Abstract
Genetic disorders of excessive salt loss from sweat glands have been observed in pseudohypoaldosteronism type I (PHA) and cystic fibrosis that result from mutations in genes encoding epithelial Na+ channel (ENaC) subunits and the transmembrane conductance regulator (CFTR), respectively. We identified a novel autosomal recessive form of isolated salt wasting in sweat, which leads to severe infantile hyponatremic dehydration. Three affected individuals from a small Bedouin clan presented with failure to thrive, hyponatremic dehydration and hyperkalemia with isolated sweat salt wasting. Using positional cloning, we identified the association of a Glu143Lys mutation in carbonic anhydrase 12 (CA12) with the disease. Carbonic anhydrase is a zinc metalloenzyme that catalyzes the reversible hydration of carbon dioxide to form a bicarbonate anion and a proton. Glu143 in CA12 is essential for zinc coordination in this metalloenzyme and lowering of the protein–metal affinity reduces its catalytic activity. This is the first presentation of an isolated loss of salt from sweat gland mimicking PHA, associated with a mutation in the CA12 gene not previously implicated in human disorders. Our data demonstrate the importance of bicarbonate anion and proton production on salt concentration in sweat and its significance for sodium homeostasis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hyponatremia is the most common electrolyte abnormality in clinical practice, and is the biochemical manifestation of a variety of illnesses (Thompson 2010). Hyponatremia is defined as a plasma sodium concentration of less than 135 mmol/L and occurs in 15–30% of hospitalized patients (Thompson 2010; Upadhyay et al. 2006). In children, it is usually associated with severe systemic disorders and is most often due to (1) intravascular volume depletion, (2) excessive salt loss or (3) hypotonic fluid overload, especially in infants. Genetic diseases resulting in excessive salt loss from sweat glands include aldosterone insensitivity syndromes of pseudohypoaldosteronism type I (PHA) (Hanukoglu 1991; Chang et al. 1996) and cystic fibrosis (CF [MIM 219700]) (O’Sullivan and Freedman 2009). We previously distinguished between two major forms of PHA (Hanukoglu 1991): A milder form of PHA [MIM 177735] results from mutations in the mineralocorticoid receptor gene (NR3C2, [MIM 600983]) (Geller et al. 2006); The severe autosomal recessive inheritance [MIM 264350] results from mutations in the genes encoding the three subunits (SCNN1A [MIM 600228]; SCNN1B [MIM 600760]; SCNN1G [MIM 600761]) of epithelial sodium channel (ENaC) (Edelheit et al. 2005, 2010). These PHA forms are characterized by severe salt wasting from aldosterone target organs including the sweat and salivary glands, distal colon, respiratory tract and kidney. Patients in early infancy present with severe hyponatremia, hyperkalemia, acidosis, dehydration with elevated sweat and saliva electrolytes, renal salt loss and markedly elevated aldosterone and plasma renin activity (Edelheit et al. 2005; Hanukoglu et al. 2008).
In this report, we present patients of three nuclear consanguineous Bedouin families from one clan who show clinical and some biochemical features similar to those observed in multi-system PHA; however, they differ from aldosterone insensitivity syndromes and CF in many aspects as described below. By positional cloning, we identified a mutation in carbonic anhydrase 12 that leads to the disorder. The data presented are consistent with a distinct clinical, biochemical and genetic entity.
Methods
Electrolytes levels in plasma and urine were measured at the biochemical laboratory of Soroka Medical Center using an Olympus ISE analysis device.
Sweat test
Qualitative test using Wescor Macroduct device was performed on all patients and family members. A positive test was defined as Cl− concentration higher than 50 mEq/L. Subjects with positive qualitative test underwent further quantitative sweat test. Sweat samples were collected by the Gibson and Cooke pilocarpine iontophoresis quantitative method. Chloride concentration was measured by titration analysis.
Hormonal assays
Blood samples for serum aldosterone and plasma renin were obtained in the morning while subjects were in a supine position and on normal sodium diet unless otherwise specified. Blood for plasma renin activity (PRA) was collected in chilled EDTA-coated vials that were kept and centrifuged at 4°C.
Aldosterone concentrations were measured by RIA (DPC Coat-a-Count aldosterone kit, Diagnostic Products Corporation, Los Angeles, CA). The sensitivity of the assay was 1.1 ng/dl, intra-assay coefficient of variation was 3.4% and inter-assay coefficient of variance was 6.5%. PRA was measured by an angiotensin I RIA kit (Diasorin, Stillwater, MN). The sensitivity of the assay was 0.3 ng/ml per h, intra-assay coefficient of variation was 5.4% and inter-assay coefficient of variance was 7.1%.
Molecular analyses
Genomic DNA was extracted from white blood cells by using standard procedures. Sequences of the coding exons and intron–exon junctions of the α, β and γ subunits of ENaC were determined as described (Edelheit et al. 2005).
