Abstract
Meckel–Gruber syndrome (MKS) is a recessively inherited, lethal disorder characterized by renal cystic dysplasia, occipital encephalocele, polydactyly and biliary dysgenesis. MKS is genetically heterogeneous with three loci mapped and two identified; MKS1 (17q23) and MKS3 (8q22.1). MKS1 is part of the Finnish disease heritage, while MKS3 has been described exclusively in consanguineous Asian families. Here we aimed to establish molecular diagnostics for MKS, determine the importance of MKS1 and MKS3 in non-consanguineous populations, and study genotype/phenotype correlations. The coding regions of MKS1 and MKS3 were screened for mutations by direct sequencing in 17 families clinically diagnosed with MKS in the US or The Netherlands. The clinical phenotype was compared to genic and allelic effects. Both mutations were identified in ten families; five MKS1 and five MKS3. All but two were compound heterozygotes, consistent with their non-consanguineous nature. The MKS1-Finmajor mutation accounted for 7/10 MKS1 mutations; two novel changes were additionally detected. Seven novel mutations were found in MKS3, including three missense changes. We concluded that MKS1 and MKS3 account for the majority of MKS in non-consanguineous populations of European origin. Polydactyly is usually found in MKS1 but rare in MKS3. Cases with no, or milder, CNS phenotypes were only found in MKS3; hypomorphic missense mutations may be associated with less severe CNS outcomes. This study is consistent with further genetic heterogeneity of MKS, but underlines the value of molecular diagnostics of the known genes to aid family planning decisions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Meckel–Gruber syndrome (MKS) is a lethal, autosomal recessive disorder in which affected fetuses present with the MKS triad; renal cystic dysplasia (RCD), occipital encephalocele, and postaxial polydactyly of the hands/feet (Alexiev et al. 2006; Salonen 1984; Salonen and Paavola 1998), as well as ductal plate malformations of the liver (Salonen 1984; Sergi et al. 2000). The initial diagnosis is typically obtained between 11 and 16 weeks gestation during routine ultrasound examinations by detection of the central nervous system (CNS) and renal phenotypes (Mittermayer et al. 2004). Phenotypic variability is evident in MKS, with polydactyly observed in 55–83% (Alexiev et al. 2006) of cases, and other CNS malformations, such as hydrocephalus and the Dandy–Walker malformation (DWM) (Cincinnati et al. 2000; Summers and Donnenfeld 1995; Walpole et al. 1991), also linked to MKS. A number of other syndromes present with similar phenotypes (Alexiev et al. 2006): trisomy 13 (holoprosencephaly, cleft lip/palate, congenital heart disease and polydactyly); trisomy 18 (choroid plexus cysts, congenital heart and kidney disease, clenched hands and rocker-bottom feet); Joubert syndrome (JS; vermis hypoplasia/dysplasia, facial abnormalities, cystic renal disease, polydactyly and cleft palate) (Maria et al. 1999), Bardet–Biedl syndrome (BBS; vision loss, mental retardation, renal disease, polydactyly and obesity) (Green et al. 1989), and Smith–Lemli–Opitz syndrome (microcephaly/mental retardation, cardia-pulmonary-renal malformations, and polydactyly). The complexity of the differential diagnoses underlines the need for molecular testing for this syndrome.
The high degree of phenotypic variability suggested genetic heterogeneity (Alexiev et al. 2006; Paavola et al. 1997; Salonen 1984), and linkage analysis of families of various origins mapped three gene loci: MKS1 (Finnish), 17q23 (Paavola et al. 1995); MKS2 (Middle Eastern/North African), 11q13 (Roume et al. 1998); and MKS3 (South Asian), 8q21-24 (Morgan et al. 2002). Positional cloning in affected families and the discovery of an animal model for MKS3 (the wpk rat) led to the identification of the MKS1 (Kyttala et al. 2006) and MKS3 (Smith et al. 2006) genes. Provisional analysis of the MKS proteins, MKS1 and meckelin, suggests involvement in the ciliary/basal body axis, similar to other disorders involving cystic kidneys (Dawe et al. 2007; Kyttala et al. 2006; Smith et al. 2006).
Identification of two MKS genes allows mutation-based molecular diagnostics. Complexity in obtaining a firm diagnosis is illustrated by the finding of BBS mutations in patients with MKS-like phenotypes (Karmous-Benailly et al. 2005) and the recent description of MKS3 (but not MKS1) mutations associated with JS (Baala et al. 2007). These highlight the utility of molecular methods for determining a clear diagnosis, and for defining the full phenotypic characteristics associated with each gene. This paper illustrates the value of molecular diagnostics for MKS1 and MKS3 in typical US/European populations.
