Abstract
Genomic imprinting is an epigenetic process in which the copy of a gene inherited from one parent (maternal or paternal) is consistently silenced or expressed at a significantly lower level than the copy from the other parent. In an effort to begin a systematic genome-wide screen for imprinted genes, we assayed differential allelic expression (DAE) at 3,877 bi-allelic protein-coding sites located in 2,625 human genes in 67 unrelated individuals using genotyping microarrays. We used the presence of both over- and under-expression of the reference allele compared to the alternate allele to identify candidate-imprinted genes. We found 61 genes with at least twofold DAE plus “flipping” of the more highly expressed allele between reference and alternate across heterozygous samples. Sixteen flipping genes were genotyped and assayed for DAE in an independent data set of lymphoblastoid cell lines from two CEPH pedigrees. We confirmed that PEG10 is paternally expressed, identified one gene (ZNF331) with multiple lines of data indicating it is imprinted, and predicted several additional imprinting candidate genes. Our findings suggest that there are at most several hundred genes in the human genome that are universally imprinted. With samples of mRNA from appropriate tissues and a collection of informative cSNPs, a genome-wide search using this methodology could expand the list of genes that undergo genomic imprinting in a tissue- or temporal-specific manner.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Imprinted genes exhibit differential allelic expression (DAE) in a parent-of-origin dependent manner. For a paternal- or maternal-imprinted gene, the allele inherited from one parent is silenced or suppressed through epigenetic mechanisms, most commonly differential methylation of regulatory sequences (Reik and Walter 2001; Murphy and Jirtle 2003). For some imprinted genes the suppressed expression of one parental allele is observed in all tissues, while for other genes imprinting is observed only in a specific tissue or at a particular developmental stage (Morison et al. 2005; Monk et al. 2006). Aberrant imprinting has been shown to result in a number of diseases, including a variety of developmental syndromes of growth and behavior, such as Prader-Willi syndrome, as well as several cancers, including Wilm’s tumor (Murphy and Jirtle 2003).
Mounting evidence indicates that DAE is widespread in populations of humans and model mammals (Storey et al. 2007; Gibson and Weir 2005; Stranger et al. 2005). To control environmental interactions and other confounding factors, several studies have examined DAE using individuals heterozygous for coding SNPs (cSNPs), where the relative expression of the two copies of a gene can be evaluated in the same cellular sample (Yan et al. 2002; Pastinen et al. 2004; Lo et al. 2003; Pant et al. 2006). A variety of different technologies were used in these studies, which also varied widely in the number of genes assayed. Nonetheless, they consistently found a large proportion of the assayed genes (20–50%) displaying DAE. A substantial proportion of this DAE appears to be due to cis-acting polymorphism resulting in the same allele being expressed at a higher level in each heterozygote (Pant et al. 2006; Spielman et al. 2007; Stranger et al. 2005). Most validation studies of such cis-effects have been based on identification of genetic variation for effects that show Mendelian inheritance and/or association with a phenotype (Tao et al. 2006). An expression pattern observed in these studies that is not compatible with simple cis-effects entails a flipping of the more highly expressed allele in different heterozygotes in a manner that is compatible with, but not fully proving, classical imprinting.
There are currently about 100 known mammalian-imprinted genes, and roughly one-third of these have been shown to be imprinted in both humans and mice, with the remaining two-third imprinted in only one of the two species, mostly mouse (Morison et al. 2005). Recently, it has been suggested that there may be hundreds of undiscovered imprinted genes in these mammalian genomes. Comparing gene expression in parthenogenote and androgenote mouse embryos, Nikaido et al. (2003) predicted 2,101 novel-imprinted transcripts. Luedi et al. (2005) used a computational predictor based on sequence features to identify 600 potentially imprinted genes in the mouse genome. However, Morison et al. (2005) pointed out methodological issues with these studies (e.g. gene expression changes associated with parthenogenote development) (Ruf et al. 2006) and argued that the small number of known imprinting-associated phenotypes indicates that there are very few as yet to be discovered imprinted genes. Indeed, experimental validation of high scoring predicted imprinted genes from these two studies have had validation rates of ∼5% (Ruf et al. 2006, 2007).
Genome-wide studies of DAE examining individuals heterozygous for cSNPs are an unbiased method to identify candidate-imprinted genes. To understand human imprinting, it is essential to conduct such studies with human samples, since imprinted genes are known to differ substantially among mammals (Morison et al. 2005; Monk et al. 2006). Lo et al. (2003) used oligonucleotide arrays to genotype seven human fetuses at 602 genes and to assess allele-specific gene expression in liver and kidney tissues. While they did not attempt to predict novel-imprinted genes from their data, Lo et al. did confirm DAE for four out of the five known imprinted genes in their study. More recently, in Pant et al. (2006) we conducted a similar study of 1,389 genes in white blood cells of 12 adults. We specifically looked at which allele was more highly expressed in genes showing DAE and predicted three novel-imprinted genes based on mono-allelic expression: ZNF463 (ZNF331), FLJ33071 (ZNF597), and PRIM2. Due to the limited number of genes and relatively small sample size in this study, it is likely that more imprinted genes remain to be discovered, although it is not clear how many.
