Abstract
A genome wide linkage analysis of nonsyndromic deafness segregating in a consanguineous Pakistani family (PKDF537) was used to identify DFNB63, a new locus for congenital profound sensorineural hearing loss. A maximum two-point lod score of 6.98 at θ = 0 was obtained for marker D11S1337 (68.55 cM). Genotyping of 550 families revealed three additional families (PKDF295, PKDF702 and PKDF817) segregating hearing loss linked to chromosome 11q13.2-q13.3. Meiotic recombination events in these four families define a critical interval of 4.81 cM bounded by markers D11S4113 (68.01 cM) and D11S4162 (72.82 cM), and SHANK2, FGF-3, TPCN2 and CTTN are among the candidate genes in this interval. Positional identification of this deafness gene should reveal a protein necessary for normal development and/or function of the auditory system.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nonsyndromic sensorineural hearing loss (NSHL) is a genetically heterogeneous neurosensory disorder (Marazita et al. 1993). So far, 52 recessive deafness loci (DFNB) have been reported in peer reviewed journals and 23 of the corresponding nuclear genes have been cloned (Petersen and Willems 2006; Morton and Nance 2006). This is not surprising since approximately 1% of the human protein-coding genes are thought to be necessary for inner ear function (Friedman and Griffith 2003).
Consanguineous families segregating deafness in the Pakistani population represent a rich genetic resource for the identification of the genes that will be helpful in understanding the molecular and cellular biology of hearing. To this end, we have ascertained approximately 800 Pakistani families segregating hearing loss and we are conducting genome wide linkage screens on families where the deafness is unlinked to reported loci for inherited hearing loss. Here we report four families in which deafness is linked to the DFNB63 locus on chromosome 11q13.2-q13.3 (Fig. 1).
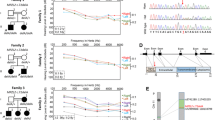
DFNB63 families a Chromosome 11 haplotypes in family PKDF537. Filled symbols represent deaf individuals. The linked haplotype is boxed in all generations. The STR markers and their relative human genetic map positions in centiMorgans (cM) according to Marshfield human genetic map are shown along with the pedigrees. Haplotype analysis of PKDF537 shows a linkage region of 13.55 cM delimited by markers D11S4191 (60.09 cM) and D11S1314 (73.64 cM). Affected individual VI:8 provided the proximal recombination breakpoint at marker D11S4191 (60.09 cM). The distal breakpoint at marker D11S1314 (73.64 cM) was provided by affected individual IX:2. b–d Pedigrees of three additional families PKDF702, PKDF295 and PKDF817 are also segregating recessive deafness linked to the DFNB63 interval on chromosome 11q13.2-q13.3
Materials and methods
Subject enrollment
Approval for this study was obtained from the Institutional Review Board (IRB) at National Center of Excellence in Molecular Biology, Lahore, Pakistan (FWA00001758) and the NIDCD/NINDS IRB at the National Institutes of Health, USA (OH-93-N-016). Written informed consent was obtained from study participants from the Punjab and Sindh provinces of Pakistan.
Clinical evaluation
Families were evaluated for hearing loss by pure tone audiometry using air and bone conductions (frequencies ranging from 250 to 8,000 Hz). Clinical histories were obtained from participating family members to rule out obvious environmental causes of hearing loss and physical evaluations were undertaken to verify that deafness was nonsyndromic. Vestibular function was evaluated in the field by testing tandem gait ability and by using the Romberg test. A funduscopic examination was performed to rule out retinitis pigmentosa.
DNA isolation, genotyping and linkage analysis
Blood was obtained through venipuncture and genomic DNA was extracted using a standard protocol (Grimberg et al. 1989). Linkage to reported DFNB loci was excluded using STR (short tandem repeat) markers (http://www.uia.ac.be/dnalab/hhh/). Genome wide scans were performed using 388 fluorescently labeled microsatellite markers spaced at an average interval of 10 cM across the genome (ABI Prism Linkage Mapping Set, v2.5 Applied Biosystems). Markers were amplified by the polymerase chain reaction (PCR) on a Gene Amp PCR system 9700, analyzed on an ABI Prism 3100 Genetic Analyzer and the alleles assigned using Genescan and Genotyper (Applied Biosystems).