Methods
Mutation screening of MKS1 and MKS3
This study was approved by relevant Institutional Review Boards or Ethics Committees, and all participants gave informed consent. Patient families were referred for research testing by genetic counselors or medical geneticists from the US and Europe. Clinical records on each pregnancy were reviewed by an experienced clinician. Blood samples were collected from each parent, and material as available from the fetus, for DNA extraction.
DNA was isolated from whole blood, cultured fibroblasts, or frozen or paraffin-embedded tissue using standard DNA extraction methods (Puregene DNA Purification System; Gentra Systems, Minneapolis, MN); paraffin-embedded tissue was digested with Proteinase K for 7 days at 55°C (Diaz-Cano and Brady 1997).
MacVector software (Accelrys Software Co.; San Diego, CA) was used to design exonic primer pairs with a minimum flanking region of 50 bp (Supplemental Tables S1, S2). PCR amplicons were generated using standard reaction methods: 50 ng DNA was amplified using 5 pmol of each primer (Illumina, Inc.; San Diego, CA), 2 pmol of each dNTP (Invitrogen), 2.0–2.5 mM MgCl2, 2.5 μl of 10x reaction buffer, and 0.5 U of Pt-Taq polymerase (Invitrogen), in a total volume of 25 μl. The PCR amplification program consisted of: 94°C, 2 min., 35 cycles of: 94°C, 30 s; 57–62°C, 30 s; 72°C, 30 s, followed by 72°C, 10 min. and 4°C, 5 min. For the GC-rich exon 1 of MKS1, a DMSO containing PCR reaction buffer (Dodé et al. 1990) was used.
PCR products were purified and sequenced at Agencourt Bioscience Corporation (Beverly, MA) utilizing the Agencourt AMPure purification reagent. Sequencing was with Big Dye Terminator V3.1, followed by purification with Agencourt CleanSeq and injection on an ABI 3730xl DNA Sequencer (Applied Biosystems; Foster City, CA). Sequence was analyzed using the Mutation Surveyor program (SoftGenetics LLC; State College, PA). The significance of possible splicing mutations was tested using the Splice Site Prediction by Neural Network website: http://www.fruitfly.org/seq_tools/splice.html. Multi-sequence alignments were generated with the ClustalW software (MacVector) employing vertebrate orthologs as previously described (Smith et al. 2006) plus other meckelin orthologous sequences: Drosophila melanogaster (D.m), Caenorhabditis elegans (C.e) and Trypanosoma brucei (T.b) obtained from NCBI or Ensembl. Previous description of SNPs was tested at GeneCards: http://www.genecards.org/info.shtml#snp.
RNA analysis
Total RNA was isolated from fibroblasts using TRIzol (Invitrogen Life Technologies; Carlsbad, CA). First-strand cDNA was synthesized from total RNA using the SuperScript III cDNA Synthesis Kit (Invitrogen). Primers within exons 3 and 8 for MKS1 and exons 1 and 4 for MKS3 (Supplemental Tables S1, S2) were designed to study putative splice site mutations. The PCR reactions were similar to those for DNA amplification.
Histology
Paraffin embedded kidney and liver samples from an MKS3 proband (family M329, R1682) were sectioned and stained with hematoxylin and eosin according to standard methods.
Results
Clinical description of the cohort
Descriptions of the 30 MKS diagnosed cases from 17 families are shown in Table 1. Twelve families were from the US (M designation) and five from Europe. Each family had a clinical diagnosis of MKS in at least one pregnancy, which was confirmed by the clinical record. Two families were consanguineous and one from an isolated population. Seven had multiple affected family members, including five in one pedigree (M361). Twenty-six patients had a CNS phenotype; 18 (14 families) had an occipital encephalocele. Other CNS phenotypes included DWM (7 cases; 3 families) and hydrocephalus (1 case). All patients of significant age had RCD that was variously described as polycystic, cystic dysplastic or multicystic. Polydactyly was seen in 10 cases (10 families), with documented involvement of hands and feet in 6. No digital abnormalities were described in 15 cases (7 families), and conclusive information was not available in 5. Histological analysis for liver abnormalities was only performed on a minority of cases, where a ductal plate malformation was noted.
