Abstract
Admixed populations present unique opportunities to discover the genetic factors underlying many multifactorial diseases. The geographical position and complex history of South Africa has led to the establishment of the unique admixed population known as the South African Coloured. Not much is known about the genetic make-up of this population, and the historical record is patchy. We genotyped 959 individuals from the Western Cape area, self-identified as belonging to this population, using the Affymetrix 500k genotyping platform. This resulted in nearly 75,000 autosomal SNPs that could be compared with populations represented in the International HapMap Project and the Human Genome Diversity Project. Analysis by means of both the admixture and linkage models in STRUCTURE revealed that the major ancestral components of this population are predominantly Khoesan (32–43%), Bantu-speaking Africans (20–36%), European (21–28%) and a smaller Asian contribution (9–11%), depending on the model used. This is consistent with historical data. While of great historical and genealogical interest, this information is also essential for future admixture mapping of disease genes in this population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
South Africa is characterized by extensive population diversity with groups originating from African (79%), Asian (2.5%) and European (9.6%) populations (http://www.statssa.gov.za). This population diversity is the result of both a multi-faceted colonization history (Mountain 2003) and South Africa’s location with respect to major trade routes from the fifteenth to the nineteenth century. The contribution of these previously continentally divided population groups from Europe, Asia and the rest of Africa, to South Africa’s diversity led to the establishment of a mixed ancestry population, predominantly in the Western Cape, known today officially as the South African Coloured population (SAC) (Adhikari 2005; Nurse et al. 1985; Van der Ross 1993). This population, which currently comprises approximately 9% of the South African population and 54% of the population of the Western Cape Province, has a complex genetic history, influenced by both the colonization history of South Africa and its historical legislature.
The South African Coloureds have their roots in the indigenous Khoesan [denoting Khoekhoe and San (Boonzaaier et al. 1996; Elphick 1985; Mountain 2003)] native to the Western Cape at the time of first colonization by European settlers of the Dutch East India Company (VOC) in 1652 (Mountain 2003; Shell 1994). After the establishment of a refreshment station at the Cape of Good Hope, now Cape Town, the VOC brought in small numbers of political exiles from Indonesia and Malaysia (Mountain 2004), and slaves from the Indian subcontinent (25.9%), the east coast of Africa (26.4%), Madagascar (25.1%) and Indonesia (22.7%) (Nurse et al. 1985; Shell 1994). These figures were calculated from the records of the slave trade (Shell 1994). The active trade in slaves began in 1658 and continued until the banning of the seaborne slave trade in 1806, with the last recorded illegal imports in 1822 (Shell 1994). In the early 1700s, the slave population in the Cape regularly outnumbered the European settlers (Mountain 2003), and men virtually always outnumbered women in both the slave and free populations (Shell 1994).
The indigenous Khoekhoe were not enslaved, but frequently served as indentured labourers or serfs on the farms (Mountain 2003; Shell 1994). A small, but significant number of women of Khoekhoe or of slave descent and their children were integrated into the colonial household, often by marriage (Mountain 2003; Shell 1994). Mixed marriages, usually between European men and women who were either Khoekhoe, manumitted (freed) slaves or of mixed parentage (Keegan 1996), and between Khoekhoe and slave (Mountain 2003) were socially acceptable in early Cape society. However, in the majority of cases, and particularly after 1700, the progeny of such mixed marriages and liaisons were assimilated into the growing group known as the “Cape Coloureds” (Keegan 1996; Mountain 2003; Nurse et al. 1985), a term used since the mid-nineteenth century (Keegan 1996). These unions were more common in the farming areas, but also occurred in the towns (Mountain 2003; Shell 1994). By the late 1700s, race-based restrictions were common, and these were formalised under the British administration from 1806 (Mountain 2003), when class was more easily overcome in society than race and ancestry (Keegan 1996).
The cohesion of the SAC population was further facilitated by both the establishment of early mission stations (from 1738) amongst Coloured and Khoekhoe populations (Mountain 2004), and by legislation. After emancipation by the British administration (1834–1838), large numbers of ex-slaves and other indigent people settled at mission stations (Mountain 2004), some of which formed the nucleus of a “Coloured group area” (Boonzaaier et al. 1996; Mountain 2003). Many of the Khoesan at these mission stations had European and/or African (particularly Xhosa) ancestry (Keegan 1996). The formalization of the racial order in society began in the late 1700s. From 1910, and particularly 1948–1994, the apartheid regime introduced legislature that outlawed inter-racial marriage and prescribed areas of residence (http://www.sahistory.org.za/pages/chronology/special-chrono/governance/apartheid-legislation.html). This separation of ethnic groups ensured further cohesion of the already established highly admixed SAC population in the Western Cape, the traditional centre of concentration of the Coloured people (Adhikari 2005; Cilliers 1963). The term “ethnic group” is used here as according to Barth (1969). Briefly, the term denotes a culturally defined group which identifies itself and is identified by others as constituting a distinguishable category.