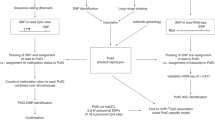
Our current study is the largest experimental scan for novel human-imprinted genes to date. Using an oligonucleotide array, we attempted to systematically assay 7,109 common cSNPs in humans. Since expression of imprinted genes can vary between tissues and developmental stages, it is difficult to identify an ideal RNA source for such a study. As a first effort, we have surveyed DAE in lymphoblastoid cell lines (LCLs) derived from 67 unrelated individuals belonging to three ethnic groups. This microarray screen was followed by a validation study of 16 candidate-imprinted genes in independent LCLs from two families and in a panel of osteoblast-like cells from 48 unrelated individuals. We identified one gene, ZNF331, for which paternal expression is the most likely explanation for the DAE we observed. Several additional genes are still candidates.
Methods
Samples, cell culture, and mRNA isolation
Sixty-seven LCLs (Supplementary Table S1) were obtained from the Coriell Cell Repositories (http://locus.umdnj.edu/ccr/). All LCLs were cultured in RPMI medium supplemented with 15% FBS in a 37°C, 5% CO2 incubator. When lymphoblasts were semi-confluent (1–1.5 million cells/ml) about 40–50 million cells were spun down and then re-suspended in 5 ml Trizol Reagent (Invitrogen) and genomic DNA (gDNA) and RNA were purified according to the manufacturer’s instruction. Each sample yielded between 200 and 400 μg of RNA and ∼1 mg of gDNA. The quality of the RNA was tested by visual inspection of intact RNA by gel electrophoresis. The RNA was then treated with DNase I, and purified again by phenol–chloroform extraction and ethanol precipitation.
Human trabecular bone samples were collected from the proximal femoral shaft from 48 patients undergoing total hip replacement at the Section of Orthopedics, Uppsala University Hospital, Uppsala, Sweden. The bone samples were minced thoroughly and washed with PBS to remove hematopoietic cells. The bone fragments were cultured in complete cell medium (α-MEM supplemented with 2 mmol/l l-glutamine, 100 U/mL penicillin, 100 mg/mL streptomycin, and 10% fetal bovine serum) and the cells were grown at 37°C with 5% CO2. At 70–80% confluence, the cells were harvested and 2 ml of Trizol Reagent (Invitrogen) was added and gDNA and RNA were purified according to the manufacturer’s instruction. High RNA quality was confirmed for all samples using Agilent 2100 BioAnalyzer (Agilent Technologies), and the concentrations were determined using Nanodrop ND-1000 (NanoDrop Technologies). The RNA was then treated with DNase I, and purified again by phenol–chloroform extraction and ethanol precipitation.
cDNA synthesis
mRNA (between 7 and 20 μg per sample) was then purified from the total RNA using a polyA isolation kit (Ambion) according to manufacturer’s instruction. cDNA was generated by reverse transcription of 1 μg of mRNA using Superscript II RT (Invitrogen) in the presence of either 50 ng of a 5′-phosphorylated random hexamer oligonucleotide or 500 ng of a 5′-phosphorylated T18V oligonucleotide (where V represents dGTP, dCTP or dATP) followed by RNaseH treatment to eliminate the RNA.
cSNP primer design
A total of 7,109 cSNPs were chosen for inclusion in our study because both alleles were observed in an independent study (Hinds et al. 2005) examining the same samples. We designed primer pairs to amplify the 7,109 cSNPs using Oligo 6 (Molecular Biology Insights), and fulfilled the following requirements: the amplicon was 50–200 bp in length; the PCR primers were between 17 and 22 nucleotides in length; and the PCR primer pairs were unique in the human genome based on BLAST analysis.
Short-range PCR, pooling and purification of samples
Samples were prepared as previously described (Pant et al. 2006). Briefly, both gDNA and cDNA were diluted to 20 ng/μl for use as templates in PCR reactions performed in 384-well-plate format in a 12 μl volume. The reaction concentrations were 1× PCR buffer, 2.75 mM MgCl2, 200 μM dNTP, 0.4 μM each primer, 0.3 U of AmpliTaq Gold DNA polymerase (Applied Biosystems), and 5 ng of either gDNA or cDNA template. Touchdown PCR was run at 95°C for 5 min; then 10 cycles of 30 s at 95°C, 30 s at 60°C with a reduction of 0.5°C for each cycle and 10 s at 72°C; and finally 40 cycles of 10 s at 95°C, 30 s at 55°C and 30 s at 72°C. For each of the 67 samples, two cDNA reactions were amplified in duplicate and the duplicates were pooled individually.
Hybridizations
One gDNA and the two duplicate cDNA hybridizations were performed for each of the 67 samples. Five micrograms of each pool of purified PCR products was labeled with Biotin-ddUTP/biotin-dUTP in total volume of 37 μl with a final concentration of 1× One-Phor-All buffer, 13.5 μM Biotin-ddUTP/Biotin-dUTP and 0.5 U of Terminal Transferase (Roche). The labeling reactions were mixed with hybridization buffer (3 M tetramethylammonium chloride, 10 mM Tris–HCl, 0.01% Triton X-100, 100 μg/ml Herring-Sperm DNA, 50 pM control oligonucleotide b948), denatured at 95°C for 10 min, and then incubated with the corresponding arrays for 16–18 h at 50°C. The arrays were washed with 6× SSPE (0.9 M NaCl, 60 mM NaH2PO4, 6 mM EDTA, 0.01% Triton X-100) and stained first with 2.5 μg/ml Streptavidin for 15 min, then with 1.25 μg/ml anti-Streptavidin antibodies for 15 min, and finally with Streptavidin-CyChrome for 15 min. Between each staining, the arrays were washed with 6× SSPE using an automated fluidics wash station. Finally, the arrays were incubated with 0.2× SSPE for 30 min and filled with 6× SSPE for scanning. The hybridization of labeled sample was detected by measuring CyChrome fluorescence using a custom built confocal laser scanner (Perlegen Sciences).