Mutation analysis of MKS1 and MKS3
Proband samples were available for screening in 10 cases (8 families); parents were screened for mutations in the remaining families. All samples were initially screened for the MKS1-Finmajor mutation (IVS15-7_35del; G470fsX562) by PCR and visualized on a 2.5% agarose gel, with wildtype and MKS1-Finmajor bands at 342 and 313 bp, respectively (Fig. 1c). Samples heterozygous for the MKS1-Finmajor deletion were sequenced for just MKS1 to identify the second mutation, while those negative for this mutation were screened for both MKS1 and MKS3.
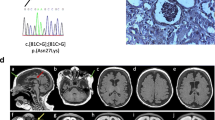
Details of MKS1 mutations. a The synonymous 417G > A mutation (family M340), showing (bottom) genomic DNA sequence (wildtype; wt and mutant; M); and (top) RT-PCR (ex 3–8) indicating that exon 4 is skipped in the proband (P) but not the wildtype (WT). b Sequence change IVS11 + 1G > A in pedigree M338. c Agarose gel electrophoresis of IVS15-Ex16 (Table S1) PCR product. The proband in M380 is homozygous and the parents heterozygous for the deletion. The larger product in the heterozygotes is a heteroduplex. The positions of the mutations are shown at the bottom against the MKS1 intron/exon structure
Five families were MKS1-Finmajor positive (M338, M340 and 55875 were heterozygous; M380 and M383 homozygous; Table 1; Fig. 1c). MKS1 sequence analysis identified two additional mutations: a typical splicing change, IVS11 + 1G > A, in M338 (Fig. 1b), and a synonymous variant, 417G > A, in M340 and 55875 (Fig. 1a; Tables 1, 2). In each family, the MKS1-Finmajor and other mutation were found to originate from different parents, consistent with recessive inheritance.
The 417G > A substitution, affecting the last nucleotide of exon 4 (Fig. 1a), was analyzed for its possible effect on splicing, which predicted the loss of the exon 4 donor site (normal score 0.14). RT-PCR of fibroblasts from the M340 proband (R1646) showed a smaller abnormal band (∼450 bp; Fig. 1a). Sequencing of the excised abnormal product confirmed exon 4 was skipped due to the 417G > A mutation. In total, five families were shown to be MKS1.
Sequencing the twelve MKS1 negative families identified seven novel MKS3 changes in five families (Fig. 2a–g; Tables 1, 2). Segregation analysis showed that all changes were derived from separate parents, consistent with their pathogenic status. Family M329 had two nonsense mutations: R208X (exon 6; Fig. 2f) and R451X (exon 13; Fig. 2g), and family 68408 had a nonsense (R208X) and a frameshifting deletion (579delA; Fig. 2e) mutation in exon 6.
Details of MKS3 mutations (a–g) as indicated. Sequence of each change is indicated (wildtype; wt and mutant; M). The amino acid sequence is shown below and the intron/exon boundary as a vertical red line in a. Reverse sequence is shown in a and so the deletion is not evident in adjacent nucleotides because of the poly-T tract. b–d Conservation of substitutions (red base) in alignment of mecklin orthologs in: human (H.s); rat (R.n); mouse (M.m); chicken (G.g); fish (Tetraodon nigroviridis; T.n); Drosophila melanogaster (D.m); nematode (Caenorhabditis elegans; C.e) and Trypanosoma brucei (T.b). The end of the seventh transmembrane domain is shown in green in d. The position of each mutation against the intron/exon structure of MKS3 is shown in the center
The proband (R1726) of family M376 had a splicing mutation (IVS1-2delA) that RT-PCR analysis and sequencing showed resulted in the skipping of exon 2 (Fig. 2a). Interestingly, the delA immediately follows a run of 24 T-nucleotides preceding exon 2. In this case, the second likely mutation was the M252T substitution in exon 8. Comparative mapping of meckelin orthologs shows M252 is highly conserved in vertebrates as well as nematode (methionine is conservatively replaced with leucine in Drosophila and Trypanosoma), supporting its pathogenic role (Fig. 2b). This conclusion was reinforced by finding the same M252T mutation in a second family (M360), which also had the R208X nonsense mutation (Table 1).
