ISCU transcript is differentially spliced in muscle, heart and liver. In patient muscle, the major part (~80%) of total ISCU mRNA was found to be of the mutant form, which is in agreement with previous findings (Sanaker et al. 2010). In patient heart, 70% of the ISCU transcript was correctly spliced, which might explain the lack of heart involvement in the disease. In liver, the correctly spliced transcript was at a level similar to that of the incorrectly spliced forms. A difference in frequency of the two mutant splice forms between different tissues was also observed, further emphasising the presence of tissue-specific splicing events. Furthermore, the mutant ISCU transcript was also present at low levels in tissues from control individuals, which, as previously reported, indicates that there is a weak splice site in the ISCU intron sequence that is strengthened by the mutation (Sanaker et al. 2010).
At the protein level, ISCU was present in heart, kidney and liver from patient, while practically no ISCU could be detected in patient muscle. Comparison of ISCU levels revealed that there was also a drastic reduction in patient heart. However, the observation that only 70% of ISCU is correctly spliced in combination with lower levels of total mitochondrial ISCU mRNA could explain the low protein levels seen in patient heart. The reduction of ISCU protein observed was still unexpected since there is no record of heart involvement in HML patients. Nevertheless, it is evident that the heart is sensitive to the loss of ISCU, since patients that are compound heterozygous for the IVS5 + 382G>C and the c.149G>A missense mutation also suffer from cardiomyopathy (Kollberg et al. 2009). Still, the ISCU levels in HML patient heart seem to be sufficient for avoiding a heart phenotype, since no sign of a defect Fe–S cluster assembly is evident based on the normal levels of SDH and aconitase and the absence of iron accumulations.
The data from this study strongly support the theory that the muscle-restricted phenotype seen in HML patients is due to the fact that more incorrectly spliced transcript is generated in muscle, leading to lower levels of functional protein as compared to other tissues. Total knock-down of ISCU and other proteins associated with the Fe–S cluster biogenesis has earlier been shown to lead to drastic phenotypes in a number of different organisms (Schilke et al. 1999; Tong and Rouault 2000; Cossee et al. 2000; Wingert et al. 2005; Smid et al. 2006). Knock-down of ISCU in mammals would therefore be expected to result in an equally serious phenotype. One can, of course, not rule out that a more complex system has evolved in higher organisms that might, in part or in whole, explain the muscle-specific phenotype. The generation of an Iscu null mouse did, however, show that ISCU is just as essential in mammals, with complete loss of ISCU resulting in early embryonic death. No homozygous null embryos were identified at E7.5–10.5, however at E3.5, two out of nine embryos were homozygous for the null allele, suggesting that ISCU is important for the implantation or the early development of the embryo. These data further support the essential role of ISCU in many vital processes. In contrast, the Iscu +/− mice showed no apparent physical impairment and were identical to their wt littermates in both behaviour and physical appearance. The Iscu null phenotype is similar to the FXN null mouse where the homozygous embryos show severe abnormalities already at E6.75, while heterozygous mice appear normal (Cossee et al. 2000). This indicates that both proteins are fundamental in mammals, likely with equally crucial roles in the Fe–S cluster assembly. Analysis of Iscu +/− mice, as expected, showed lower levels of ISCU protein as compared to wt littermates. The reduction was however less than 50%, indicating an up-regulation of the functional allele. Furthermore, aconitase and SDHB levels were the same as in wt mice and there was no iron accumulation in any of the tissues analysed. These data suggest that the ISCU levels in the heterozygous mice are high enough to sustain the Fe–S cluster formation needed for a functional aerobic energy metabolism and to avoid problems in the iron metabolism. It would, however, be interesting to see how the system would react when put under stress, either by extensive exercise or by extreme diets.
In conclusion, we report differences in the splicing patterns of ISCU mRNA in different energy-demanding organs of patients with HML. The differences are also observed at the protein level, where more functional protein is generated in non-skeletal muscle tissues. Furthermore, we show that complete loss of ISCU results in early embryonic death in mice. This provides further support of alternative splicing as a mechanism responsible for the relatively mild phenotype in HML patients. The nature of the differential splicing described in this report is yet unknown. However, alternative splicing has been shown to be much more common as a regulatory mechanism than earlier believed and can either be tissue-specific or induced by changes in the microenvironment. It has, for example, been shown that alternative splicing correlates with ATP depletion in neurodegenerative diseases and is regulated by hypoxia in breast cancer (Maracchioni et al. 2007; Hirschfeld et al. 2009). The alternative splicing in HML could be tissue-specific, but may also depend on general or disease induced differences in the metabolic state.