Data processing
Data for the 7,109 cSNPs were processed, quality filtered, and screened for expression in LCLs following the methods of Pant et al. (2006). The intensities from each pair of replicate cDNA arrays were averaged. This pre-processing produces an estimate of the reference allele frequency \( (\hat{p}) \) in both cDNA and gDNA for each cSNP in every sample. gDNA values of \( \hat{p} \) were used to genotype each sample at each cSNP as described in Pant et al. (2006). For each cSNP, we computed the mean value of \( \hat{p} \) in cDNA for samples in each genotype. We removed from further analysis all cSNPs for which the mean cDNA \( \hat{p} \) for either homozygous reference or homozygous alternative individuals fell significantly outside the range of corresponding gDNA \( \hat{p} \) values, suggesting hybridization problems since cDNA and gDNA values should be similar in homozygotes. We also removed any cSNP that did not show significant differences in mean cDNA reference allele frequencies between genotypes based on an F test performed at level α = 0.05. P values were corrected for multiple comparisons using the false discovery rate (FDR) controlling procedure of Benjamini and Hochberg (1995). Finally, cSNPs with fewer than two heterozygous individuals were excluded from analysis. After filtering, the data set contained 3,877 cSNPs located in 2,625 unique genes.
Differential allelic expression
To quantify allele-specific differences in expression, we computed the ratio of reference to alternate allele cDNA intensity at each cSNP in heterozygous individuals. Expression ratios were truncated below at 0.1 and above at 10, representing tenfold differential expression relative to the reference (approximately the maximum reliably detectable value from these arrays). We identified all heterozygotes with two, four or tenfold differential expression between alleles at each cSNP.
Flipping
We defined “flipping” as the presence of both over- and under-expression of the reference cSNP allele relative to the alternate allele in different heterozygous individuals. We required at least one heterozygous individual with a cDNA intensity ratio > k and at least one with a ratio < 1/k in order to label a gene as flipping at the k-fold level (k = 2, 4, or 10). The sparseness of replication precluded formal statistical tests of significance of differences of allelic expression, but the test of flipping at a k-fold expression level proved to be a robust exploratory test to nominate candidates for pedigree testing.
CEPH pedigrees
Twenty-seven genes were selected for experimental validation in an independent data set of LCLs from two three-generation CEPH families (pedigrees 1420 and 1444). Pedigree 1420 contains 7 individuals in the third generation and 1444 contains 8 individuals. The pedigree tests allow robust rejection of copy number variation (CNV) or uniparental disomy as an explanation for the allelic flipping, since they confirm a parent-of-origin effect not expected if CNV or uniparental disomy were acting.
Quantitative sequencing
We used HapMap data for the CEPH population to select a common and potentially informative exonic SNP (i.e. coding or in the UTR) for each candidate-imprinted gene. We designed primers surrounding each SNP and at least 20 bp away from the intron–exon boundaries. For each cell line, we separately amplified genomic DNA and cDNA (obtained from RT-PCR by random hexamers) and sequenced the PCR products. Genotypes were inferred directly from the gDNA sequencing data. The allelic ratio in cDNA was estimated from the peak height in the trace files after normalization of the peak intensity (Ge et al. 2005). High-quality data were obtained for 16 genes. For each gene, allele-specific expression was mapped onto the pedigrees and analyzed for imprinting based on evidence of parent-of-origin dependent DAE.
Validation assays for human bone samples were carried out using the same method as described above. High-quality data was obtained for four genes (PVR, ZNF331, TBC1D4, BMP8A).
Results
We designed high-density oligonucleotide arrays to assay for DAE at 7,109 exonic SNPs, in LCLs generated from 67 unrelated individuals belonging to three populations (20 African Americans, 24 CEPH European American, and 23 Han Chinese American) (Supplementary Table S1). Genomic DNA and cDNA samples from each LCL were amplified with PCR primers surrounding each cSNP. The PCR products were then labeled and hybridized to the high-density oligonucleotide arrays (Pant et al. 2006). We extracted the fluorescence intensities for all probes corresponding to each SNP allele and estimated the concentration of each allele in the DNA and cDNA samples. We then used the estimates to genotype the SNPs in each genomic DNA sample and to quantify the ratio of reference to alternate SNP alleles in the cDNA samples.
We analyzed 3,877 cSNPs located in 2,625 genes for DAE. These SNPs were filtered from the 7,109 assayed cSNPs based on stringent quality control procedures. For the purpose of identifying novel-imprinted genes, we looked for “flipping” of the over-expressed allele between reference and alternate in different heterozygous individuals, a defining characteristic of genomic imprinting.