Two missense changes were found in family M361: R440Q (exon 13) and L966P (exon 27). R440 is located in the N-terminal extracellular domain, ∼15 aa before the start of the first transmembrane domain. Homology mapping showed that this residue is conserved in all species save Drosophila, where arginine is replaced by lysine (Fig. 2c). L966, the last amino acid of the final transmembrane domain of meckelin, is conserved to chicken, and is substituted to a similarly hydrophobic valine or alanine in other species (Fig. 2d). Proline is a kinking amino acid and may disrupt the α-helical transmembrane tertiary structure of this domain. Further support that these two missense mutations are pathogenic was provided by direct sequencing segregation analysis using genomic DNA from three terminated fetuses of this family (R1730–R1732).
A number of intronic and exonic polymorphisms were also identified in MKS1 and MKS3 and their frequencies determined in the US and Dutch populations (Table S3). Four were common, described SNPs (Table S3). Two others were splicing changes distant from the splice sites, which were not predicted to alter splicing (Reese et al. 1997). The one rare change, IVS12 + 19G > A, was heterozygous in a consanguineous family and therefore unlikely disease related. One silent MKS3 change, 2397T/C, was found in a parent in M345. As no other MKS3 change was found in the other parent, and the change was synonymous we judged it unlikely to be pathogenic. One rare missense change, D/E71, in MKS1: is highly conservative; at a residue that is glutamic acid (E) in all other tested mammalian species; and was found in a patient with a more likely MKS1 mutation (IVS11 + 1G > A).
MKS3 kidney and liver histology
Material from a 20 week MKS3 fetus allowed the renal and liver phenotypes associated with this genotype to be analyzed. The renal architecture was distorted by numerous, haphazardly arranged and variably-sized cortical and medullary cysts with intervening, loose edematous stroma that was variably cellular, typical of RCD. A subcapsular nephrogenic zone was present only focally, and showed normal maturation; however, nephron number appeared markedly decreased (Fig. 3a). Cysts were variably sized, with smaller cysts lined by more cuboidal cells with eosinophilic cytoplasm, while larger cysts were lined by flattened epithelium. Within a number of cysts the lining epithelium transitioned from cells with eosinophilic or cleared cytoplasm to more flattened cells with basophilic cytoplasm.
The liver architecture was intact without grossly evident cysts. The portal regions were expanded by loose connective tissue stroma with peripherally arranged proliferating bile ducts (Fig. 3b). The bile ducts were focally ectatic, but only minimally dilated. There was no significant portal inflammation and the portal vein and hepatic artery appeared normal.
Discussion
We have developed a molecular diagnostic screen for MKS that couples PCR screening of the MKS1-Finmajor founder mutation with direct sequence analysis of MKS1 and MKS3. Most genetic and epidemiological studies of MKS have concentrated on isolated populations (i.e. Finnish), or ones with a high level of consanguinity. In this study, most families were typical (non-consanguineous) and diagnosed in the US or The Netherlands. The out-bred nature of the population was reflected in the fact that all but two of the ten mutation proven cases were compound heterozygotes. The prevalence of MKS in Finland has been estimated at 1:9,000 (Salonen and Norio 1984), but figures have varied widely from 1:13,250 (Holmes et al. 1976) to 1:140,000 (Seller 1978) for the US or other European populations. A relatively high rate is supported by the fact that MKS is the most common syndromic form of neural tube defects and polydactyly (Castilla et al. 1998; Simpson et al. 1991; Stevenson et al. 2000). Recent data showing the prevalence of MKS in fetuses diagnosed with hyperechoic cystic kidneys at about one-third the incidence of autosomal recessive polycystic kidney disease (ARPKD) in the same population (frequency ∼1:20,000) (Chaumoitre et al. 2006) suggests a level of ∼1:60,000 could be used for estimating risk, with a carrier rate of ∼1:250. This may be higher when JS cases associated with MKS3 mutations (Baala et al. 2007) are included, indicating a moderate level of demand for molecular diagnostics.
Of the ten families molecularly diagnosed with MKS, 5 were MKS1 and 5 MKS3, indicating equal representation of the two diseases in this population. All MKS1 cases had at least one MKS1-Finmajor allele (7/10 mutant alleles; 2 were homozygous). This level is similar to the non-Finnish families in the previous MKS1 study (6/8) (Kyttala et al. 2006), indicating this is a common mutation in European derived populations, and prescreening patients for this readily detectible change is helpful in a diagnostics setting. Two novel MKS1 mutations were also identified: 417G > A in 2 patients, and IVS11 + 1G > A in an African American patient.
