Abstract
Pathogenic mutations in TMPRSS3, which encodes a transmembrane serine protease, cause non-syndromic deafness DFNB8/10. Missense mutations map in the low density-lipoprotein receptor A (LDLRA), scavenger-receptor cysteine-rich (SRCR), and protease domains of the protein, indicating that all domains are important for its function. TMPRSS3 undergoes proteolytic cleavage and activates the ENaC sodium channel in a Xenopus oocyte model system. To assess the importance of this gene in non-syndromic childhood or congenital deafness in Turkey, we screened for mutations affected members of 25 unrelated Turkish families. The three families with the highest LOD score for linkage to chromosome 21q22.3 were shown to harbor P404L, R216L, or Q398X mutations, suggesting that mutations in TMPRSS3 are a considerable contributor to non-syndromic deafness in the Turkish population. The mutant TMPRSS3 harboring the novel R216L missense mutation within the predicted cleavage site of the protein fails to undergo proteolytic cleavage and is unable to activate ENaC, thus providing evidence that pre-cleavage of TMPRSS3 is mandatory for normal function.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Congenital hearing loss is the most common sensory defect in humans, with an incidence of about one in 1000 births. One additional child in 1000 becomes deaf before adulthood. Hearing loss is classified as syndromic if deafness is associated with other manifestations, or as non-syndromic if deafness is the isolated phenotype. Approximately half of childhood deafness cases have a genetic origin (reviewed in Petit 1996). Pathogenic mutations in 38 different genes have already been described for non-syndromic deafness and 43 other loci have been described (http://www.uia.ac.be/dnalab/hhh/). Despite this extensive heterogeneity, mutations involving a single locus, DFNB1, account for most cases of genetic non-syndromic deafness in most populations (Nance and Kearsey 2004). DFNB1 corresponds to mutations in GJB2 encoding connexin26. We have described the genetic cause of the recessive non-syndromic deafness DFNB8/10 that map on chromosome 21 (Bonne-Tamir et al. 1996; Veske et al. 1996), which is due to pathogenic mutations in the TMPRSS3 gene (Scott et al. 2001). TMPRSS3 encodes a type II transmembrane serine protease (TTSP) expressed in fetal cochlea. It can activate the sodium channel ENaC and has been shown to undergo proteolytic cleavage (Guipponi et al. 2002). We and others have identified ten different TMPRSS3 mutations in deaf patients. The missense mutations affect all functional domains of the protein (Ben-Yosef et al. 2001; Masmoudi et al. 2001; Scott et al. 2001; Wattenhofer et al. 2002; Ahmed et al. 2004).
In the present study, we report the analysis of 25 Turkish families with non-syndromic autosomal recessive childhood deafness without mutations in GJB2. We found pathogenic mutations in TMPRSS3 in three families, indicating that mutations in this gene are a significant cause of childhood non-syndromic deafness in the Turkish population (12% among the families negative for GJB2 mutations). Two of the three families harbor novel mutations. Interestingly, one of the novel mutations identified in this study maps in the predicted TMPRSS3 cleavage site. We show that this mutant protein does not undergo proteolytic cleavage and fails to activate ENaC.
Materials and methods
Patients
Twenty-five Turkish families from Trabzon, Rize, and Ordu, which segregated either congenital or childhood deafness, with at least two affected members, were included in this study. A high frequency of consanguinity (77% of the families) suggests an autosomal recessive mode of inheritance in the majority of the families. There was no evidence for an autosomal dominant or X-linked mode of inheritance, or any obvious syndrome. Audiometric results of unaffected sibs were normal. Affected family members showed severe to profound sensorineural childhood non-syndromic deafness, with both males and females being affected. No information was available regarding walking age. Informed consent was obtained for all participating family members. Peripheral blood was obtained from members of all 25 families, and genomic DNA was isolated from blood lymphocytes (Grimberg et al. 1989). The Tunisian patients S17, S19, and S42 are members of family S, previously described (Masmoudi et al. 2001).
Audiometric ISO values for patients with a TMPRSS3 mutation can be found in Table 2.