The majority of people who self-identify as Coloured are Afrikaans speaking. According to the 2001 census, 81.0% of the Coloured people in the Western Cape were Afrikaans speaking, and 18.6% English speaking, while in our study area of Ravensmead/Uitsig these figures were 90.1 and 9.3% respectively. The population of Ravensmead/Uitsig is 91% Christian, and only 1.5% Muslim (2001 SA census), which raises an important distinction with another population group in South Africa known as the Cape Malays. The latter have their origins in the political exiles brought from the Dutch East Indies (mainly Indonesia) in the 1700s (Mountain 2004). They brought the religion of Islam to South Africa, which served as a unifying force in the community and may have created a genetic subgroup, which is not the focus of our study. The term “Malay”, or alternatively “Cape Muslim” is used by members of the group to denote their affiliation with Islam. The Malay from a minority group (10.3% of the SAC in the Western Cape) which has not been incorporated into the core structure of the South African Coloured people (Nurse et al. 1985).
The SAC population, characterized by extensive admixture of multiple population sources, provides a unique opportunity to investigate genomic patterns of population admixture. Given that Africa (Conrad et al. 2006; Tishkoff and Williams 2002; Tishkoff and Kidd 2004), and in particular South Africa (Tishkoff et al. 2009), has the most diverse human populations it is imperative that large-scale genome studies of both human demographic history and disease association are carried out using African samples (Campbell and Tishkoff 2008; Tishkoff et al. 2009). Knowledge of the nature of admixture in a population is also important when considering disease association studies on a population such as the SAC (Babb et al. 2007; Barreiro et al. 2006; Cooke et al. 2008; Hoal et al. 2004; Möller et al. 2007, 2009; Rossouw et al. 2003), and comparing these associations with the results found in other ethnic groups. We present the results of a large genome-wide analysis consisting of 959 individuals from the SAC group, genotyped with a panel of 500,000 single-nucleotide polymorphism (SNP) markers, of which nearly 75,000 markers are shared with both the International HapMap Consortium (Frazer et al. 2007; The International HapMap Consortium 2005) and the Human Genome Diversity Project (HGDP) (Cann et al. 2002). This is the first high-resolution SNP study of a large and representative sample of this unique population. Although understanding the demographic history of the SAC population is of interest in its own right, we anticipate that this highly diverse admixed population will provide opportunities for the identification of genes associated with complex diseases in this population and its ancestral source populations. Characterizing the pattern of genetic variation in this study population will provide valuable baseline data for subsequent analysis of disease association.
Materials and methods
Study site and subjects
Study subjects, self-identified as SAC, were enrolled from Ravensmead and Uitsig, two suburbs of Cape Town, which are contiguous and which we subsequently found to be genetically indistinguishable (F st = 0.001). In 1962, these suburbs were declared an area for habitation by Coloureds only, under the Group Areas Act of the apartheid government. Although this act was repealed in 1991, 98% of people in this suburb self-identified themselves as “Coloured” in the 2001 South African census. Informed consent was obtained from all study participants. The study was approved by the Institutional Review Board of Stellenbosch University, Tygerberg, South Africa. Blood was taken and DNA extracted by standard methods.
Sampling, genotyping and genotype calling
All samples and CEU (Utah residents with ancestry from Northern and Western Europe) controls from the International HapMap Project (Frazer et al. 2007; The International HapMap Consortium 2005) were genotyped using the Affymetrix 500k genotyping platform. SNP genotypes were called using the Affymetrix Power Tools pipeline (V1.10.0). First, samples that had a reported NSP/STY concordance rate of <90% were discarded. The dynamic model (DM) algorithm’s call rate was used as an initial quality control measure. CEL files with a call rate of 93% or higher were selected, and used to train probe-specific models using the BRLMM algorithm (Affymetrix 2006). These models were then saved, and used to call all samples with STY and NSP call rates of 70% or higher. Genotype calling performance was determined by measuring concordance of the included HapMap cell line samples with the genotypes of these individuals from the HapMap project (The International HapMap Consortium 2005), and was found to be >99%. Furthermore, four duplicate SAC samples were included in the SNP genotyping experiment, which allowed for validation of the genotype-calling algorithm on SAC samples. Genotype concordance of these SAC samples was >97%. In addition to data generated in this study, we obtained genome-wide SNP data from two additional public data sources: the International HapMap Project (Frazer et al. 2007; The International HapMap Consortium 2005) (http://www.hapmap.org) and the Human Genome Diversity Project (Cann et al. 2002) (HGDP; http://hagsc.org/hgdp/files.html). Populations were chosen from these public data sources to represent putative ancestral populations that may have contributed through admixture to the SAC population. The populations chosen were representative of four major groups, namely (1) European (2) non-Khoesan African (including East African, Bantu and Pygmy populations) (3) Khoesan and (4) Asian (Table 1). We reduced the SNPs genotyped in this study to a subset (n = 74,889) shared between SAC and the public data sources (Table 1).