Lod score calculations
Lod scores were calculated using the FASTLINK computer package (Schaffer 1996) and MLINK was used for two-point lod scores. A fully penetrant recessive model with no phenocopies and a disease allele frequency of 0.001 was assumed. Microsatellite markers positions and map distances were derived from the Marshfield genetic map (http://www.research.marshfieldclinic.org/). Meiotic recombination frequencies were considered to be equal for males and females and allele frequencies for microsatellite markers were calculated by genotyping 100 unrelated unaffected individuals from the same population.
Candidate gene screening
Candidate genes in the DFNB63 interval on chromosome 11q13.2-q13.3 were identified using the UCSC Genome Bioinformatics web browser (UCSC Genome Bioinformatics: http://www.genome.ucsc.edu/) and were initially selected for mutation screening on the basis of their expression or function in the inner ear. Primers used for PCR amplification and subsequent sequencing of SHANK2 were designed from the flanking region of each exon using Primer3 web utility (http://www.frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi/). Amplification, sequencing reactions and mutation analysis were carried out as described previously (Shabbir et al. 2006).
Results
All affected individuals in four Pakistani families reported here displayed congenital bilateral profound hearing loss. Although not rigorously tested using posturography and a rotary chair, vestibular function appeared to be normal in affected individuals and clinical evaluation suggested no skin or renal anomalies. Funduscopic examinations of affected individuals showed no signs of retinitis pigmentosa. We therefore concluded that no other disorder was co-segregating with hearing loss in these four families.
Deafness segregating in family PKDF537 was not linked to the reported recessive deafness loci. Therefore, a genome wide linkage analysis was undertaken and deafness segregating in family PKDF537 showed initial evidence of linkage to chromosome 11q12.2-q13.4. Additional markers were genotyped and haplotype analysis revealed a 13.55 cM interval of homozygosity delimited by markers D11S4191 (60.09 cM) and D11S1314 (73.64 cM; Fig. 1). A maximum two-point lod score (Z max) of 6.98 at recombination fraction θ = 0 was obtained for the marker D11S1337 (68.55 cM; Table 1). DFNB63 linked STR markers were used to screen 550 Pakistani families segregating recessive deafness. During this screening, deafness in three additional families (PKDF295, PKDF702 and PKDF817) was found to be linked to DFNB63 (Fig. 1; Table 1). The proximal boundary was given by the marker D11S4113 (68.01 cM) in PKDF817, whereas PKDF702 provided a distal breakpoint at marker D11S4162 (72.82 cM) defining a critical interval of 4.81 cM (2.13 Mb; Fig. 2).
DFNB63 linkage intervals of hearing loss in families PKDF295, PKDF537, PPKDF702 and PKDF817 on human chromosome 11q12.2-q13.4. STR markers are represented by filled circles. The sex averaged recombination distances in cM and in Mb are indicated along with STR markers. Cytogenetic localizations of some candidate genes in this interval are indicated. The genetic linkage distances are from the Center for Medical Genetics, Marshfield Medical Research Foundation Web site (http://www.research.marshfieldclinic.org/genetics). The Mb positions of markers and genes are from the May 2004 NCBI build 35 at http://www.genome.cse.ucsc.edu/cgi-bin/hgGateway
Discussion
Haplotype analysis of four chromosome 11q13.2-q13.3 linked families revealed a 4.81 cM region of homozygosity for DFNB63. Families PKDF702 and PKDF817 were presumed to be unrelated but have the same haplotype across this interval. Families PKDF295 and PKDF537 each have unique haplotypes across this region, and probably harbor different mutant deafness alleles. Thus there are three distinct haplotypes across the four families. The maximum two point lod scores were obtained for different markers except for PKDF702 and PKDF817, indicating a possible involvement of more than one deafness causing genes in DFNB63 linked families. The DFNB63 locus does not overlap with reported chromosome 11 deafness loci that include DFNB2/USH1B/DFNA11 (Weil et al. 1997), DFNB18/USH1C (Ahmad et al. 2002), DFNB20 (Moynihan et al. 1999), DFNB21/DFNA8 (Mustafa et al. 1999), DFNB24 (R. Smith, personal communication), DFNB51 (Shaikh et al. 2005) and Jervell and Lange-Nielsen syndrome type 1 (JLNS1) (Neyroud et al. 1997).