Although MKS3 was previously only described in consanguineous families from South Asia and the Middle East, it is clear that it is a common cause of MKS in Western populations. A total of seven novel mutations were identified, two of which were found on five alleles. There is strong evidence that the three MKS3 missense changes are pathogenic: they were not found on 50 control chromosomes from the same populations; they are at highly conserved sites (Fig. 2); M252T was found in two separate families with a MKS3 nonsense or typical splicing change; L966P and R440Q segregated with the disease in three affected offspring; one previously described homozygous missense change, Q376P, was found in a consanguineous MKS3 family (Smith et al. 2006); and only two very conservative amino acid polymorphisms were found in MKS3 (Table S3). While there is clearly more allelic heterogeneity in this population, the finding of some ancestral changes may prioritize exons that are initially analyzed; 4/10 mutant alleles were in exon 6.
All patients where data was available had RCD. Analysis of the MKS3 phenotype showed cortical and medullary cysts with just focal regions of nephrogenic development, similar to previous descriptions of RCD in MKS kidneys (Fig. 3a) (Bernstein et al. 1974). Only a minority of patients had histological data available on the liver, but each of those showed ductal plate malformations. The illustrated MKS3 liver shows the ductal plate malformation, with bile duct proliferation, characteristic of MKS (Sergi et al. 2000), although little dilatation is seen in this case (Fig. 3b).
In all the mutation characterized families, at least one case had an occipital encephalocele. Previous studies have indicated an encephalocele in ∼90% of MKS cases (Sergi et al. 2000), but that it may be less common in MKS3 families (Morgan et al. 2002). Here, this abnormality was found in all MKS1 fetuses (6), but four MKS3 cases from two families did not have an encephalocele, one having DWM, two having no CNS defect, and one described as having an elongated head. Interestingly, three of these were from the same family (M361), which had two MKS3 missense mutations. This raises the possibility that allelic effects may be important to the severity of the CNS phenotype, and that missense changes may be hypomorphic alleles. This is analogous to ARPKD where all cases with two truncating mutations die by the neonatal period, but those with one or more missense change more often survive (Bergmann et al. 2003). A finding that further strengthens this possibility is that MKS3 mutations can also cause JS, a non-lethal disorder with a CNS phenotype of cerebellar vermis hypoplasia (Baala et al. 2007). In JS cases, most mutations are missense or atypical splicing, which might be hypomorphic alleles and explain the milder phenotype; although, oligogenic inheritance could also play a role in the phenotypic expression (Baala et al. 2007).
The phenotype that was most clearly different between MKS1 and MKS3 was the digital abnormalities. Of the five MKS1 cases where data was available, all showed polydactyly, many of both fingers and toes (Table 1). In contrast, of the 11 MKS3 fetuses where digits could be visualized, only 1 had polydactyly; six toes on one foot. Previous studies suggested polydactyly to be less common in MKS3 (Morgan et al. 2002; Smith et al. 2006), and this is now clearly demonstrated. Such is the phenotypic difference that this feature provides an early clue to the gene involved before molecular analysis is initiated. Although roles in the ciliary/basal body axis are suspected for the MKS proteins (Dawe et al. 2007; Kyttala et al. 2006; Smith et al. 2006), it seems from the phenotypic differences between MKS1 and MKS3 that some differences in protein function can be expected.
Mutations at either gene were not identified in seven families. While it is possible that these represent missed mutations (i.e. large deletions or splicing mutations distant from the intron/exon boundaries), the fact that two mutations were identified in all our molecularly diagnosed cases makes this unlikely. There was some doubt regarding the MKS diagnosis in four of these families. In family 68420, no frank neural tube defect or polydactyly was present, plus other congenital abnormalities (such as genital abnormalities) were present. In M337, DWM was observed in three cases, with no clear polydactyly; and one fetus was diagnosed with trisomy 18 (the other two were karyotypically normal). A third family of Ashkenazi Jewish decent had three cases presenting with DWM and no polydactyly. M345 was atypical in that cleft palate was detected. The remaining mutation negative cases, two consanguineous families (63531 and 68281) and one non-consanguineous family (M381), presented with the typical MKS triad. A further MKS locus (such as MKS2) is therefore supported by these cases, but family samples were not available for linkage analysis.
The schema that we have described here will be useful for determining a firm diagnosis in typical MKS cases, as well as allowing testing of atypical phenotypes, such as JS. The devastating nature of MKS indicates that preimplantation genetic diagnostics (PGD) is an important option for these families, which a firm molecular diagnosis makes possible.