Our ability to observe flipping depends on the number of heterozygotes for a given cSNP, which is a function of sample size (n = 67) and minor allele frequency. For each assayed cSNP, both alleles were observed in a previous study of the same populations (Hinds et al. 2005), suggesting relatively high minor allele frequencies for these polymorphic sites. Table 1 shows the number of heterozygous individuals per successfully assayed cSNP. Ninety-five percent of cSNPs have more than five heterozygotes, and 84% have more than ten. In a sample of ten unrelated heterozygotes, the probability of not observing the reference allele both maternally and paternally inherited is less than 1 in 1,000. This probability is 0.031 for a sample of five heterozygotes. Thus, in the absence of measurement error, we should be able to observe flipping in the vast majority of the 3,877 cSNPs that are located in genes that are imprinted in LCLs. Nonetheless, genes will vary substantially in terms of our power to detect both DAE and flipping due to experimental noise in the microarray data as well as differences in the number of heterozygotes per cSNP and the number of cSNPs per gene.
Based on examining heterozygous individuals for the presence of large fold differences between reference and alternate alleles we categorized each cSNP as (1) no DAE, (2) DAE without flipping, or (3) DAE with flipping (Fig. 1). In order to categorize cSNPs based on the magnitude of the observed expression differences, we considered three different thresholds for absolute fold changes: two, four, and tenfold.
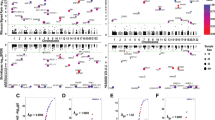
Typical gene expression patterns for three types of genes. Distributions of the expression ratio R = log2 (reference/alternate) across heterozygous samples for three genes with different patterns of differential allelic expression (DAE). Larger ratios indicate higher expression of the reference allele. DAE corresponds with expression ratios significantly above or below zero [twofold DAE = |R| > 1, fourfold DAE = |R| > 2, and tenfold DAE = |R| > log2(10) = 3.32]. Red The gene LRAP has expression ratios centered at zero and does not show DAE in any heterozygotes. Blue The gene DTX2 shows DAE (as much as tenfold in some heterozygotes). All expression ratios are positive, indicating that the reference allele is always expressed higher than the alternate allele. Thus, DTX2 does not flip. Yellow The known human-imprinted gene PEG10 shows tenfold DAE and flipping (both over- and under-expression of the reference allele). Purple indicates overlap of the red (LRAP) and blue (DTX2) distributions, and green indicates overlap of blue (DTX2) with yellow (PEG10)
Of the 3,877 cSNPs successfully assayed 87% showed no evidence of DAE. At the least stringent twofold threshold, 496 cSNPs present in 460 unique genes have DAE in at least one heterozygote (Supplemental Table S2). Of these genes, 59 (64 cSNPs) show DAE in at least one heterozygote at the most stringent tenfold level, and an additional 59 (73 cSNPs) do so at the fourfold level. Only three genes, PEG10, hypothetical protein LOC400036, and pseudogene LOC285647, show mono-allelic expression, defined as tenfold DAE in all heterozygotes. PEG10 is the only one of these three genes that shows flipping.
The majority of the 496 cSNPs with twofold DAE do not display the flipping expression pattern. The 69 cSNPs that flip (14%) have at least one heterozygote showing over-expression and at least one showing under-expression of the reference allele. While we define flipping as a single case of over-expression and a single case of under-expression at the appropriate fold threshold, most of the flipping cSNPs (51 of 69) had more than these two requisite cases of DAE. For cSNPs with twofold flipping, on average 27.3% of heterozygotes showed DAE. This proportion rises to 48.9% for fourfold flipping and 62.6% for tenfold flipping. The 69 cSNPs with twofold flipping are located in 61 unique genes, ten of which show flipping at the fourfold level and an additional six of which do so at the tenfold level (Supplementary Table S3).
As a quality control measure, we examined consistency of DAE and flipping calls between cSNPs in the same gene. We report results for a twofold threshold. Calls at four and tenfold thresholds are generally more consistent. Thirty percent of the 2,625 assayed genes are represented by more than one cSNP on the array (Table 2, max = 12 cSNPs in MKI67). Of these 810 genes with multiple cSNPs, 82.2% had perfectly consistent DAE calls (613 with no cSNPs showing DAE, 53 with all cSNPs showing DAE). Among the 197 genes with at least one cSNP showing DAE, 74.6% showed DAE in half or more the cSNPs (37.1% with more than half). Flipping calls were even more consistent. The vast majority (96.9%) of genes with multiple cSNPs consistently show no flipping in any cSNP. Of the 25 genes that do show flipping in at least one cSNP, 80% show flipping in half or more of the cSNPs, and 20% consistently show flipping in all cSNPs. Thus, our filtered gene list shows high concordance between cSNPs in the same gene. The moderate level of variability we observe is certainly expected in microarray data and reminds us of the possibility of some false positive and false negative calls for genes assayed with a single cSNP. Some variability may also be explained by alternative splicing.
Of the 2,625 genes examined in our study, three are protein-coding genes that are known to be imprinted in humans (PLAGL1/ZAC, ATP10A, PEG10) (Morison et al. 2005). These genes provide an opportunity to test the ability of our methods to detect imprinting. ATP10A and PEG10 show DAE and flipping, at the fourfold and tenfold levels, respectively. PLAGL1/ZAC does not show DAE or flipping. Interestingly, our observation is consistent with the previous finding of Kamiya et al. (2000) that PLAGL1/ZAC is imprinted in several fetal tissues but is biallelically expressed in white blood cells. Further, Valleley et al. (2007) identified an alternative unmethylated CpG island promotor from which biallelic PLAGL1/ZAC transcripts are derived in peripheral blood leukocytes.