Genotyping was done using Affymetrix (Santa Clara, CA) GeneChip® Human Mapping 250K Sty arrays. The genotype calls were determined using Affymetrix Genotyping Console Software. Dedicated software (KinSNP) was used to automatically search the microarray results for homozygous regions consistent with linkage (Amir el et al. 2010). Linkage analysis of polymorphic markers in the linkage interval and neighboring the CFTR gene was done according to Parvari et al. (1998). In addition to testing known polymorphic markers to confirm the linkage interval, additional markers were developed and tested using the Tandem Repeats Finder (Benson 1999) and PCR primers designed with the “Primer3” Web site.
Two-point and multipoint Lod scores were calculated using Superlink v1.4 at the PedTool server. Linkage analysis was performed assuming recessive inheritance with 99% penetrance and an incidence of 0.01 or 0.001 for the disease allele in the population.
RNA of lymphoblastoid cells was extracted using the EZ-RNA II kit of Biological Industries (Israel) and cDNA was synthesized by the Reverse-iT 1st Strand Synthesis Kit (ABgene, UK), using 1.9 μg of RNA. A portion (1/20) of the reverse transcription reaction was used for PCR amplifications with specific primers. The cDNA coding regions for the candidate genes: SLC24A1, RAB8B, RAB11A, HERC1 and CA12 were PCR amplified in overlapping fragments. The free PCR primers were digested by a combination of shrimp alkaline phosphatase 0.6 and 6 U Exonuclease I (both from Fermentas, Lithuania) and directly sequenced on an ABI PRISM® 3100 DNA Analyzer using the BigDye™ Terminator v. 1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, USA). The primers that revealed the mutation in the CA12 cDNA were: forward primer: 5′-GCTCCTGCTGGTGATCTTAAA-3′, reverse primer: 5′-GAAATGATCAACAACTTCCGG-3′. PCR conditions were 40 cycles of 95°, 60° and 72°. To verify the mutation on genomic DNA, the following primers were used for exon 4: forward primer: 5′-GGAAAGGGAACATTCTCAGG-3′, reverse primer: 5′-CCAGCAGCATCACTCAGTAAA-3′. PCR conditions were as above. The verification of the prevalence of the Glu143Lys mutation in CA12 in the Bedouin population was done by PCR amplification of the genomic DNA followed by restriction analysis with Eco130 I (Fermentas, Lithuania) that cuts the mutated sequence, but not the normal sequence.
Modeling the structural effect of the mutation
The crystal structure data of the wild-type CA12 (Whittington et al. 2001) (atom coordinates) was taken from RSCB PDB, I.D.: 1JCZ. The putative structure of Glu143Lys CA12 mutant was modeled using FoldX program (http://foldx.crg.es/), version 3.0, using “RepairPDB” followed by “BuildModel” commands with options set according to the recommended examples.
Results
Case reports
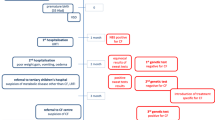
Three nuclear consanguineous Bedouin families from one clan were identified (Supplementary Fig. 1, Table 1). The patients were born after normal pregnancies and had normal birth weight.
Patient III-4
A 1-month-old Bedouin baby boy was admitted to the pediatric ward due to vomiting and diarrhea for 3 days. On admission, signs of moderate dehydration and failure to thrive (FTT) were noted. Laboratory findings showed severe hyponatremia and hyperkalemia with normal blood pH, but mildly reduced plasma bicarbonate. Urinary sodium excretion and urinary Na/K ratio were low. Fluid resuscitation with i.v. NaCl resulted in significant clinical improvement. Hydrocortisone therapy was also initiated with the presumed diagnosis of congenital adrenal hyperplasia. Both PRA and serum aldosterone level were elevated, as was sweat chloride concentration (Table 1). Endocrine investigations by basal and ACTH stimulation testing ruled out adrenal insufficiency. After clinical improvement was achieved, oral sodium chloride supplementation (3.42 mEq/kg/day) was initiated resulting in normalization of serum sodium, potassium, aldosterone levels and PRA. The transtubular potassium concentration gradient (TTKG) calculated as U K /Bl K X Bl Osm /U Osm at 3 months of age was 11.2 (normal values are >6 in hyperkalemia (Choi and Ziyadeh 2008) when serum K+ levels were 5.4 mEq/L and blood osmolality was 279 mOsmol/L. Catch up growth was noted during follow-up. At 6 years of age, the patient has been growing well, but salt craving is still reported. Repeated sweat test revealed high chloride secretion.
Patient III-5
A 4-month-old Bedouin baby boy was admitted with a history of restlessness for 5 days and signs of dehydration and FTT. Laboratory findings revealed hyponatremia, with hyperkalemia. Urinary sodium excretion and urinary Na/K ratio were low. Both PRA and serum aldosterone levels were elevated. Sweat chloride concentration was abnormally high (Table 1). After clinical improvement, a supplement of oral sodium chloride (4.28 mEq/kg/day) was initiated and resulted in serum sodium normalization. The TTKG, calculated at 11 months of age, was 11.6 (when serum K+ levels were 5.0 mEq/L and blood osmolality was 285 mOsmol/L). At 1 year of age, serum sodium, aldosterone concentration and PRA were normalized. Catch up growth was also evident. At 5.5 years of age, the patient still has salt craving.