References
Agar JN, Krebs C, Frazzon J, Huynh BH, Dean DR, Johnson MK (2000) IscU as a scaffold for iron–sulfur cluster biosynthesis: sequential assembly of [2Fe–2S] and [4Fe–4S] clusters in IscU. Biochemistry 39 (27):7856–7862 bi000931n [pii]
Cossee M, Puccio H, Gansmuller A, Koutnikova H, Dierich A, LeMeur M, Fischbeck K, Dolle P, Koenig M (2000) Inactivation of the Friedreich ataxia mouse gene leads to early embryonic lethality without iron accumulation. Hum Mol Genet 9 (8):1219–1226 ddd132 [pii]
Hall RE, Henriksson KG, Lewis SF, Haller RG, Kennaway NG (1993) Mitochondrial myopathy with succinate dehydrogenase and aconitase deficiency. Abnormalities of several iron–sulfur proteins. J Clin Invest 92(6):2660–2666
Haller RG, Henriksson KG, Jorfeldt L, Hultman E, Wibom R, Sahlin K, Areskog NH, Gunder M, Ayyad K, Blomqvist CG et al (1991) Deficiency of skeletal muscle succinate dehydrogenase and aconitase. Pathophysiology of exercise in a novel human muscle oxidative defect. J Clin Invest 88(4):1197–1206
Hirschfeld M, zur Hausen A, Bettendorf H, Jager M, Stickeler E (2009) Alternative splicing of Cyr61 is regulated by hypoxia and significantly changed in breast cancer. Cancer Res 69(5):2082–2090. doi:10.1158/0008-5472
Kollberg G, Tulinius M, Melberg A, Darin N, Andersen O, Holmgren D, Oldfors A, Holme E (2009) Clinical manifestation and a new ISCU mutation in iron-sulphur cluster deficiency myopathy. Brain doi:10.1093/brain/awp152
Larsson LE, Linderholm H, Mueller R, Ringqvist T, Soernaes R (1964) Hereditary metabolic myopathy with paroxysmal myoglobinuria due to abnormal glycolysis. J Neurol Neurosurg Psychiatry 27:361–380
Lill R, Muhlenhoff U (2008) Maturation of iron–sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu Rev Biochem 77:669–700. doi:10.1146/annurev.biochem.76.052705.162653
Linderholm H, Muller R, Ringqvist T, Sornas R (1969) Hereditary abnormal muscle metabolism with hyperkinetic circulation during exercise. Acta Med Scand 185(3):153–166
Linderholm H, Essen-Gustavsson B, Thornell LE (1990) Low succinate dehydrogenase (SDH) activity in a patient with a hereditary myopathy with paroxysmal myoglobinuria. J Intern Med 228(1):43–52
Maracchioni A, Totaro A, Angelini DF, Di Penta A, Bernardi G, Carri MT, Achsel T (2007) Mitochondrial damage modulates alternative splicing in neuronal cells: implications for neurodegeneration. J Neurochem 100(1):142–153. doi:10.1111/j.1471-4159.2006.04204.x
Mochel F, Knight MA, Tong WH, Hernandez D, Ayyad K, Taivassalo T, Andersen PM, Singleton A, Rouault TA, Fischbeck KH, Haller RG (2008) Splice mutation in the iron-sulfur cluster scaffold protein ISCU causes myopathy with exercise intolerance. Am J Hum Genet 82(3):652–660. doi:10.1016/j.ajhg.2007.12.012
Muhlenhoff U, Richhardt N, Ristow M, Kispal G, Lill R (2002) The yeast frataxin homolog Yfh1p plays a specific role in the maturation of cellular Fe/S proteins. Hum Mol Genet 11(17):2025–2036
Olsson A, Lind L, LE Thornell, Holmberg M (2008) Myopathy with lactic acidosis is linked to chromosome 12q23.3–24.11 and caused by an intron mutation in the ISCU gene resulting in a splicing defect. Hum Mol Genet 17(11):1666–1672. doi:10.1093/hmg/ddn057
Rodriguez-Manzaneque MT, Tamarit J, Belli G, Ros J, Herrero E (2002) Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol Biol Cell 13(4):1109–1121. doi:10.1091/mbc.01-10-0517
Sanaker PS, Toompuu M, Hogan VE, He L, Tzoulis C, Chrzanowska-Lightowlers ZM, Taylor RW, Bindoff LA (2010) Differences in RNA processing underlie the tissue specific phenotype of ISCU myopathy. Biochim Biophys Acta 1802(6):539–544. doi:10.1016/j.bbadis.2010.02.010
Schilke B, Voisine C, Beinert H, Craig E (1999) Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 96(18):10206–10211
Smid O, Horakova E, Vilimova V, Hrdy I, Cammack R, Horvath A, Lukes J, Tachezy J (2006) Knock-downs of iron-sulfur cluster assembly proteins IscS and IscU down-regulate the active mitochondrion of procyclic Trypanosoma brucei. J Biol Chem 281(39):28679–28686. doi:10.1074/jbc.M513781200
Strom AL, Forsgren L, Holmberg M (2005) Identification and characterization of spinocerebellar ataxia type 7 (SCA7) isoform SCA7b in mice. Biochim Biophys Acta 1731(3):149–153. doi:10.1016/j.bbaexp.2005.10.004
Tong WH, Rouault T (2000) Distinct iron-sulfur cluster assembly complexes exist in the cytosol and mitochondria of human cells. EMBO J 19(21):5692–5700. doi:10.1093/emboj/19.21.5692
Wingert RA, Galloway JL, Barut B, Foott H, Fraenkel P, Axe JL, Weber GJ, Dooley K, Davidson AJ, Schmid B, Paw BH, Shaw GC, Kingsley P, Palis J, Schubert H, Chen O, Kaplan J, Zon LI (2005) Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature 436(7053):1035–1039. doi:10.1038/nature03887
Yang J, Bitoun JP, Ding H (2006) Interplay of IscA and IscU in biogenesis of iron–sulfur clusters. J Biol Chem 281(38):27956–27963. doi:10.1074/jbc.M601356200
Acknowledgments
We thank the families for their invaluable participation in the study. This project was supported by the Swedish Research Council and the Marcus Borgström Foundation. The experiments comply with the current laws of Sweden, in which the experiments were performed.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nordin, A., Larsson, E., Thornell, LE. et al. Tissue-specific splicing of ISCU results in a skeletal muscle phenotype in myopathy with lactic acidosis, while complete loss of ISCU results in early embryonic death in mice. Hum Genet 129, 371–378 (2011). https://doi.org/10.1007/s00439-010-0931-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-010-0931-3