Among the candidate deafness genes in the critical DFNB63 interval are SHANK2, FGF-3, TPCN2 and CTTN. The scaffold protein SHANK2, which has a SH3 (Src homology 3) domain, a PDZ domain and seven ankyrin repeats, is involved in intracellular signaling cascades and protein–protein interactions (Han et al. 2006). We sequenced the 24 coding and noncoding exons of SHANK2 in four families, but a mutation was not found. Based on their domain content and expression in the inner ear additional genes will be screened for mutant alleles associated with DFNB63.
In the mouse embryo Fgf3 is expressed in the inner ear, cerebellum, retina and teeth of mouse embryo (Wilkinson et al. 1989) and mice homozygous for a targeted disruption of Fgf3 have developmental defects of the inner ear and tail (Mansour et al. 1993). Another candidate, two-pore segment channel 2 (TPCN2), encodes a Ca2+/Na+ cation channel (Huang et al. 2006). Several ion channels and pumps are necessary to maintain the endocochlear potential and the unique ionic composition of the endolymph (Kubish et al. 1999; Kharkovets et al. 2000; Marcus et al. 2002; Wangemann et al. 1995).
Mapping a new locus for nonsyndromic recessive deafness to chromosome 11 in four Pakistani families emphasizes the genetic heterogeneity of this disorder. The identification of the gene responsible for deafness linked to DFNB63 is expected to deepen our understanding of the hearing process.
Electronic database information
URLs for data in this article are as follows:
Center for Medical Genetics, Marshfield Medical Research Foundation: http://www.research.marshfieldclinic.org/genetics/
Hereditary Hearing Loss Homepage: http://www.uia.ac.be/dnalab/hhh/
Primer3 Web-Based Server (primer3_www.cgi v 0.2): http://www.frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi
UCSC Genome Bioinformatics: http://www.genome.ucsc.edu/
References
Ahmed ZM, Smith TN, Riazuddin S, Makishima T, Ghosh M, Bokhari S, Menon PSN, Deshmukh D, Griffith AJ, Riazuddin S, Friedman TB, Wilcox ER (2002) Nonsyndromic recessive deafness DFNB18 and Usher syndrome type IC are allelic mutations of USHIC. Hum Genet 110:527–531
Friedman TB, Griffith AJ (2003) Human nonsyndromic sensorineural deafness. Annu Rev Genomics Hum Genet 4:341–402
Grimberg J, Nawoschik S, Belluscio L, McKee R, Turck A, Eisenberg A (1989) A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res 17:8390
Han W, Kim KH, Jo MJ, Lee JH, Yang J, Doctor RB, Moe OW, Lee J, Kim E, Lee MG (2006) Shank2 associates with and regulates Na+/H+ exchanger 3. J Biol Chem 281:1461–1469
Huang X, Godfrey TE, Gooding WE, McCarty KS Jr, Gollin SM (2006) Comprehensive genome and transcriptome analysis of the 11q13 amplicon in human oral cancer and synteny to the 7F5 amplicon in murine oral carcinoma. Genes Chromosomes Cancer 45:1058–1069
Kharkovets T, Hardelin JP, Safieddine S, Schweizer M, El-Amraoui A, Petit C, Jentsch Tj (2000) KCNQ4, a K+ channel mutated in a form of dominant deafness, is exoressed in the inner ear and central auditory pathway. Proc Natl Acad Sci USA 97:4333–4338
Kubisch C, Schroeder BC, Friedrich T, Lutjohann B, El-Amraoui A, Marlin S, Petit C, Jentsch TJ (1999) KCNQ4, a novel potassium channel expressed in sensory outer hair cells, is mutated in dominant deafness. Cell 96:437–446
Mansour SL, Goddard JM, Capecchi MR (1993) Mice homozygous for a targeted disruption of the proto-oncogene int-2 have developmental defects in the tail and inner ear. Development 117:13–28