Population structure
Population structure analyses were performed to characterize the genetic contributions to the SAC population. We used STRUCTURE (Falush et al. 2003; Pritchard et al. 2000), which identifies population structure without prior assignment of individuals to populations. STRUCTURE has an upper limit on the number of SNPs that can be analysed, and assumes both Hardy–Weinberg equilibrium and complete linkage equilibrium between adjacent markers (Falush et al. 2003; Pritchard et al. 2000). The selection of highly informative markers reduces the number of genotypes required for the accurate inference of ancestry. Therefore, we selected SNPs from the set of shared markers (n = 74,889) that were ancestry informative for the putative contributions to the SAC, and that were putatively unlinked. We used Rosenberg’s Ancestry Informative Markers (AIMs) selection method (Rosenberg et al. 2003), taking potential linkage into account by selecting AIMs separated by a physical distance of at least 1 MB. Alternative marker selection strategies including random selection, random selection accounting for linkage disequilibrium and AIMs not accounting for linkage disequilibrium were also tested (Table S1).
We used the admixture model with correlated allele frequencies to investigate the number of populations evident in the combined SAC-HapMap-HGDP dataset (Table 1). Convergence of MCMC chains was assessed with five independent runs (burn-in = 1,000, chain length = 2,500) for each K between 1 and 8. The number of populations (K) was estimated as the number that maximized the probability of the data, and minimized the variance in this probability over successive iterations (Pritchard et al. 2000). For each SNP subset, we estimated the proportions of inferred ancestry for each individual using the optimal number of ancestral populations (K), and plotted these proportions using DISTRUCT (Rosenberg 2004). A potential limitation in estimating proportions of ancestry for SAC in these analyses is that the standard implementation of the admixture model used does not account for linkage disequilibrium due to admixture (Falush et al. 2003), known to be a feature of our study population (Nurse et al. 1985). An alternative model, accounting for linkage disequilibrium due to admixture provides more accurate estimates of statistical uncertainty in admixed populations, but has runtimes that scale exponentially with the number of ancestral subpopulations (Falush et al. 2003). Therefore, we also estimated ancestral proportions using the linkage model in STRUCTURE, but only for the optimal number of ancestral subpopulations identified in the previous analyses. In this case, we used a larger sample of 10,000 SNPs, since linkage due to admixture is incorporated into the model, but we still maintained a physical distance of at least 10 Kb between adjacent SNPs to limit the effect of background linkage disequilibrium. We performed principal component analysis using SMARTPCA in the EIGENSOFT package (Patterson et al. 2006; Price et al. 2006) and included all SNP markers shared between the populations analyzed (n = 74,889). Finally, we used FRAPPE (Tang et al. 2005; Li et al. 2008), which models background linkage equilibrium and thus allows for the inclusion of physically linked SNPs. All 74,889 SNPs were used in a FRAPPE analysis that comprised 10,000 EM iterations with a convergence threshold of 10,000.