Genotyping, linkage analysis, and mutation search
PCR amplification of polymorphic microsatellite markers D21S212 and D21S1225 was carried out in 12.5 μl with 100 ng genomic DNA and 2 pmol of each primer. Amplification products were run on a 6% (19:1, acrylamide:bisacrylamide) denaturing polyacrylamide gel. Haplotypes were reconstructed for all persons in the 25 pedigrees by using chromosome 21 markers D21S212 and D21S1225 adjacent to TMPRSS3 on 21q22.3. Linkage analyses were performed using the MLINK software. The disease was modeled as an autosomal recessive trait with complete penetrance at birth. Two-point LOD-scores (Z) in these families were calculated. Inbreeding loops were preserved in the pedigrees. Haplotype analysis of polymorphic markers adjacent to the TMPRSS3 gene on chromosome 21q22.3 on DNA of Turkish patients 53-1, 53-2, and 53-3 and on the Tunisian patients S17, S19, and S42 was performed as previously described (Berry et al. 2000). All 12 coding exons of TMPRSS3 and their splice junctions were PCR amplified as described elsewhere (Wattenhofer et al. 2002) and sequenced.
Functional analysis of TMPRSS3 missense mutations in Xenopus oocytes
The construction of plasmids pSD5TMPRSS3 Wt and pSD5TMPRSS3 P404L is described elsewhere (Guipponi et al. 2002). Mutant R216L was generated using the QuickChange mutagenesis system following the manufacturer’s instructions (Stratagene). This construct was verified by sequencing. Transcription was performed on 1 μg vector linearized with ScaI, using 0.5 IU/μl SP6 RNA polymerase (Promega) in a reaction mix of: 40 mM Tris–HCl (pH 7.5), 6 mM MgCl2, 10 mM NaCl, 2 mM spermidine, 1.5 mM each ATP, CTP, UTP, 0.3 mM GTP, 0.5 mM GpppG, 10 mM DTT, 1 IU/μl RNAsin, and 8 ng/μl BSA, 1 h at 40°C. Template was removed with RNAse-free DNAse I, and the cRNA purified on RNeasy mini spin-columns (Quiagen). Oocytes in stage V/VI from Xenopus laevis (Noerdhoek, South Africa) were injected with 0.25 ng of each cRNA coding for the rat α-, β-, and γ-ENaC subunits in the presence or absence of 2.5 ng of wild type or mutant TMPRSS3 cRNA in a total volume of 100 nl. Oocytes were incubated in modified Barth saline (MBS) solution, and 24 h after injection, electrophysiological measurements were performed. The amiloride-sensitive current (INa) was measured by two-electrode voltage clamp in the presence of 120 mM Na+ in Frog Ringer with 5 μM amiloride at a holding potential of −100 mV. Three series of experiments were performed, each with four to eight oocytes per condition. In each series, the individual current values measured were normalized to the average of the ENaC+H2O values of that series. The results are reported as the means of all normalized values for each condition, ±SEM. Kruskal–Wallis test followed by a Dunn’s multiple comparison post-test were performed to determine significance. The means of absolute INa values for ENaC+H2O in the three series were 337±50.6, 446±46.4, and 4533±980.8 nA. These values for ENaC+TMPRSS3 wild type were 4386±1642, 2117±717.4, and 13,517±2414 nA.
Western blot analysis
Injected Xenopus oocytes were incubated in MBS for 24 h. Microsomal membrane protein extracts were obtained from oocytes lysed as previously described (Geering et al. 1989). Protein extract corresponding to 25% of the total protein content of a single oocyte was loaded in each lane. Proteins were separated on a 10% SDS–polyacrylamide gel under reducing and denaturing conditions and transferred to a nitrocellulose membrane (Protran from Schleicher & Schuell). The membrane was processed using rabbit anti-TMPRSS3 serum (Covalab, dilution 1/1000) and anti-rabbit IgG horseradish peroxidase linked antibody (Amersham Biosciences, dilution 1/10,000) according to standard procedures.