References
Alexiev BA, Lin X, Sun CC, Brenner DS (2006) Meckel–Gruber syndrome: pathologic manifestations, minimal diagnostic criteria, and differential diagnosis. Arch Pathol Lab Med 130:1236–1238
Baala L, Romano S, Khaddour R, Saunier S, Smith UM, Audollent S, Ozilou C, Faivre L, Laurent N, Foliguet B, Munnich A, Lyonnet S, Salomon R, Encha-Razavi F, Gubler MC, Boddaert N, Lonlay P, Johnson CA, Vekemans M, Antignac C, Attie-Bitach T (2007) The Meckel–Gruber syndrome gene, MKS3, is mutated in Joubert syndrome. Am J Hum Genet 80:186–194
Bergmann C, Senderek J, Sedlacek B, Pegiazoglou I, Puglia P, Eggermann T, Rudnik-Schneborn S, Furu L, Onuchic LF, De Baca M, Germino GG, Guay-Woodford L, Somlo S, Moser M, Buttner R, Zerres K (2003) Spectrum of mutations in the gene for autosomal recessive polycystic kidney disease (ARPKD/PKHD1). J Am Soc Nephrol 14:76–89
Bernstein J, Brough AJ, McAdams AJ (1974) The renal lesion in syndromes of multiple congenital malformations. Cerebrohepatorenal syndrome; Jeune asphyxiating thoracic dystrophy; tuberous sclerosis; Meckel syndrome. Birth Defects Orig Artic Ser 10:35–43
Castilla EE, Lugarinho R, da Graca Dutra M, Salgado LJ (1998) Associated anomalies in individuals with polydactyly. Am J Med Genet 80:459–465
Chaumoitre K, Brun M, Cassart M, Maugey-Laulom B, Eurin D, Didier F, Avni EF (2006) Differential diagnosis of fetal hyperechogenic cystic kidneys unrelated to renal tract anomalies: a multicenter study. Ultrasound Obstet Gynecol 28:911–917
Cincinnati P, Neri ME, Valentini A (2000) Dandy–Walker anomaly in Meckel–Gruber syndrome. Clin Dysmorphol 9:35–38
Dawe HR, Smith UM, Cullinane AR, Gerrelli D, Cox P, Badano JL, Blair-Reid S, Sriram N, Katsanis N, Attie-Bitach T, Afford SC, Copp AJ, Kelly DA, Gull K, Johnson CA (2007) The Meckel–Gruber syndrome proteins MKS1 and meckelin interact and are required for primary cilium formation. Hum Mol Genet 16:173–186
Diaz-Cano SJ, Brady SP (1997) DNA extraction from formalin-fixed, paraffin-embedded tissues: protein digestion as a limiting step for retrieval of high-quality DNA. Diagn Mol Pathol 6:342–346
Dodé C, Rochette J, Krishnamoorthy R (1990) Locus assignment of human α globin mutations by selective amplification and direct sequencing. Br J Haematol 76:275–281
Green JS, Parfrey PS, Harnett JD, Farid NR, Cramer BC, Johnson G, Heath O, McManamon PJ, O’Leary E, Pryse-Phillips W (1989) The cardinal manifestations of Bardet–Biedl syndrome, a form of Laurence–Moon–Biedl syndrome. N Engl J Med 321:1002–1009
Holmes LB, Driscoll SG, Atkins L (1976) Etiologic heterogeneity of neural-tube defects. N Engl J Med 294:365–369
Karmous-Benailly H, Martinovic J, Gubler MC, Sirot Y, Clech L, Ozilou C, Auge J, Brahimi N, Etchevers H, Detrait E, Esculpavit C, Audollent S, Goudefroye G, Gonzales M, Tantau J, Loget P, Joubert M, Gaillard D, Jeanne-Pasquier C, Delezoide AL, Peter MO, Plessis G, Simon-Bouy B, Dollfus H, Le Merrer M, Munnich A, Encha-Razavi F, Vekemans M, Attie-Bitach T (2005) Antenatal presentation of Bardet–Biedl syndrome may mimic Meckel syndrome. Am J Hum Genet 76:493–504
Kyttala M, Tallila J, Salonen R, Kopra O, Kohlschmidt N, Paavola-Sakki P, Peltonen L, Kestila M (2006) MKS1, encoding a component of the flagellar apparatus basal body proteome, is mutated in Meckel syndrome. Nat Genet 38:155–157
Maria BL, Boltshauser E, Palmer SC, Tran TX (1999) Clinical features and revised diagnostic criteria in Joubert syndrome. J Child Neurol 14:583–590; discussion 590–581