Results
Population structure
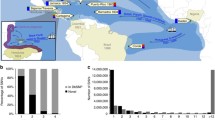
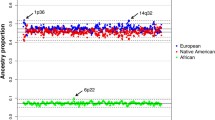
In the STRUCTURE analyses, including both SAC data and data from potential ancestral populations derived from public sources; the number of populations was estimated as between 4 and 7 (Fig. S1). Not accounting for background linkage disequilibrium between SNPs, by including putatively linked markers (Fig. S1 A, C), resulted in higher estimates of K. The inferred major contributions to the South African Coloured population were consistent (Figs. 1, S2, S3, S4), although the estimate for the number of ancestral populations varied with each subset of SNPs. Consistent with historical data, the four major inferred contributions to SAC were Khoesan Africans, non-Khoesan Africans, Europeans and a smaller Asian contribution. Of these, the Khoesan contribution is the largest under both the linkage model in STRUCTURE and the FRAPPE analyses, followed by European, African and Asian (Fig. 2; Tables 2, S2). A large contribution from Khoesan was inferred despite the fact that data for only a small number of Khoesan individuals (n = 5) were present in the publicly available datasets. Although the Khoesan sample size is small, this is compensated for by the relatively large proportion of Khoesan ancestry in SAC individuals, and by the large sample size of SAC. The STRUCTURE method estimates ancestral population allele frequencies and further allows for individual’s genomes to be drawn from multiple ancestral populations, thus accounting for admixture. Therefore, the estimation of ancestral population allele frequencies is not based on only a small sample of Khoesan, but on the entire sample of SAC that have Khoesan ancestry. Whilst the small sample of Khoesan assists in the clustering and allele frequency estimation, it is most useful in identifying which of ancestral populations identified using STRUCTURE are Khoesan (Fig. 1). Indeed, a STRUCTURE analysis without putative parental populations and only using SAC individuals reveals three of the four ancestral contributions to SAC (results not shown). It is likely that the minor Asian contribution is difficult to detect without pure Asian samples. Estimates of Khoesan ancestry proportions in SAC, obtained from the STRUCTURE model which takes admixture linkage disequilibrium into account, were lower than from the admixture model without accounting for admixture linkage disequilibrium (Fig. 2; Table 2). FRAPPE, which uses all loci and accounts for background linkage equilibrium, provided estimates consistent with the admixture model in STRUCTURE. Therefore, differences between the admixture and linkage models in STRUCTURE may be due to the increased computational complexity of the latter.
Proportion of each individual’s ancestry for the number of ancestral populations from K = 2 to the estimated number of ancestral populations with greatest probability (Fig. S2). Plots shown are for unlinked Ancestry Informative Markers and admixture model. Plots for additional datasets/models are available as Supplementary material (Figs. S2, S3, S4, S5). Population labels are as in Table 1
Mean, range and 95% confidence limits (notches) on estimated proportions of ancestry for SAC individuals using either an admixture or linkage model. This figure is based on inclusion of all ancestral populations in Table 1
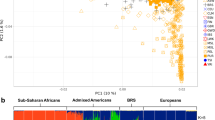
Principal component analyses showed SAC spanning the variation between Africans and non-Africans along the first pair of eigenvectors (Fig. 4). The PCA results suggest that the ancestral Asian population that contributed to SAC is more closely related to the contemporary Gujarati Indian population, than to the Chinese (CHB) and Japanese (JPT) populations from HapMap 3 (Fig. 4), as has been shown previously (Tishkoff et al. 2009).
The 959 individuals investigated in our study have a greater proportion of Khoesan ancestry and lower proportion of both European and Indian ancestry than the 39 individuals genotyped by Tishkoff et al. (2009) who showed approximately equal ancestries of Khoesan, European, Black African and Indian (19–25%), with 8% attributed to East Asian. It is possible that their group of 39 contained a proportion of people from the Cape Malay group, who may have a genetic make-up higher in Indian and lower in Khoesan ancestry, due to greater Indonesian or Malaysian ancestry. The samples used by Tishkoff et al. (2009) were collected from volunteers and blood donors residing (some temporarily) in the Western Cape (MJ Kotze, personal communication), and not from a specific area. Individuals from the SAC group sampled in other areas of South Africa could have a different genetic make-up. An early study of blood group gene frequencies in Cape Town found similar ancestral contributions from European, Black and Asian, but the criteria for inclusion were not clear (Botha 1972).
Discussion
This is the first genome-wide analysis of a large, well-defined set of individuals from the SAC population. Our results illustrate the very high degree of admixture in the SAC population, comprising input from mainly four geographically distant populations. We have genotyped 959 individuals from the SAC population, and selected almost 75,000 markers for population structure analyses. The results that we inferred using STRUCTURE, which suggest the SAC population group to have four major ancestral components, are consistent with the historical record. As expected, Khoesan, European, African and Asian (Indian) populations have contributed to SAC, the proportions of which are dependent on the statistical model used in inference (Fig. 2; Table S2). Differences between the admixture and linkage model are to be expected, since each accounts for different components of linkage disequilibrium. The admixture model ignores linkage disequilibrium along chromosomes as a result of admixture, whereas the linkage model does not (Falush et al. 2003), and thus the latter is a better approximation of the population history of SAC. Nonetheless, the inferred ancestry proportions indicate a substantial contribution from the Khoesan, and considerable variation in ancestry proportions between individuals (Figs. 1, 3). The degree of Khoesan ancestry reflects the role of indigenous Khoesan in the early establishment of the SAC population (Mountain 2003). It could be argued that the rather small Khoesan sample size contributes to uncertainty with respect to estimating ancestral proportions. However, these results are consistent with an independent study with a slightly larger population of Khoesan (Tishkoff et al. 2009). A recent report by Quintana-Murci et al. (2010) comparing maternal and paternal contributions to the SAC has put the Khoesan contribution at over 70%, and about 40%, respectively.