Patient III-8
A 3-month-old Bedouin baby boy was admitted to the pediatric ward with fever and diarrhea for 1 day. His past medical history was significant for severe FTT, neonatal urinary tract infection and a previous episode of dehydration. Laboratory findings revealed hyponatremia, hyperkalemia and normal blood pH, but mildly reduced plasma bicarbonate. Urinary sodium excretion and urinary Na/K ratio were low. PRA and serum aldosterone levels were elevated. Sweat chloride concentration was highly elevated (Table 1). He was supplemented with oral sodium chloride (4.28 mEq/kg per day), but during the first year of follow-up he was admitted several times due to recurrent episodes of diarrhea due to cow milk allergy and lung infections, none of which resulted in subsequent respiratory sequels. His plasma sodium levels during these hospitalizations ranged between 118 and 137 mEq/L. The TTKG, calculated at 5 months of age, was 8.8 when he was severely hyponatremic (118 mEq/L). Serum K+ level was 5.2 mEq/L and blood and urine osmolalities were 255 and 709 mOsmol/L, respectively. His blood osmolality on salt supplementation was within the normal range (283 mOsmol/L).
At 2 years of age, his oral sodium chloride treatment was discontinued by the parents due to low compliance and he was readmitted due to weakness and loss of appetite. He had poor weight gain (Table 1). Laboratory evaluation disclosed hyponatremia, prerenal azotemia (urea 101 mg/dL and creatinine 0.6 mg/dL), compensated metabolic acidosis with normal anion gap, but normal serum K+ levels. Urinary sodium was 20 mEq/L with appropriately low fractional excretion of sodium (0.12%). Oral sodium chloride treatment was reinstituted. This treatment improved his linear growth and weight gain.
Blood pressure and serum urea and creatinine were within the normal range in all the patients during follow-up visits. Salivary electrolytes concentrations were measured in patients III-4 and III-8 at 4 and 32 months of age, respectively: sodium levels were 22/13 mmol/L, potassium 24.3/18.2 mmol/L and chloride 31/25 mmol/L representing normal concentrations for both patients. No evidence for chronic renal, respiratory or gastrointestinal abnormalities was found in any of the patients.
The study was approved by the Soroka Medical University Center Institutional Review Board, and all patients or their parents provided written informed consent before participating.
Genotyping the family members for the Glu143Lys mutation in CA12 revealed two apparently healthy male adults, who were found to be homozygous: patient II-3 had normal serum sodium (142 mEq/L) but elevated sweat chloride (71 mmol/L); his brother patient II-4 had normal serum sodium concentration (139 mEq/L) and sweat chloride concentrations (25 mEq/L). In addition, patient III-6’s 8-month-old brother had several measurements consistent with mild hyponatremia (130–132 mEq/l) during an episode of urinary tract infection. He had normal renal anatomy and his renal function was within normal values. Chloride concentrations in sweat could not be measured due to inadequate amounts of sweat obtained on two occasions. The association of the father of patients II-3 and II-4 to the family is known and presented in Supplementary Fig. 1. His mother’s exact link could not be traced. The patients’ pedigree suggests an autosomal recessive mode of inheritance (Supplementary Fig. 1).
Genetic studies
The excessive salt loss from sweat observed in the patients could be due to multi-system PHA or cystic fibrosis. Sequencing of the coding regions of the α, β and γ subunits of ENaC did not show any mutation in these genes. Assuming disease by homozygosity of a mutation from a common ancestor, linkage to CFTR was excluded by the finding of heterozygosity of alleles at polymorphic markers adjacent to the gene. We further searched for homozygous regions consistent with linkage by genotyping the three infant patients and their parents with the Affymetrix (Santa Clara, CA) GeneChip Human Mapping 250K Sty arrays. A single large homozygous region (>16 cM and 12.3 Mb) was found to be shared by the three affected patients. To confirm linkage to this region, all family members were tested with both known polymorphic microsatellite markers and with additional markers developed for this purpose. A linkage was identified to the chromosomal locus 15q21.3-q23, chr15:54877571-67190226 (NCBI Build 36.1) (Supplementary Fig. 1) with a Lod score of 2.81 for D15S1507 and a multipoint Lod score of 3.65. Linkage analysis was performed assuming recessive inheritance with 99% penetrance and an incidence of 0.01 or 0.001 for the disease allele in the population. The 12.3-Mb interval contains 103 genes. To identify the mutation in the cDNA of the patients’ genes in the interval, total RNA was extracted from lymphoblastoid cells of patient III-4, reverse transcribed, PCR amplified in overlapping fragments and the PCR products were sequenced. To identify the mutations among the many genes in the interval, they were prioritized for further sequencing according to their site of expression and function. Assuming first that the defect could affect an ion exchanger or transporter, the only relevant gene SLC24A1, a potassium-dependent sodium/calcium exchanger present in the interval, was analyzed but no deleterious mutations were identified. This interval contains genes like: transcription factors (e.g., HTF4), receptors (e.g., NARG2), numerous enzymes (e.g., ALDH1A2, LIPC, ADAM10) and signal transduction (MAP2K1). Among these, the next prioritized were the candidate genes RAB8B, RAB11A and HERC1, which encode proteins that are associated with and regulate transporters. Again, no deleterious mutation was identified in any of these genes. Since we hypothesized that the production of protons may affect sodium transport through the sodium proton exchangers, we next sequenced the Carbonic Anhydrase 12 gene (CA12, EC_number = “4.2.1.1”, [MIM 603263]). A non-synonymous variation replacing glutamate at position 143 with lysine (NM_001218: c.583G>A, NP_001209.1: p.Glu143Lys) was found in exon 4 (Supplementary Fig. 2a). This result was confirmed by direct sequencing of the PCR products amplified from the genomic DNA of all family members, showing that the variation is present in all patients in the homozygous state, but not in the healthy young siblings (Supplementary Fig. 2a). This variation was not reported in the human genome variation database (dbSNP). The glutamic acid at position 143 in CA12 is invariant in all CA isoenzymes (Supplementary Fig. 2c).