Results
To evaluate the importance of mutations in TMPRSS3 in childhood non-syndromic deafness in Turkey, we collected pedigrees with at least two affected individuals. We have ascertained 25 Turkish families comprising members likely to be affected by non-syndromic childhood hearing impairment. Affected and unaffected members of each family were examined by clinical evaluation and pure tone audiometry. The pedigrees were compatible with congenital/childhood onset, autosomal recessive, severe to profound sensorineural deafness. No additional phenotypes were observed.
The segregation of D21S212 and D21S1225, two highly polymorphic microsatellite markers flanking TMPRSS3, was studied for linkage with the phenotype. We reconstructed the haplotypes of all available members of the 25 families. No recombinants were found between the two tested markers and the deafness phenotype in six of the families (Table 1). At θ=0, the LOD score at D21S212, which is 0.5 Mb centromeric to TMPRSS3, was between 0.125 and 1.646 in the six families with potential linkage to TMPRSS3. The DNA from one affected member of these six families was used for mutation analysis. We identified TMPRSS3 pathogenic mutations in three patients and confirmed co-segregation of the mutation and the phenotype in these families by sequencing DNA of other family members. Consistently, linkage analysis in these families (families 40, 53, and 88) showed the highest LOD score (Table 1).
In family 53 (Fig. 1a), the proband 53-1 is homozygous for the transition c.1211C→T, a mutation previously identified in a Tunisian family that substitutes P404 by Leu (Masmoudi et al. 2001). The deaf father (53-2) and deaf father’s cousin (53-9) are also homozygous for this transition, whereas the hearing mother (53-3) is heterozygous. All affected members of family 53 were hearing until the age of 6 or 7 years, whereas in the Tunisian family, deafness was congenital (Table 2). To assess if the P404L mutation in both families has a single origin, we analyzed microsatellite markers around the TMPRSS3 locus in both families. The haplotypes in the two families are different at both the 5′ and 3′ of the mutation favoring a different origin hypothesis (Fig. 2).
Pedigrees of the three families harboring mutations in TMPRSS3. Black symbols represent deaf patients whereas white symbols represent hearing individuals. The haplotypes of each tested individual is indicated. Black bars indicate the haplotypes linked to the deafness phenotype. a Family 53. b Family 88. c Family 40
Haplotype analysis of polymorphic markers on 21q22.3 in Tunisian family S (left) and Turkish family 53 (right). Complete black symbols represent deaf patients, whereas complete white or black and white symbols represent hearing individuals, homozygous or heterozygous at the TMPRSS3 locus, respectively. Black bars indicate the haplotypes linked to the deafness phenotype. At the left, position of the polymorphic marker in the UCSC Genome Browser (http://genome.ucsc.edu/) is indicated (assembly of May 2004)
In family 88 (Fig. 1b), patient 88-1 is homozygous for the transition c.1192C→T. This newly identified nucleotide change leads to a Q398X nonsense codon. In addition, patient 88-1 is heterozygous for the c.1367G→A SNP, excluding a large deletion as the second mutant allele in 88-1. In this family, the deafness phenotype is congenital for all affected members.
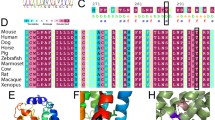
We identified another novel mutation in family 40 (Fig. 1c), a transversion c.647G→T. This nucleotide substitution modifies an arginine-to-leucine codon at position 216 (R216L). The proband (40-1) and his deaf brother (40-4) are homozygous for this mutation, while both parents (40-2 and 40-3) are heterozygous. The hearing sister (40-5) is homozygous for the normal allele. Amino acid residue R216 is well conserved among serine proteases and is usually R or K (Fig. 3). This substitution was not observed in 10 and 383 Turkish and European Mediterranean unrelated individuals, respectively, either by SSCP or sequencing (Wattenhofer et al. 2002). These results are concordant with c.647G→T being a recessive pathogenic mutation. Deafness in patient 40-1 was detected when he was 1.5 years old because of speech delay. In contrast, deafness in his younger brother, 40-4, was diagnosed at birth. Because patient 40-1 was the first case of a deafness-affected child in this family and his parents were thus not aware of a deafness possibility, we cannot exclude the hypothesis that 40-1 was indeed deaf at birth.