We next examined expression patterns for human orthologs of known and predicted mouse-imprinted genes present in our set of 2,625 genes. We assayed two orthologs, COPG2 and ASB4, of known mouse-imprinted genes that show no prior evidence of being imprinted in humans (Morison et al. 2005). Neither of these two genes display DAE in our sample set. Among the 600 putative mouse-imprinted genes predicted by Luedi et al. (2005), 58 have a human ortholog in our data set. Of these genes, 16 show DAE, 4 with flipping (ATP10A, STK32C, UNC13B, and STIM2) and 12 without flipping. The remaining 42 genes are not differentially expressed in our samples.
From our list of 61 candidate-imprinted genes that contain cSNPs displaying DAE with flipping, we selected 25 genes for validation in an independent data set of LCLs derived from individuals belonging to two three-generation CEPH families (pedigrees 1420 and 1444). These 25 genes were selected based on the magnitude of expression fold changes, the number of heterozygotes, proportion of heterozygotes showing DAE, and genomic proximity to known imprinted genes. Three genes that met our criteria for inclusion in the validation study (ATF5, EPHX2 and BTN3A2) were excluded, because they had been shown previously to be statistically associated with haplotypes in the HapMap CEPH individuals (Pastinen et al. 2005). Hence, these genes are very likely to be regulated genetically, rather than epigenetically. We note that one selected gene, ZNF331/ZNF463 (henceforth ZNF331), also showed tenfold DAE and flipping in white blood cells in Pant et al. (2006). In addition to the 25 genes selected from our microarray screen, we also included two candidates that were not assayed on the microarray but did show tenfold DAE and flipping in Pant et al. (2006): ZNF597/FLJ33071 and PRIM2A.
Twenty-seven genes were genotyped and analyzed for DAE in the CEPH pedigrees in order to validate imprinting status. The data for eight of these genes (ATP10A, BMP8A, LOC389814, PLAG2G4C, PRIM2A, STK32C, TSPAN4, and ZNF228) were not analyzable due to low expression in the LCLs, multiple bands after amplification, or poor quality sequences after optimization. Three additional genes (KIF21A, STIM2, and ZNF597/FLJ33071) had too few heterozygotes to enable inference regarding uni-parental expression. Expression patterns across the two CEPH pedigrees were analyzed for the remaining 16 genes (Table 3, Supplementary Table S4). In order to be fully informative, the pedigrees must show inheritance of both alleles maternally and paternally into heterozygotes where DAE can be assessed. The informativeness of the pedigree data for each gene is summarized in Supplementary Table S5.
Our pedigree analysis included one positive control, PEG10, a known imprinted gene that displays tenfold DAE and flipping in our microarray study. PEG10 is the only gene in the screen with completely monoallelic expression and flipping. The pedigree analysis confirmed paternal-imprinted expression of PEG10 (Fig. 2).
Quantitative sequencing of candidate-imprinted genes. Differential allelic expression for CEPH pedigrees 1420 and 1444, consisting of four grandparents (top), two parents (middle) and seven or eight offspring (bottom). Males are squares, and females are circles. For each individual, the DAE determined by quantitative sequencing of RT-PCR product is represented by the histogram inside the square/circle. Homozygous individuals are shown with two columns of identical height and color (red reference allele, blue alternate allele). For heterozygous individuals, the relative expression of each allele (red and blue) is displayed by the height of the columns. PEG10 is imprinted. PVR, TBC1D4 and ZNF331 are consistent with imprinting
We identified two genes for which the pedigree analyses are compatible with imprinting: TBC1D4 (maternal expression) and ZNF331 (paternal expression). For each of these genes, only one of the two CEPH families has heterozygous offspring (pedigree 1420 for TBC1D4, pedigree 1444 for ZNF331; Supplemental Table S5). In the informative families, TBC1D4 and ZNF331 show strong over-expression of the alternate allele in a manner that is completely consistent with uni-parental expression (Fig. 2). These data coupled with over-expression of the reference allele in multiple unrelated individuals in our array data, indicates that both alleles can be expressed at a high level and suggests that these two genes are imprinted.
Data for PVR are largely consistent with imprinting, but the presence of several heterozygotes without DAE indicates that PVR is not classically imprinted. Only pedigree 1420 is informative. In that family, we observe transmission of both alleles maternally and paternally in the second generation (Fig. 2), and the allelic expression data are consistent with paternal expression. Since both parents in pedigree 1420 are heterozygous at the cSNP used to assay PVR (rs714948), it is not possible to analyze the expression patterns in the third generation with regard to parent-of-origin effects. However, the three heterozygous offspring do show moderate to strong DAE.
Data for an additional four genes (CUZD1, DFKZ/ZNF772, GALNTL4, and PLK2) are largely consistent with imprinting, but are not conclusive (Supplementary Table S5). CUZD1 has heterozygous offspring in both pedigrees and shows transmission of both alleles through both parents. However, in each case both individuals in the second generation are heterozygous (like PVR), making it difficult to assess parent-of-origin effects. In both families, one trio (two grandparents and a parent) shows a pattern of DAE consistent with imprinting, while the other parent does not show DAE (Supplementary Figure S1). Thus, the data for CUZD1 are consistent with maternal expression, but further studies are needed to determine if the gene is truly imprinted. DFKZ/ZNF772 and GALANTL4 both show over-expression of a maternally inherited reference allele in heterozygous offspring, but we cannot conclusively say these genes are imprinted (Supplementary Figure S2, Supplementary Figure S3). Pedigree data for PLK2 are not highly informative, but are consistent with maternal expression (Supplementary Figure S4).