To examine the prevalence of this variation in the Bedouin population of the same geographic region, 192 controls were tested by PCR amplification of genomic DNA and restriction with Eco130I, which cuts only the non-synonymous variation sequence (Supplementary Fig. 2b). Three heterozygotes but no homozygotes were identified, thus excluding this variation as a common polymorphism and establishing its frequency to 0.78% in the highly inbred Bedouin population.
Carbonic anhydrases and the expected result of the replacement of glutamate 143 by lysine
CA12 belongs to the carbonic anhydrases family (CAs; also known as carbonate dehydratases EC 4.2.1.1). CAs are ubiquitous metalloenzymes present in prokaryotes and eukaryotes, and are encoded by four evolutionarily unrelated gene families. In mammals, 16 α-CA isozymes or CA-related proteins have been described, with different catalytic activity, subcellular localization and tissue distribution (Supuran 2008). CAs catalyze the reversible hydration of carbon dioxide to form a bicarbonate anion and a proton.
The widely accepted mechanism for this enzymatic reaction (Supuran 2008; Lesburg and Christianson 1995; Kiefer et al. 1995) is that a hydroxide coordinated to a bivalent zinc ion performs a nucleophilic attack on CO2. This results in a bicarbonate ion, which is displaced from its association to zinc by an incoming water molecule. Subsequent deprotonation of this water and removal of the proton regenerate the nucleophilic zinc-bound hydroxide ion, and prepare the enzyme for the next catalytic cycle. All components of this catalytic system must be correctly positioned to provide the precision required for the hydroxide’s nucleophilic attack on CO2 in aspects of electron negativity, distance and orientation. In the catalytic center of CA12, four direct ligands hold the zinc in place: the hydroxide ion and three histidine residues (Whittington et al. 2001). These four direct ligands are maintained in optimal position by a “second coordination shell” comprising four additional residues, each of which is hydrogen bonded to its corresponding direct ligand (Fig. 1a) (Lesburg and Christianson 1995; Kiefer et al. 1995; Whittington et al. 2001). This setup of the catalytic cleft is typical of the CA family and is highly conserved, as shown both by amino acid alignment to six other types of CA and comparison of crystallographic structures to CA2 (Whittington et al. 2001). Alteration of these hydrogen bonds in CA2 (Kiefer et al. 1995) and their subsequent crystallographic structure (Lesburg and Christianson 1995) revealed the importance of these hydrogen bond networks to the enzyme’s function in multiple aspects.
The effect of the mutation on the CA12 catalytic site with its direct and second shell ligands: the zinc 2+ ion is coordinated by four direct ligands (coordination shown by arrows). Four additional, second shell ligands hold the direct ligands in place and orientation by hydrogen bonds (shown by dashed lines). The amino acid side chains are detailed, and their respective counterparts in CA2 (same type, different number) are detailed in parenthesis. The fifth coordinating factor is an acetate introduced by the crystallization (coordinating arrow not shown). Hydrogens are omitted for clarity; some portions of the backbone are shown. Gray carbon, blue nitrogen, red oxygen, pink zinc II. a Crystal structure of wild-type CA12 (Whittington et al. 2001). The hydrogen bond between His145 and Glu143 contributing to the optimal coordination setup for catalytic activity is highlighted in green. Crystal structure data (atom coordinates) have been taken from RSCB PDB, I.D.: 1JCZ. b Putative structure of Glu143Lys CA12 mutant, modeled using FoldX program. The hydrogen bond between Lys143 and His145 is absent. The lysine is repelled by the side chain of His145, and is oriented away from histidine’s side chain (highlighted in green), which in turn does not fixate the latter in its optimal coordinating position to zinc2+ (color figure online)
In our homozygous patients Glu143 in CA12, a second shell residue, is replaced by lysine (Fig. 1b). This residue, corresponding to Glu117 in CA2, has been shown to be the most influential on the protein’s zinc affinity when compared with other second shell residues (Lesburg and Christianson 1995). Glutamate is negatively charged (Lesburg and Christianson 1995) and therefore has a strong hydrogen bond to His145 that is a direct ligand to zinc (equivalent to His119 in CA2). In contrast, the positively charged lysine would be repelled from histidine’s side chain, precluding the hydrogen bond between these two residues. This is demonstrated by a FoldX simulation (Pey et al. 2007): the distance measured between the nitrogens of histidine 145 and of lysine 143 is too large to enable a hydrogen bond (6.6 Å) (Fig. 1b). In mutations in CA2 where hydrogen bonds between the second shell and direct ligands were interrupted (Lesburg and Christianson 1995; Kiefer et al. 1995), the enzyme’s zinc affinity was lowered by 4.5- to 15-fold (Kiefer et al. 1995). An unprotonated lysine may theoretically form a hydrogen bond with His145 (lysine’s neutral nitrogen acting as the hydrogen bond acceptor), but the resultant bond would be significantly weaker than that formed by glutamate, due to lysine’s neutrality and its longer side chain. The importance of the length of this side chain was demonstrated by the change of Glu117 to Asp in CA2 that results in a threefold decrease in zinc affinity (Lesburg and Christianson 1995; Kiefer et al. 1995). Therefore, the mutation changing glutamate 143 into lysine would disrupt the coordination shells to the zinc and thus impair the enzyme’s catalytic activity.