Some authors have proposed that the Khoesan people in South Africa are becoming extinct (Mountain 2003). The San in particular endured bouts of genocide from all other groups (Mountain 2003; Shell 1994), and the Khoekhoe society had collapsed completely before 1713 (Elphick 1985), the time of a devastating smallpox epidemic (Nurse et al. 1985). Although many members of the Khoesan existed on the fringes of colonial society (Mountain 2003), many others, particularly the women, were part of the household of the pioneer farmers, in a patriarchal societal system that had elements of slavery, indentured labour and authoritarian family life (Shell 1994). Often Khoekhoe men were bonded labourers on the farms (Keegan 1996; Shell 1994) and integrated into European colonial society (Elphick 1985). The near extinction of the Khoesan, however, is not apparent from our results; given that some SAC individuals harbour large proportions of Khoesan ancestry (Figs. 1, 2), and assuming that the HGDP Khoesan population is a sufficiently pure source of ancestral Khoesan diversity.
In addition to the strong Khoesan contribution to SAC, a large proportion of their ancestry is derived from non-Khoesan Africans (Fig. 1), in particular Bantu-speaking populations. The East African contribution expected was not detected, probably since the populations used here are Bantu-speaking (LWK), admixed with European (MKK as evident in Figs. 1, 4), or since the SNPs used provide insufficient resolution to resolve this contribution. Furthermore, many imported male slaves did not reproduce (Shell 1994), making this expected contribution minor in comparison to southern Bantu-speaking individuals. Although we did not have samples for southern African Bantu, these groups are themselves admixed with the Khoesan (Nurse et al. 1985; Thorp 2000; Tishkoff et al. 2009), which is also evidenced by click consonants in the Xhosa language. Khoesan ancestry was, therefore, assumed to be derived primarily from the Khoesan populations, and in addition from the Bantu-speaking ancestors who also had Khoesan ancestry. Furthermore, substantial input from the European settlers (mainly Dutch, German, British and French), and a smaller contribution from Asia, is evident in the SAC (Fig. 1). This Asian contribution is consistent with 26% of imported slaves originating in East India, mainly Bengal (Shell 1994), and the apparent shared ancestry between populations from East Asia and Gujarati Indians from the Indian subcontinent (Fig. 1). The use of HapMap Gujarati Indians as a proxy for the Indian populations that were the actual ancestral populations in the SAC is supported by the genetic homogeneity of Gujarati and Bengali populations (Tishkoff et al. 2009). One analysis we performed, which included a random subset of SNPs, did detect the Gujarati Indian contribution to SAC (Fig. S2). However, this result may be influenced by the larger proportion of linked SNPs in that analysis, and the inability of STRUCTURE to account for background linkage disequilibrium. Nonetheless, the Indian contribution to SAC is supported by PCA analysis of all 75,000 markers (Fig. 4). Low levels of ancestry from East Asia (CHB/JPT in HapMap) may be ascribed partly to the Chinese who formed part of the “free blacks” (Keegan 1996), a group forming 9% of the Cape Town population by 1821 (Shell 1994). Free blacks were free persons not of European origin, and comprised manumitted slaves, a few political exiles, and several hundred Chinese convicts (Mountain 2004; Shell 1994). Chinese, Indian and Cape-born slaves have also been found to contribute to the Afrikaner population, apart from the predominant European component (Greeff 2007; Heese 1971).
The limitations of this study include the use of the Affymetrix 500k SNP chip, containing markers primarily designed for use in Europeans. This could have led to a strong ascertainment bias that may well have influenced the quantitative details of the analyses performed. The resolution of inferred ancestral contributions could certainly be improved with the addition of both more suitable ancestral population samples from Malaysia and Indonesia, an appropriate Bantu-speaking population, and with a larger sample of San, currently not publicly available. The genotype results concur with the historical record, but in addition provide quantitative information of the extent of the contribution of putative ancestral groups, not obtainable by conventional historical research. The contributions of the parent populations to the present-day SAC population were made at different periods in the past. Our estimates, therefore, reflect the result of their past contributions (after drift or variance in reproductive success), and not the absolute contribution of these different source populations.