Marazita ML, Ploughman LM, Rawlings B, Remington E, Arnos KS, Nance WE (1993) Genetic Epedimiological studies of early onset deafness in U. S. School-age population. Am J Med Genet 46:486–491
Marcus DC, Wu T, Wangemann P, Kofuji P (2002) KCNJ10 (Kir4.1) potassium channel knockout abolishes endocochlear potential. Am J Physiol 282:C403–C407
Morton CC, Nance WE (2006) Newborn hearing screening—a silent revolution. N Engl J Med 18:2151–2164
Moynihan L, Houseman M, Newton V, Mueller R, Lench N (1999) DFNB20, a novel locus for autosomal recessive, non-syndromal sensorineural hearing loss maps to chromosome 11q25-qter. Eur J Hum Genet 7:243–246
Mustapha M, Weil D, Chardenoux S, Elias S, El-Zir E, Beckmann JS, Loiselet J, Petit C (1999) An alpha-tectorin gene defect causes a newly identified autosomal recessive form of sensorineural pre-lingual non-syndromic deafness, DFNB21. Hum Mol Genet 8:409–412
Neyroud N, Tesson F, Denjoy I, Leibovici M, Donger C, Barhanin J, Faure S, Gary F, Coumel P, Petit C, Schwartz K, Guicheney P (1997) A novel mutation in the potassium channel gene KVLQT1 causes the Jervell and Lange-Nielsen cardioauditory syndrome. Nat Genet 15:186–189
Petersen MB, Willems PJ (2006) Nonsyndromic, autosomal-recessive deafness. Clin Genet 69:371–392
Schaffer AA (1996) Faster linkage analysis computations for pedigrees with loops or unused alleles. Hum Hered 46:226–235
Shabbir MI, Ahmed ZM, Khan SY, Riazuddin S, Waryah AM, Khan SN, Camps RD, Gosh M, Kabra M, Belyantseva IA, Friedman TB, Riazuddin S (2006) Mutations of human TMHS cause recessively inherited non-syndromic hearing loss. J Med Genet 43(8):634–640
Shaikh RS, Ramzan K, Nazli S, Sattar S, Khan SN, Riazuddin S, Ahmed ZM, Friedman TB, Riazuddin S (2005) A new locus for nonsyndromic deafness DFNB51 maps to chromosome 11p13-p12. Am J Med Genet 138(4):392–395
Wangemann P, Liu J, Marcus DC (1995) Ion transport mechanisms responsible for K+ secretion and the transepithelial voltage across marginal cells of stria vascularis in vitro. Hear Res 84:19–29
Weil D, Kussel P, Blanchard S, Levy G, Levi-Acobas F, Drira M, Ayadi H, Petit C (1997) The autosomal recessive isolated deafness, DFNB2, and the Usher 1B syndrome are allelic defects of the myosin-VIIA gene. Nat Genet 16:191–193
Wilkinson DG, Bhatt S, McMahon AP (1989) Expression pattern of the FGF-related proto-oncogene int-2 suggest multiple roles in fetal development. Development 105:131–136
Acknowledgments
The authors are grateful to the families who made this research possible. We are also thankful to Julie Schultz, Linda Peters, Byung-Yoon Choi, Rob Morell and Dennis Drayna for their suggestions regarding this manuscript. This study was supported by the Higher Education Commission, Islamabad, Pakistan; Ministry of Science and Technology, Islamabad, Pakistan and by the International Center for Genetic Engineering and Biotechnology, Trieste, Italy under project CRP/PAK02-01 (contract no. 02/013) and by intramural funds from the National Institute on Deafness and Other Communication Disorders, NIH (1 ZO1 DC000039-09).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Khan, S.Y., Riazuddin, S., Tariq, M. et al. Autosomal recessive nonsyndromic deafness locus DFNB63 at chromosome 11q13.2–q13.3. Hum Genet 120, 789–793 (2007). https://doi.org/10.1007/s00439-006-0275-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-006-0275-1