Discussion
In this study, we have presented the clinical and biochemical findings of a new syndrome of hereditary sodium loss from sweat glands. We assume that the salt loss seems to be confined to the exocrine sweat glands, although GI sodium losses were not definitely ruled out. By genotyping and gene sequencing, we have identified that this syndrome is associated with a mutation in the gene that encodes carbonic anhydrase 12 on chromosome 15.
Although CA12 is highly expressed in the kidney (Ivanov et al. 2001; Parkkila et al. 2000), colon (Ivanov et al. 2001; Kivela et al. 2000) and pancreas (Ivanov et al. 2001; Kivelä et al. 2000), the finding that our patients do not present sodium loss in all these organs suggests that other carbonic anhydrases compensate for its enzymatic activity in all tissues except for the sweat glands. Specifically, the finding that there is no loss of salt in the kidney may be explained by a possible compensation by CA4 and CA14, which are located in the basolateral membrane of the proximal tubule with the catalytic side facing externally, similarly to CA12 (Parkkila et al. 2000; Brown et al. 1990; Schwartz et al. 2000; Whittington et al. 2001). Moreover, in the kidney, carbonic anhydrase is active in the proximal tubule and thus excessive loss of salt can be compensated by reabsorption in the distal tubule, particularly through the raised aldosterone levels that are seen in the patients. Indeed, mutations affecting the aldosterone-mediated sodium transport have a profound effect as they represent the last point of regulation in a spatial context in the kidney.
The presumed phenotypic effect of Glu143Lys mutation in CA12 is clearly different from the multi-system PHA in that the patients do not display salt loss from the kidney and salivary glands, nor do they suffer from persistent hyperkalemia. Unlike patients with PHA, the normal TTKG in our patients are consistent with normal renal potassium handling.
Hyponatremic hypochloremic dehydration with metabolic alkalosis is a rare but known presentation of CF in infants. Urinary sodium excretion is appropriately low in these patients (Ballestero et al. 2006). Unlike hyponatremic CF patients, our patients presented with mild compensated metabolic acidosis.
In addition to CF patients, we traced a single report in the literature of a female PHA patient with salt loss confined to the sweat and salivary glands during the first 19 months of life (Anand et al. 1976), but the molecular etiology was not investigated.
We observed a phenotypic heterogeneity ranging from severe infantile hyponatremia to apparently healthy adult subjects with or without excessive salt loss in the sweat, among individuals homozygous for the Glu143Lys mutation in CA12. Although all our patients were male, we consider this finding to be a coincidence due to the small number of patients. An independent study (Feldshtein et al. 2010, noted in the addendum) on a different clan in Israel reports female subjects affected with the same disease. Phenotypic heterogeneity among members of the same kindred is a well-recognized but not always an explained phenomenon in diverse genetic disorders, such as glucocorticoid remediable aldosteronism (Stowasser et al. 1999) and PHA type I with mutations in the MR gene (Geller et al. 2006). Precipitating events such as intercurrent infections accompanied by anorexia, vomiting and diarrhea could play a significant role during infancy in inducing hyponatremic dehydration in susceptible individuals homozygous for C12 mutation. The prevalence of such infections declines with age. Furthermore, behavioral adaptations such as salt craving and age-dependent developmental changes of other enzymes or transporters in the sweat gland may ameliorate the severe infantile phenotype observed in CA12-deficient activity. The above-mentioned possibilities may explain the asymptomatic clinical picture of homozygous adults with a mutated CA12 gene.
However, intense physical activity in hot temperatures, gastroenteritis and inadequate salt intake may expose patients who excrete abnormally high amounts of NaCl in their sweat to hyponatremia, as has previously been described in patients with CF (Epaud et al. 2005; Priou-Guesdon et al. 2010).
Ca12 and salt secretion in the sweat gland
The Glu143Lys mutation we identified in CA12 and the phenotypes of the patients indicate that this enzyme has a significant role in salt uptake in sweat glands. Carbonic anhydrases in general are involved in the regulation of acid–base balance, extracellular and intracellular pH and electrolyte transport (Chiche et al. 2009). CA12 is expressed in many cell types in various tissues including kidney, mesothelial cells lining body cavities and sweat and salivary glands (Ivanov et al. 2001; Liao et al. 2009). Immunohistochemical studies show that CA12 is located in the basolateral membranes of epithelial cells (Ivanov et al. 2001; Halmi et al. 2004; Karhumaa et al. 2000; Hermo et al. 2005; Hynninen et al. 2004; Kyllonen et al. 2003) with its active site located outside the cell (Whittington et al. 2001).