In addition to the results presented here being of historical interest, the inferred ancestral contributions are highly relevant for mapping of disease genes. The SAC population in the Western Cape suffers from one of the highest incidence rates of tuberculosis (TB) ever recorded (Kritzinger et al. 2009), and knowledge of their population structure, and ancestry could be used to search for TB susceptibility loci through admixture mapping (McKeigue 1997; Montana and Pritchard 2004; Seldin 2007; Zhu et al. 2006, 2008). The SAC population in this study, of which we have genotyped approximately 3%, constitutes an excellent study population for the mapping of TB susceptibility genes, because their ancestral populations have substantially different rates of TB infection and disease (Stead et al. 1990). An essential requirement for admixture mapping is the elucidation of ancestral proportions of the populations involved. Thus, the results reported here will enable the investigation of the impact of admixture on TB susceptibility for example, and potentially explain the apparent high vulnerability of this population to disease. Furthermore, given the unique composition of this population, novel susceptibility alleles to complex diseases could be identified.
References
Adhikari M (2005) Not white enough, not black enough: racial identity in the South African Coloured community. Ohio University Press
Affymetrix (2006) BRLMM: an improved genotype calling method for the GeneChip Human Mapping 500K Array Set. Affymetrix
Babb C, van der Merwe L, Beyers N, Pheiffer C, Walzl G, Duncan K, van Helden P, Hoal EG (2007) Vitamin D receptor gene polymorphisms and sputum conversion time in pulmonary tuberculosis patients. Tuberculosis 87:295–302
Barreiro LB, Neyrolles O, Babb CL, Tailleux L, Quach H, McElreavey K, Helden PD, Hoal EG, Gicquel B, Quintana-Murci L (2006) Promoter variation in the DC-SIGN encoding gene CD209 is associated with tuberculosis. PLoS Med 3:e20
Barth F (1969) Ethnic groups and boundaries: the social organization of culture difference. Little, Brown and company, Boston
Boonzaaier E, Malherbe C, Smith A, Berens P (1996) The Cape Herders: a history of the Khoikhoi of Southern Africa. David Philip Publishers, Cape Town
Botha MC (1972) Blood group gene frequencies. An indication of the genetic constitution of population samples in Cape Town. Am J Roentgenol Radium Ther Nucl Med 115:Suppl 27
Campbell MC, Tishkoff SA (2008) African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet 9:403–433
Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A, Chen Z, Chu J, Carcassi C, Contu L, Du R, Excoffier L, Ferrara GB, Friedlaender JS, Groot H, Gurwitz D, Jenkins T, Herrera RJ, Huang X, Kidd J, Kidd KK, Langaney A, Lin AA, Mehdi SQ, Parham P, Piazza A, Pistillo MP, Qian Y, Shu Q, Xu J, Zhu S, Weber JL, Greely HT, Feldman MW, Thomas G, Dausset J, Cavalli-Sforza LL (2002) A human genome diversity cell line panel. Science 296:261–262
Cilliers SP (1963) The Coloureds of South Africa: a factual survey. Banier Publishers (Pty) Ltd, Cape Town
Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA, Pritchard JK (2006) A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet 38:1251–1260
Cooke GS, Campbell SJ, Bennett S, Lienhardt C, McAdam KP, Sirugo G, Sow O, Gustafson P, Mwangulu F, van HP, Fine P, Hoal EG, Hill AV (2008) Mapping of a novel susceptibility locus suggests a role for MC3R and CTSZ in human tuberculosis. Am J Respir Crit Care Med 178:203–207
Elphick R (1985) Khoikhoi and the founding of White South Africa. Ravan Press, Johannesburg