The remaining eight genes (ARL4C, CAECAM21, KIAA1466, LGALS14, LOC400451, NALP2, UNC13B, and ZNF589) conclusively did not have parent-of-origin effects in LCLs (Supplementary Figure S5), although they did show DAE and flipping in the array data.
Based on the results of our pedigree validation studies, we selected four genes for additional validation in a panel of human osteoblasts (HOBs) from 48 unrelated individuals. We used primary cells to minimize potential artifacts induced by prolonged cell culture and multiple cell passaging. In HOBs, we observed monoallelic expression and flipping completely consistent with classical imprinting for the known imprinted genes SNRPN and MEST (data not shown). We selected ZNF331, TBC1D4, and PVR for validation based on their expression patterns consistent with imprinting in the pedigree analysis. We also included BMP8A, which did not produce high-quality quantitative sequencing data in the pedigree analysis, because this gene was one of our top candidates in the microarray screen. Data for all four genes is in Table S6. PVR did not show DAE in the HOBs, suggesting that the pattern of differential expression we observed in LCLs is tissue specific and may not reflect imprinting. TBC1D4 and BMP8A both show DAE in some heterozygotes (6 out of 17 for TBC1D4; 10 out of 15 for BMP8A). Each gene has one heterozygote with an allelic expression ratio that flips relative to the other individuals with DAE. This pattern of expression is more consistent with heritable cis-acting variation than with imprinting. We assayed two SNPs for ZNF331: a relatively rare SNP (rs1351) at the 3′UTR and a more common SNP (rs16984961) at the 5′UTR of the RefSeq annotation of ZNF331. Interestingly, the 3′ and 5′ SNPs in ZNF331 give different results. Despite having only three heterozygotes, rs1351 shows monoallelic expression and flipping, strongly suggestive of imprinting. In contrast, rs16984961 shows DAE in only four out of six heterozygotes, although it does show flipping. These findings are consistent with the complex isoform annotation of ZNF331 and the known examples of isoform-specific imprinting in human genes, such as MEST.
Discussion
This is the first study to conduct a genome-scale assessment of imprinting in humans. We present a novel approach that uses genotyping microarrays to assay a large set of genes for expression patterns consistent with imprinting. A smaller set of high confidence candidate-imprinted genes can then be validated with more labor-intensive assays, such as quantitative sequencing. Using this method, we systematically scanned 2,625 human protein-coding genes for differential expression of the two parental alleles. Our results confirm previous findings that DAE is common in the human genome. While many genes display departures from perfectly stoichiometric expression of the two alleles, very few are consistently differentially expressed across all heterozygotes and only three show truly mono-allelic expression patterns. Sixty-one genes showed a pattern of DAE and flipping of the over-expressed allele that is suggestive of imprinting. To identify novel-imprinted loci, we focused on these 61 genes that show both alleles over-expressed. Their differential expression patterns are less likely to be due to cis-acting genetic effects than those of genes that do not flip.
We compared our microarray results to the literature on mammalian-imprinted genes. Despite looking at only one cell type and being somewhat limited by sample size, our findings are completely consistent with current knowledge of genomic imprinting in humans. We did not, however, detect DAE in the human orthologs of two known mouse-imprinted genes. Among 58 orthologs of predicted mouse-imprinted genes (Luedi et al. 2005), 16 showed DAE in our LCLs (4 with flipping). These findings are consistent with either a high false positive rate in Luedi et al. (2005) or with the fact that imprinting status is known to differ substantially between the mouse and human genomes (Morison et al. 2005; Monk et al. 2006). Loss of imprinting regulation in immortalized LCLs is another possible explanation.
The most promising candidates for imprinted genes from our microarray screen were selected for validation in LCLs from two CEPH pedigrees and a panel of unrelated HOBs. The known imprinted gene, PEG10, showed monoallelic expression and flipping in the microarray data, which was confirmed in the pedigree analysis. These findings confirm paternal expression of PEG10 and indicate that our assays are sensitive enough to detect a truly imprinted gene. None of the genes we assayed showed as strong and consistent a picture of genomic imprinting as that observed for PEG10. However, our screen did uncover one gene that is very likely to be imprinted in humans: ZNF331. Our data suggest that ZNF331 is probably imprinted in an isoform-specific manner. ZNF331 is a krueppel C2H2-type zinc-finger protein that may be rearranged in thyroid tumors. We are not aware of any previous evidence that this gene is imprinted.
Interestingly, most of the candidate genes from our microarray screen were not classically imprinted in our validation samples, although they generally did show DAE.
The expression patterns for these genes are most likely due to a regulatory variant that is not in LD with the cSNP we assayed due to recombination between the two SNPs. This could make the assayed SNP appear to flip in relative expression level in a manner not related to the parent-of-origin. Another possible explanation is random monoallelic expression through differential methylation, as documented for IL1A (Pastinen et al. 2004). Also, among sample variation could result from differences in LCL culturing. These results demonstrate the importance of performing validation tests of candidate-imprinted genes in family pedigrees and multiple tissues, because DAE and flipping of cSNPs can occur through multiple molecular mechanisms.