The extracellular localization of the active site of CA12 on the basolateral side raises questions about the mechanisms by which CA12 regulates salt loss on the luminal side of sweat gland ducts. Initial secretion of sweat is isotonic. This sweat flows through the ductal portion of the gland, in the dermis and epidermis of the skin, where it is modified by the reabsorption of solutes such as NaCl, lactate and bicarbonate, leaving hypotonic sweat (Quinton 2007). The final composition of the fluids excreted by the sweat glands is modified by the concerted activities of ENaC and CFTR, which are enriched in the apical membrane of the sweat duct (Reddy et al. 1999). Both ENaC and CFTR operate under wide changes in luminal pH conditions and are inhibited by acidic pH in the cytosol of the sweat duct cells (Reddy et al. 1998, 2008). We speculate that CA12 exerts its effect by contributing to the maintenance of close-to-normal intracellular pH in the sweat duct epithelial cells, similar to its suggested mode of action in tumor cells (Ivanov et al. 2001; Chiche et al. 2009). CA12 is an exclusive carbonic anhydrase in this organ (Ivanov et al. 2001), other membrane-bound carbonic anhydrases are not expressed in the sweat gland (Mastrolorenzo et al. 2003) and, thus, cannot compensate for its activity. Our finding highlights the importance of pH on the salt concentration in sweat and its significance on sodium homeostasis.
Addendum
After submission of this manuscript, a study by Feldshtein et al. (Am J Hum Genet. 87:713–720, Epub date Oct 28, 2010.) reported the same homozygous mutation in CA12 in members of a different Israeli-Bedouin clan living in a different village. In vitro studies of the mutated enzyme showed that relative to the wild-type enzyme, its activity is only 70% and it is profoundly inhibited by chloride ions.
Web resources
ClustalW. http://www.ebi.ac.uk/Tools/clustalw2/index.html
FoldX. http://foldx.crg.es/
KinSNP. http://bioinfo.bgu.ac.il/bsu/software/KinSNP/
Online Mendelian inheritance in man (OMIM) http://www.ncbi.nlm.nih.gov/OMIM
PedTool. http://bioinfo.cs.technion.ac.il/superlink/
Primer3. http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi
Tandem repeat. http://tandem.bu.edu/trf/trf.basic.submit.html
The human genome variation database. (SNP: http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?locusId=771)
References
Amir el-AD, Bartal O, Morad E, Nagar T, Sheynin J, Parvari R, Chalifa-Caspi V (2010) KinSNP software for homozygosity mapping of disease genes using SNP microarrays. Hum Genomics 4:394–401
Anand SK, Froberg L, Northway JD, Weinberger M, Wright JC (1976) Pseudohypoaldosteronism due to sweat gland dysfunction. Pediatr Res 10:677–682
Ballestero Y, Hernandez MI, Rojo P, Manzanares J, Nebreda V, Carbajosa H, Infante E, Baro M (2006) Hyponatremic dehydration as a presentation of cystic fibrosis. Pediatr Emerg Care 22:725–727
Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580
Brown D, Zhu XL, Sly WS (1990) Localization of membrane-associated models of CA IV and CA XIV. Carbonic anhydrase type IV in kidney epithelial cells. Proc Natl Acad Sci USA 87:7457–7461
Chang SS, Grunder S, Hanukoglu A, Rösler A, Mathew PM, Hanukoglu I, Schild L, Lu Y, Shimkets RA, Nelson-Williams C (1996) Mutations in subunits of the epithelial sodium channel cause salt wasting with hyperkalaemic acidosis, pseudohypoaldosteronism type 1. Nat Genet 12:248–253
Chiche J, Ilc K, Laferriere J, Trottier E, Dayan F, Mazure NM, Brahimi-Horn MC, Pouyssegur J (2009) Hypoxia-inducible carbonic anhydrase IX and XII promote tumor cell growth by counteracting acidosis through the regulation of the intracellular pH. Cancer Res 69:358–368
Choi MJ, Ziyadeh FN (2008) The utility of the transtubular potassium gradient in the evaluation of hyperkalemia. J Am Soc Nephrol 19:424–426
Edelheit O, Hanukoglu I, Gizewska M, Kandemir N, Tenenbaum-Rakover Y, Yurdakok M, Zajaczek S, Hanukoglu A (2005) Novel mutations in epithelial sodium channel (ENaC) subunit genes and phenotypic expression of multisystem pseudohypoaldosteronism. Clinil Endocrinol 62:547–553
Edelheit O, Hanukoglu I, Shriki Y, Tfilin M, Dascal N, Gillis D, Hanukoglu A (2010) Truncated beta epithelial sodium channel (ENaC) subunits responsible for multi-system pseudohypoaldosteronism support partial activity of ENaC. J Steroid Biochem Mol Biol 119:84–88