Mittermayer C, Lee A, Brugger PC (2004) Prenatal diagnosis of the Meckel–Gruber syndrome from 11th to 20th gestational week. Ultraschall Med 25:275–279
Morgan NV, Gissen P, Sharif SM, Baumber L, Sutherland J, Kelly DA, Aminu K, Bennett CP, Woods CG, Mueller RF, Trembath RC, Maher ER, Johnson CA (2002) A novel locus for Meckel–Gruber syndrome, MKS3, maps to chromosome 8q24. Hum Genet 111:456–461
Paavola P, Salonen R, Weissenbach J, Peltonen L (1995) The locus for Meckel syndrome with multiple congenital anomalies maps to chromosome 17q21-q24. Nat Genet 11:213–215
Paavola P, Salonen R, Baumer A, Schinzel A, Boyd PA, Gould S, Meusburger H, Tenconi R, Barnicoat A, Winter R, Peltonen L (1997) Clinical and genetic heterogeneity in Meckel syndrome. Hum Genet 101:88–92
Reese MG, Eeckman FH, Kulp D, Haussler D (1997) Improved splice site detection in Genie. J Comput Biol 4:311–323
Roume J, Genin E, Cormier-Daire V, Ma HW, Mehaye B, Attie T, Razavi-Encha F, Fallet-Bianco C, Buenerd A, Clerget-Darpoux F, Munnich A, Le Merrer M (1998) A gene for Meckel syndrome maps to chromosome 11q13. Am J Hum Genet 63:1095–1101
Salonen R (1984) The Meckel syndrome: clinicopathological findings in 67 patients. Am J Med Genet 18:671–689
Salonen R, Norio R (1984) The Meckel syndrome in Finland: epidemiologic and genetic aspects. Am J Med Genet 18:691–698
Salonen R, Paavola P (1998) Meckel syndrome. J Med Genet 35:497–501
Seller MJ (1978) Meckel syndrome and the prenatal diagnosis of neural tube defects. J Med Genet 15:462–465
Sergi C, Adam S, Kahl P, Otto HF (2000) Study of the malformation of ductal plate of the liver in Meckel syndrome and review of other syndromes presenting with this anomaly. Pediatr Dev Pathol 3:568–583
Simpson JL, Mills J, Rhoads GG, Cunningham GC, Conley MR, Hoffman HJ (1991) Genetic heterogeneity in neural tube defects. Ann Genet 34:279–286
Smith UM, Consugar M, Tee LJ, McKee BM, Maina EN, Whelan S, Morgan NV, Goranson E, Gissen P, Lilliquist S, Aligianis IA, Ward CJ, Pasha S, Punyashthiti R, Malik Sharif S, Batman PA, Bennett CP, Woods CG, McKeown C, Bucourt M, Miller CA, Cox P, Algazali L, Trembath RC, Torres VE, Attie-Bitach T, Kelly DA, Maher ER, Gattone VH, Harris PC, Johnson CA (2006) The transmembrane protein meckelin (MKS3) is mutated in Meckel–Gruber syndrome and the wpk rat. Nat Genet 38:191–196
Stevenson RE, Allen WP, Pai GS, Best R, Seaver LH, Dean J, Thompson S (2000) Decline in prevalence of neural tube defects in a high-risk region of the United States. Pediatrics 106:677–683
Summers MC, Donnenfeld AE (1995) Dandy–Walker malformation in the Meckel syndrome. Am J Med Genet 55:57–61
Walpole IR, Goldblatt J, Hockey A, Knowles S (1991) Dandy–Walker malformation (variant), cystic dysplastic kidneys, and hepatic fibrosis: a distinct entity or Meckel syndrome? Am J Med Genet 39:294–298
Acknowledgments
The many physicians, genetic counselors, and families are thanked for their involvement in this study, and for providing comprehensive clinical records. This work was supported by NIDDK grant DK68581.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Consugar, M.B., Kubly, V.J., Lager, D.J. et al. Molecular diagnostics of Meckel–Gruber syndrome highlights phenotypic differences between MKS1 and MKS3. Hum Genet 121, 591–599 (2007). https://doi.org/10.1007/s00439-007-0341-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-007-0341-3