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Shen Y, Sun W, Wang H, Wang Y, Wang Y, Xiong X, Xu L, Waye MMY, Tsui SKW, Xue H, Wong JT-F, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallée C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PKH, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, Deloukas P, Bird CP, Delgado M, Dermitzakis ET, Gwilliam R, Hunt S, Morrison J, Powell D, Stranger BE, Whittaker P, Bentley DR, Daly MJ, de Bakker PIW, Barrett J, Chretien YR, Maller J, McCarroll S, Patterson N, Pe’er I, Price A, Purcell S, Richter DJ, Sabeti P, Saxena R, Schaffner SF, Sham PC, Varilly P, Altshuler D, Stein LD, Krishnan L, Smith AV, Tello-Ruiz MK, Thorisson GA, Chakravarti A, Chen PE, Cutler DJ, Kashuk CS, Lin S, Abecasis GR, Guan W, Li Y, Munro HM, Qin ZS, Thomas DJ, McVean G, Auton A, Bottolo L, Cardin N, Eyheramendy S, Freeman C, Marchini J, Myers S, Spencer C, Stephens M, Donnelly P, Cardon LR, Clarke G, Evans DM, Morris AP, Weir BS, Tsunoda T, Mullikin JC, Sherry ST, Feolo M, Skol A, Zhang H, Zeng C, Zhao H, Matsuda I, Fukushima Y, Macer DR, Suda E, Rotimi CN, Adebamowo CA, Ajayi I, Aniagwu T, Marshall PA, Nkwodimmah C, Royal CDM, Leppert MF, Dixon M, Peiffer A, Qiu R, Kent A, Kato K, Niikawa N, Adewole IF, Knoppers BM, Foster MW, Clayton EW, Watkin J, Gibbs RA, Belmont JW, Muzny D, Nazareth L, Sodergren E, Weinstock GM, Wheeler DA, Yakub I, Gabriel SB, Onofrio RC, Richter DJ, Ziaugra L, Birren BW, Daly MJ, Altshuler D, Wilson RK, Fulton LL, Rogers J, Burton J, Carter NP, Clee CM, Griffiths M, Jones MC, McLay K, Plumb RW, Ross MT, Sims SK, Willey DL, Chen Z, Han H, Kang L, Godbout M, Wallenburg JC, L’Archevêque P, Bellemare G, Saeki K, Wang H, An D, Fu H, Li Q, Wang Z, Wang R, Holden AL, Brooks LD, McEwen JE, Guyer MS, Wang VO, Peterson JL, Shi M, Spiegel J, Sung LM, Zacharia LF, Collins FS, Kennedy K, Jamieson R, Stewart J (2007) A second generation human haplotype map of over 3.1 million SNPs. Nature 449:851–861
Greeff JM (2007) Deconstructing Jaco: genetic heritage of an Afrikaner. Ann Hum Genet 71:674–688
Heese JA (1971) Die Herkoms van die Afrikaner, 1657–1867. A Balkema, Cape Town
Hoal EG, Lewis L-A, Jamieson SE, Tanzer F, Rossouw M, Victor T, Hillerman R, Beyers N, Blackwell JM, van Helden PD (2004) SLC11A1 (NRAMP1) but not SLC11A2 (NRAMP2) polymorphisms are associated with susceptibility to tuberculosis in a high-incidence community in South Africa. Int J Tuberc Lung Dis 8:1464–1471
Keegan T (1996) Colonial South Africa and the origins of the racial order. David Philip Publishers, Cape Town
Kritzinger FE, den BS, Verver S, Enarson DA, Lombard CJ, Borgdorff MW, Gie RP, Beyers N (2009) No decrease in annual risk of tuberculosis infection in endemic area in Cape Town, South Africa. Trop Med Int Health 14:136–142
Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM (2008) Worldwide human relationships inferred from genome-wide patterns of variation. Science 319:1100–1104
McKeigue PM (1997) Mapping genes underlying ethnic differences in disease risk by linkage disequilibrium in recently admixed populations. Am J Hum Genet 60:188–196
Möller M, Kwiatkowski R, Nebel A, van Helden PD, Hoal EG, Schreiber S (2007) Allelic variation in BTNL2 and susceptibility to tuberculosis in a South African population. Microbes Infect 9:522–528
Möller M, Nebel A, Valentonyte R, van Helden PD, Schreiber S, Hoal EG (2009) Investigation of chromosome 17 candidate genes in susceptibility to TB in a South African population. Tuberculosis (Edinb) 89:189–194
Montana G, Pritchard JK (2004) Statistical tests for admixture mapping with case–control and cases-only data. Am J Hum Genet 75:771–789
Mountain A (2003) The first people of the Cape, 1st edn. David Philips Publishers, Cape Town
Mountain A (2004) An unsung heritage. David Philip Publishers, Cape Town
Nurse GT, Weiner JS, Jenkins T (1985) The peoples of Southern Africa and their affinities. Clarendon Press, Oxford
Patterson N, Price AL, Reich D (2006) Population structure and eigenanalysis. PLoS Genet 2:e190
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Quintana-Murci L, Harmant C, Quach H, Balanovsky O, Zaporozhchenko V, Bormans C, van Helden PD, Hoal EG, Behar DM (2010) Strong maternal Khoisan contribution to the South African Coloured population: a case of gender-biased admixture. Am J Hum Genet 86:611–620
Rosenberg N (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Rosenberg NA, Li LM, Ward R, Pritchard JK (2003) Informativeness of genetic markers for inference of ancestry. Am J Hum Genet 73:1402–1422
Rossouw M, Nel HJ, Cooke GS, van Helden PD, Hoal EG (2003) Association between tuberculosis and a polymorphic NFkappaB binding site in the interferon gamma gene. Lancet 361:1871–1872
Seldin MF (2007) Admixture mapping as a tool in gene discovery. Curr Opin Genet Dev 17:177–181