Epaud R, Girodon E, Corvol H, Niel F, Guigonis V, Clement A, Feldmann D, Bensman A, Ulinski T (2005) Mild cystic fibrosis revealed by persistent hyponatremia during the French 2003 heat wave, associated with the S1455X C-terminus CFTR mutation. Clin Genet 68:552–553
Feldshtein M, Elkrinawi S, Yerushalmi B, Marcus B, Vullo D, Romi H, Ofir R, Landau D, Sivan S, Supuran CT, Birk OS (2010) Hyperchlorhidrosis caused by homozygous mutation in CA12, encoding carbonic anhydrase XII. Am J Hum Genet 87:713–720
Geller DS, Zhang J, Zennaro MC, Vallo-Boado A, Rodriguez-Soriano J, Furu L, Haws R, Metzger D, Botelho B, Karaviti L (2006) Autosomal dominant pseudohypoaldosteronism type 1: mechanisms, evidence for neonatal lethality, and phenotypic expression in adults. J Am Soc Nephrol 17:1429–1436
Halmi P, Lehtonen J, Waheed A, Sly WS, Parkkila S (2004) Expression of hypoxia-inducible, membrane-bound carbonic anhydrase isozyme XII in mouse tissues. Anat Rec A Discov Mol Cell Evol Biol 277:171–177
Hanukoglu A (1991) Type I pseudohypoaldosteronism includes two clinically and genetically distinct entities with either renal or multiple target organ defects. J Clin Endocrinol Metab 73:936–944
Hanukoglu A, Edelheit O, Shriki Y, Gizewska M, Dascal N, Hanukoglu I (2008) Renin–aldosterone response, urinary Na/K ratio and growth in pseudohypoaldosteronism patients with mutations in epithelial sodium channel (ENaC) subunit genes. J Steroid Biochem Mol Biol 111:268–274
Hermo L, Chong DL, Moffatt P, Sly WS, Waheed A, Smith CE (2005) Region- and cell-specific differences in the distribution of carbonic anhydrases II, III, XII, and XIV in the adult rat epididymis. J Histochem Cytochem 53:699–713
Hynninen P, Hamalainen JM, Pastorekova S, Pastorek J, Waheed A, Sly WS, Tomas E, Kirkinen P, Parkkila S (2004) Transmembrane carbonic anhydrase isozymes IX and XII in the female mouse reproductive organs. Reprod Biol Endocrinol 2:73
Ivanov S, Liao SY, Ivanova A, Danilkovitch-Miagkova A, Tarasova N, Weirich G, Merrill MJ, Proescholdt MA, Oldfield EH, Lee J (2001) Expression of hypoxia-inducible cell-surface transmembrane carbonic anhydrases in human cancer. Am J Pathol 158:905–919
Karhumaa P, Parkkila S, Tureci O, Waheed A, Grubb JH, Shah G, Parkkila AK, Kaunisto K, Tapanainen J, Sly WS (2000) Identification of carbonic anhydrase XII as the membrane isozyme expressed in the normal human endometrial epithelium. Mol Hum Reprod 6:68–74
Kiefer LL, Paterno SA, Fierke CA (1995) Hydrogen bond network in the metal binding site of carbonic anhydrase enhances zinc affinity and catalytic efficiency. J Am Chem Soc 117:6831–6837
Kivela A, Parkkila S, Saarnio J, Karttunen TJ, Kivela J, Parkkila AK, Waheed A, Sly WS, Grubb JH, Shah G (2000) Expression of a novel transmembrane carbonic anhydrase isozyme XII in normal human gut and colorectal tumors. Am J Pathol 156:577–584
Kivelä AJ, Parkkila S, Saarnio J, Karttunen TJ, Kivelä J, Parkkila AK, Pastoreková S, Pastorek J, Waheed A, Sly WS (2000) Expression of transmembrane carbonic anhydrase isoenzymes IX and XII in normal human pancreas and pancreatic tumours. Histochem Cell Biol 114:197–204
Kyllonen MS, Parkkila S, Rajaniemi H, Waheed A, Grubb JH, Shah GN, Sly WS, Kaunisto K (2003) Localization of carbonic anhydrase XII to the basolateral membrane of H-secreting cells of mouse and rat kidney. J Histochem Cytochem 51:1217–1224
Lesburg CA, Christianson DW (1995) X-ray crystallographic studies of engineered hydrogen bond networks in a protein–zinc binding site. J Am Chem Soc 117:6838–6844
Liao SY, Lerman M, Stanbridge E (2009) Expression of transmembrane carbonic anhydrases, CAIX and CAXII, in human development. BMC Dev Biol 9:22
Mastrolorenzo A, Zuccati G, Massi D, Gabrielli MG, Casini A, Scozzafava A, Supuran CT (2003) Immunohistochemical study of carbonic anhydrase isozymes in human skin. Eur J Dermatol. 13:440–444
O’Sullivan BP, Freedman SD (2009) Cystic fibrosis. Lancet 373:1891–1904
Parkkila S, Parkkila AK, Saarnio J, Kivela J, Karttunen TJ, Kaunisto K, Waheed A, Sly WS, Tureci O, Virtanen I (2000) Expression of the membrane-associated carbonic anhydrase isozyme XII in the human kidney and renal tumors. J Histochem Cytochem 48:1601–1608
Parvari R, Hershkovitz E, Kanis A, Gorodischer R, Shalitin S, Sheffield VC, Carmi R (1998) Homozygosity and linkage-disequilibrium mapping of the syndrome of congenital hypoparathyroidism, growth and mental retardation, and dysmorphism to a 1-cM interval on chromosome 1q42–43. Am J Hum Genet 63:163–169