With a sample size of 67 LCLs, this is the best-powered genome-wide survey of genomic imprinting in humans to date. Nonetheless, failure to observe flipping does not rule out imprinting, particularly for the 199 genes with 5 or fewer heterozygotes in our sample. Hence, of the 2,625 genes we assayed a few may be imprinted but missed by our screen. We scanned only ∼10% of known protein-coding genes and no non-coding RNA (ncRNA) genes. Recent studies suggest that 30% or more of imprinted genes are ncRNAs (Nikaido et al. 2003; Morison et al. 2005), so that a complete survey of imprinting should include both coding and non-coding transcripts. Thus, our prediction of one new imprinted gene, plus just a handful of additional candidates, is clearly an underestimate of the true number of novel-imprinted genes in humans. Nonetheless, our low yield of novel-imprinted genes is consistent with the results of Ruf et al. (2006, 2007) and indicates that it is unlikely that the number of as yet to be discovered imprinted genes is in the range of 500–1,000, as predicted by Nikaido et al. (2003) and Luedi et al. (2005). Based on our validation rates and given that our scan covered approximately 10% of the protein-coding genes in humans, a rough calculation suggests that a full genome screen would be unlikely to detect more than a few hundred additional genes that undergo genomic imprinting detectable in LCLs.
One limitation to our approach is the use of a single cultured cell type (LCLs) when genomic imprinting of many genes is restricted to brain and/or placenta (Morison et al. 2005) and imprinted genes may be expressed at very low levels in blood cells. Epigenetic drift in established cell lines is also a concern. It is fortunate that several known human-imprinted genes retain their epigenetic signatures in LCLs, as shown by two of three known imprinted genes tested in this study as well as findings from several other recent studies. These include reports of genes that exhibit parental modes of transmission in LCLs from related individuals (MEST: Pastinen et al. 2004), as well as other epigenetic signatures, such as random monoallelic imbalance (IL1A: Pastinen et al. 2004) and X-inactivation. In addition, molecular evidence that epigenetic marks are retained in LCLs has been shown for many genes (SNRPN: Schweizer et al. 1999, Fulmer-Smentek and Francke 2001; SGCE: Grabowski et al. 2003). Interestingly, genes that show monoallelic expression in muscle, but biallelic expression in lymphocytes and fibroblasts, show biallelic expression in LCLs (Zhou et al. 2006). This suggests that LCLs will not always be a good model for genes that exhibit tissue-specific imprinting, but that they are a valid model of what occurs in the tissue from which the cells originate (i.e. lymphocytes). Importantly, LCLs do not appear to produce false positive cases of classical imprinting in this or other reports. Our study validates that LCLs can be used to identify and determine the mode of inheritance of the subset of imprinted genes that are expressed in lymphoblastoid cells. Nevertheless, the most exhaustive survey of genomic imprinting in humans would derive mRNA from a variety of tissues, including placenta and fetal brain. Thus, it is beyond the scope of this study to accurately estimate how many novel-imprinted genes are yet to be discovered.
Another limitation of our approach is the availability of cSNPs with high enough heterozygosity to be informative about DAE with practical sample sizes. We assayed 7,109 common cSNPs in 67 individuals and obtained high-quality data for 3,877 cSNPs in 2,625 genes. With a larger collection of common cSNPs, our microarray approach could be used for a truly genome-wide screen of imprinting in humans.
Our study is a step towards developing an exhaustive catalog of mammalian-imprinted genes. Such a list will facilitate the study of genetic diseases associated with aberrant imprinting and the exposure of deleterious alleles through gene silencing. Once the function of all imprinted genes is better understood, we will be able to answer the hotly debated question of what roles imprinting plays in mammalian biology. While it is clear that imprinted genes are important for development in utero (Murphy and Jirtle 2003), the evolutionary forces that led to imprinting despite the cost of the resulting hemizygosity remain an open question. Genetic conflict resulting from multiple paternities is a popular hypothesis (Reik and Walter 2001; Morison et al. 2005), but imprinting can evolve under a wide range of different molecular evolutionary models (Spencer et al. 1998). With the availability of an unbiased list of imprinted genes and whole genome sequences from many vertebrates, we will be better able to evaluate the evolutionary forces that have shaped imprinted regions of the human genome.