Shell R (1994) Children of bondage. Witwatersrand University Press, Johannesburg
Stead WW, Senner JW, Reddick WT, Lofgren JP (1990) Racial differences in susceptibility to infection by Mycobacterium tuberculosis. N Engl J Med 322:422–427
Tang H, Peng J, Wang P, Risch NJ (2005) Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol 28:289–301
The International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437:1299–1320
Thorp CR (2000) Hunter-Gatherers and farmers: an enduring frontier in the Caledon Valley, South Africa. Publishers of British Archaeological Reports
Tishkoff SA, Kidd KK (2004) Implications of biogeography of human populations for ‘race’ and medicine. Nat Genet 36:S21–S27
Tishkoff SA, Williams SM (2002) Genetic analysis of African populations: human evolution and complex disease. Nat Rev Genet 3:611–621
Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo JM, Doumbo O, Ibrahim M, Juma AT, Kotze MJ, Lema G, Moore JH, Mortensen H, Nyambo TB, Omar SA, Powell K, Pretorius GS, Smith MW, Thera MA, Wambebe C, Weber JL, Williams SM (2009) The genetic structure and history of Africans and African Americans. Science 324:1035–1044
Van der Ross RE (1993) 100 questions about Coloured South Africans. UWC Printing Department, Cape Town
Zhu X, Zhang S, Tang H, Cooper R (2006) A classical likelihood based approach for admixture mapping using EM algorithm. Hum Genet 120:431–445
Zhu X, Tang H, Risch N (2008) Admixture mapping and the role of population structure for localizing disease genes. Adv Genet 60:547–569
Acknowledgments
We thank all the participants on this study, Alan Morris for helpful discussions, and The Meraka Institute (http://www.meraka.org.za/) for computational resources. We are grateful to Karen Small of Strategic Development Information and GIS, City of Cape Town for provision of demographic data. We thank GlaxoSmithKline for the donation of the GeneChip Human Mapping 500K Array Sets from Affymetrix. The experiments performed in this study comply with the current laws of South Africa.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
The collective term for people of mixed ancestry in southern Africa is “Coloured” and is recognized and used officially in South Africa. Whilst we acknowledge that in some cultures this term may have acquired a derogatory connotation, this is certainly not intended here.
Electronic supplementary material
Below is the link to the electronic supplementary material.
439_2010_836_MOESM1_ESM.tif
Figure S1 Estimates of the number of ancestral populations (K) for the SAC and combined HapMap and HGDP samples under an admixture model using the STRUCTURE software. The estimated probability of the data given the model is plotted against increasing K for each of the subsets of SNP data used (see Methods) (TIFF 6 kb)
439_2010_836_MOESM2_ESM.tif
Figure S2 Proportion of each individual’s ancestry for the number of ancestral populations from k = 2 to the estimated number of ancestral populations with greatest probability (Fig S1). Plots shown are for random linked SNPs (TIFF 600 kb)
439_2010_836_MOESM3_ESM.tif
Figure S3 Proportion of each individual’s ancestry for the number of ancestral populations from k = 2 to the estimated number of ancestral populations with greatest probability (Fig S1). Plots shown are for unlinked random SNPs (TIFF 256 kb)
439_2010_836_MOESM4_ESM.tif
Figure S4 Proportion of each individual’s ancestry for the number of ancestral populations from k = 2 to the estimated number of ancestral populations with greatest probability (Fig S1). Plots shown are for linked Ancestry Informative Markers (TIFF 356 kb)
439_2010_836_MOESM5_ESM.tif
Figure S5: Proportion of each individual’s ancestry derived using the linkage model in STRUCTURE for the optimal number of ancestral populations (K = 4) (TIFF 102 kb)
Rights and permissions
About this article
Cite this article
de Wit, E., Delport, W., Rugamika, C.E. et al. Genome-wide analysis of the structure of the South African Coloured Population in the Western Cape. Hum Genet 128, 145–153 (2010). https://doi.org/10.1007/s00439-010-0836-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-010-0836-1