Pey AL, Stricher F, Serrano L, Martinez A (2007) Predicted effects of missense mutations on native-state stability account for phenotypic outcome in phenylketonuria, a paradigm of misfolding diseases. Am J Hum Genet 81:1006–1024
Priou-Guesdon M, Malinge MC, Augusto JF, Rodien P, Subra JF, Bonneau D, Rohmer V (2010) Hypochloremia and hyponatremia as the initial presentation of cystic fibrosis in three adults. Ann Endocrinol (Paris) 71:46–50
Quinton PM (2007) Cystic fibrosis: lessons from the sweat gland. Physiology 22:212–225
Reddy MM, Kopito RR, Quinton PM (1998) Cytosolic pH regulates GCl through control of phosphorylation states of CFTR. Am J Physiol 275:C1040–C1047
Reddy MM, Light MJ, Quinton PM (1999) Activation of the epithelial Na+ channel (ENaC) requires CFTR Cl-channel function. Nature 402:301–304
Reddy MM, Wang XF, Quinton PM (2008) Effect of cytosolic pH on epithelial Na channel in normal and cystic fibrosis sweat ducts. J Membr Biol 225:1–11
Schwartz GJ, Kittelberger AM, Barnhart DA, Vijayakumar S (2000) Carbonic anhydrase IV is expressed in H(+)-secreting cells of rabbit kidney. Am J Physiol (Renal Physiol) 278:F894–F904
Stowasser M, Huggard PR, Rossetti TR, Bachmann AW, Gordon RD (1999) Biochemical evidence of aldosterone overproduction and abnormal regulation in normotensive individuals with familial hyperaldosteronism type I. J Clin Endocrinol Metab 84:4031–4036
Supuran CT (2008) Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat Rev Drug Disc 7:168–181
Thompson CJ (2010) Hyponatraemia: new associations and new treatments. Eur J Endocrinol 162:S1–S3
Upadhyay A, Jaber BL, Madias NE (2006) Incidence and prevalence of hyponatremia. Am J Med 119:S30–S35
Whittington DA, Waheed A, Ulmasov B, Shah GN, Grubb JH, Sly WS, Christianson DW (2001) Crystal structure of the dimeric extracellular domain of human carbonic anhydrase XII, a bitopic membrane protein overexpressed in certain cancer tumor cells. Proc Natl Acad Sci USA 98:9545–9550
Acknowledgments
Grants or fellowships supporting the writing of the paper: the research was supported by grant 300000-5152 of the Chief Scientist of the Israeli Ministry of Health to RP and EH, and by a grant of the Chief Scientist of the Israel Ministry of Health to AH and IH.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
The authors E. Muhammad, N. Leventhal, E. Hershkovitz and R. Parvari contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
439_2010_930_MOESM1_ESM.tif
Family pedigree and haplotype analysis. The haplotype analysis based on microsatellite markers from 15q21.3-q23 revealed a founder haplotype (gray bar) for which the patients are homozygous in the critical region harboring the CA12 gene. The numbers of the patients correspond to those presented in Table 1.Patient II-3 was identified by analysis of the mutation. Filled symbols represent homozygote for the mutation and half filled symbols heterozygotes. The two lines indicate the crossing events delimiting the homozygous interval shared by all patients (TIF Image 681 KB)
439_2010_930_MOESM2_ESM.tif
Identification of the Glu143Lys Mutation in the CA12 gene at the Functionally Conserved Glu143 Amino Acid. a. Sequence of the genomic DNA corresponding c.583G>A mutation resulting in Glu143Lys. Patients were homozygous for the mutation, parents and siblings carrying the founder haplotype were heterozygous, and the healthy siblings without the founder haplotype were normal.b. Restriction analysis with Eco 130I enables to identify the mutation by cleavage of the mutated sequence. Normal- 600 bp PCR product of control. Homozygote presents 254bp and 346bp products of a patient. Heterozygote-presents 600bp, 254bp and 346bp products of a parent. At the left is marker X of Roche.c. Functional conservation of the replaced amino acid Glu143 mutated to Lys. The multiple sequence alignment, carried out by ClustalW represents the alignment of CA 12 to the 16 α-CA isozymes.The numbering relates to the positions of CAII, the best characterized enzyme, according to Whittington et al. (2001). Asterisk (*), identical; colon (:), strictly conserved (TIF Image 1,679 KB)
Rights and permissions
About this article
Cite this article
Muhammad, E., Leventhal, N., Parvari, G. et al. Autosomal recessive hyponatremia due to isolated salt wasting in sweat associated with a mutation in the active site of Carbonic Anhydrase 12 . Hum Genet 129, 397–405 (2011). https://doi.org/10.1007/s00439-010-0930-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-010-0930-4