References
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300
Fulmer-Smentek SB, Francke U (2001) Association of acetylated histones with paternally expressed genes in the Prader-Willi deletion region. Hum Mol Genet 10:645–652
Ge B, Gurd S, Gaudin T, Dore C, Lepage P, Harmsen E, Hudson TJ, Pastinen T (2005) Survey of allelic expression using EST mining. Genome Res 15:1584–1591
Gibson G, Weir B (2005) The quantitative genetics of transcription. Trends Genet 21:616–623
Grabowski M, Zimprich A, Lorenz-Depiereux B, Kalscheuer V, Asmus F, Gasser T, Meitinger T, Strom TM (2003) The epsilon-sarcoglycan gene (SGCE), mutated in myoclonus-dystonia syndrome, is maternally imprinted. Eur J Hum Genet 11:138–144
Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR (2005) Whole-genome patterns of common DNA variation in three human populations. Science 307:1072–1079
Kamiya M, Judson H, Okazaki Y, Kusakabe M, Muramatsu M, Takada S, Takagi N, Arima T, Wake N, Kamimura K, Satomura K, Hermann R, Bonthron DT, Hayashizaki Y (2000) The cell cycle control gene ZAC/PLAGL1 is imprinted—a strong candidate gene for transient neonatal diabetes. Hum Mol Genet 9:453–460
Lo HS, Wang Z, Hu Y, Yang HH, Gere S, Buetow KH, Lee MP (2003) Genetic variation in gene expression is common in the human genome. Genome Res 13:1855–1862
Luedi PP, Hartemink AJ, Jirtle RL (2005) Genome-wide prediction of imprinted murine genes. Genome Res 15:875–884
Monk D, Arnaud P, Apostolidou S, Hills FA, Kelsey G, Stanier P, Feil R, Moore GE (2006) Limited evolutionary conservation of imprinting in the human placenta. Proc Natl Acad Sci USA 103:6623–6628
Morison IM, Ramsay JP, Spencer HG (2005) Evolution of mammalian imprinting. Trends Genet 21:457–465
Murphy SK, Jirtle RL (2003) Imprinting evolution and the price of silence. BioEssays 25:577–588
Nikaido I, Saito C, Mizuno Y, Meguro M, Bono H, Kadomura M, Kono T, Morris GA, Lyons PA, Oshimura M, RIKEN GER Group, GSL Members, Hayashizaki Y, Okazaki Y (2003) Discovery of imprinted transcripts in the mouse transcriptome using large-scale expression profiling. Genome Res 13:1402–1409
Pant PV, Tao H, Beilharz EJ, Ballinger DG, Cox DR, Frazer KA (2006) Analysis of allelic differential expression in human white blood cells. Genome Res 16:331–339
Pastinen T, Sladek R, Gurd S, Sammak A, Ge B, Lepage P, Lavergne K, Villeneuve A, Gaudin T, Brandstrom H, Beck A, Verner A, Kingsley J, Harmsen E, Labuda D, Morgan K, Vohl MC, Naumova AK, Sinnett D, Hudson TJ (2004) A survey of genetic and epigenetic variation affecting human gene expression. Physiol Genomics 16:184–193
Pastinen T, Ge B, Gurd S, Gaudin T, Dore C, Lemire M, Lepage P, Harmsen E, Hudson TJ (2005) Mapping common regulatory variants to human haplotypes. Hum Mol Genet 14:3963–3971
Reik W, Walter J (2001) Evolution of imprinting mechanisms: the battle of the sexes begins in the zygote. Nat Genet 27:255–256
Ruf N, Dunzinger U, Brinckmann A, Haaf T, Nurnberg P, Zechner U (2006) Expression profiling of uniparental mouse embryos is inefficient in identifying novel imprinted genes. Genomics 87:509–519
Ruf N, Bahring S, Galetska D, Pliushch G, Luft F, Nurnberg P, Haaf T, Kelsey G, Zechner U (2007) Sequence-based bioinformatic prediction and QUASEP identify genomic imprinting of the KCNK9 potassium channel gene in mouse and human. Hum Mol Genet 16:2591–2599
Schweizer J, Zynger D, Francke U (1999) In vivo nuclease hypersensitivity studies reveal multiple sites of parental origin-dependent differential chromatin conformation in the 150 kb SNRPN transcription unit. Hum Mol Genet 8:555–566
Spencer HG, Feldman MW, Clark AG (1998) Genetic conflicts, multiple paternity and the evolution of genomic imprinting. Genetics 148:893–904
Spielman RS, Bastone LA, BurdickJT, Morely M, Ewens WJ, Cheung VG (2007) Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet 39:226–231
Storey JD, Madeoy J, Strout JL, Wurfel M, Ronald J, Akey JM (2007) Gene-expression variation within and among human populations. Am J Hum Genet 80:502–509
Stranger BE, Forrest MS, Clark AG, Minichiello MJ, Deutsch S, Lyle R, Hunt S, Kahl B, Antonarakis SE, Tavaré S, Deloukas P, Dermitzakis ET (2005) Genome-wide associations of gene expression variation in humans. PLoS Genet 1:e78
Tao H, Cox DR, Frazer KA (2006) Allele-specific KRT1 expression is a complex trait. PLoS Genet 2:e93
Valleley EM, Cordery SF, Bonthron DT (2007) Tissue-specific imprinting of the ZAC/PLAGL1 tumour suppressor gene results from variable utilization of monoallelic and biallelic promoters. Hum Mol Genet 16:972–981
Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW (2002) Allelic variation in human gene expression. Science 297:1143
Zhou H, Brockington M, Jungbluth H, Monk D, Stanier P, Sewry CA, Moore GE, Muntoni F (2006) Epigenetic allele silencing unveils recessive RYR1 mutations in core myopathies. Am J Hum Genet 79:859–868
Acknowledgments
At Perlegen Sciences we thank Erica J. Beilharz for project management assistance, Geoff B. Nilsen for designing the high-density array, and P.V. Pant for assistance with data analysis. At McGill University we thank Scott Gurd for technical assistance, Tomi M. Pastinen for supporting the osteoblast work and helpful discussions, and Olof Nilsson at Uppsala University, Sweden for collecting the bone samples for the osteoblast panel. T.J. Hudson is the recipient of a Clinician-Scientist Award in Translational Research by the Burroughs Wellcome Fund. This work was supported by an NHGRI grant to K.A. Frazer and by Genome Canada and Genome Quebec grants to T.J. Hudson. K.A. Frazer is a former employee of Perlegen Sciences, Inc.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pollard, K.S., Serre, D., Wang, X. et al. A genome-wide approach to identifying novel-imprinted genes. Hum Genet 122, 625–634 (2008). https://doi.org/10.1007/s00439-007-0440-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-007-0440-1