In order to characterize more precisely the R216L mutant, we functionally characterized the predicted encoded protein. It has been suggested that TMPRSS3 is cleaved at the RIVGG zymogen activation site, between amino acids R216 and I217 (Guipponi et al. 2002). This sequence is well conserved among serine proteases (Fig. 3). Hence, we postulated that the transversion present in family 40 would result in a TMPRSS3 mutant protein that cannot be cleaved. To test this assumption, we expressed both wild type and mutant TMPRSS3 in Xenopus oocytes, isolated the membrane fraction, and performed a Western blot using anti-TMPRSS3 serum (Guipponi et al. 2002). The P404L mutant was previously shown not to undergo proteolytic cleavage (Guipponi et al. 2002). In contrast to the wild type TMPRSS3, and similarly to the P404L mutant, the R216L variant fails to undergo proteolytic cleavage (Fig. 4a). To investigate whether this non-cleaved zymogen was functionally active, we tested its capacity to activate ENaC. As expected (Guipponi et al. 2002), co-expression of the human wild type TMPRSS3 and rat ENaC subunits led to an increase in ENaC mediated current (INa) compared to oocytes expressing only the ENaC subunits (P<0.01) (Fig. 4b, lane 2 versus lane 1). In contrast, and similarly to the P404L mutation, the R216L mutant was totally inactive (Fig. 4b). Thus, we can conclude that the R216L missense mutation impairs TMPRSS3 proteolytic activation, thereby resulting in an inactive protein.
Functional analysis of the R216L mutant in Xenopus oocytes. Oocytes were injected with rat ENaC subunits in the presence of water (H2O, lane 1), TMPRSS3 wild type (wt, lane 2), or missense mutations (R216L, lane 3 and P404L, lane 4). Ni non-injected oocytes. a Biochemical analysis of wild type and mutant TMPRSS3 by Western blot. Proteolytic cleavage of TMPRSS3 is partial. Both mutants R216L and P404L proteins did not show the cleaved product indicated by an arrow. b Comparison of the effect of Wt-TMPRSS3 and mutants on INa (sodium current) in Xenopus oocytes. *P<0.05 versus lane 1; **P<0.01 versus lane 1; ns non-significant versus lane 1. n=15–18 for each condition, performed in a total of three series of experiments
Discussion
TMPRSS3 encodes a serine protease with LDLRA and SRCR domains and is the cause of deafness in DFNB8/10 families (Ben-Yosef et al. 2001; Masmoudi et al. 2001; Scott et al. 2001; Wattenhofer et al. 2002). Mutations identified so far map in all TMPRSS3 domains (reviewed in Wattenhofer et al. 2002), and all three domains have been shown to be important for the ability of TMPRSS3 to activate ENaC and for its catalytic activity (Guipponi et al. 2002; Lee et al. 2003). ENaC is a sodium channel known to be regulated by serine protease activity (Vallet et al. 1997). It is expressed in various tissues, including the inner ear, where it has been proposed to be involved in maintaining a low Na+ concentration in the endolymph (Couloigner et al. 2001). By RT-PCR, we observed co-expression of TMPRSS3 and the three ENaC genes both in the stria vascularis and in the modiolus of P5 rats. In addition, in a Xenopus oocyte model, TMPRSS3 wild type, but not all mutants identified in DFNB8/10, is able to activate ENaC (Guipponi et al. 2002). Based on these results, we have proposed ENaC as a potential substrate for TMPRSS3 in the inner ear (Guipponi et al. 2002). Here, we report the characterization of a new deafness-causing mutation, R216L, which fails to activate the ENaC channel although harboring non-mutated LDLRA, SRCR, and serine protease domains. In addition, R216L does not undergo proteolytic cleavage. We postulated that TMPRSS3 requires a proteolytic cleavage in order to become active (Guipponi et al. 2002). Similar to other serine proteases, the predicted cleavage site is between R216 and I217 in the context of the R/K-I-V-G-G consensus sequence. The engineered mutant S401A of TMPRSS3 catalytic site, which is unable to undergo proteolytic cleavage (Guipponi et al. 2002), showed that this cleavage is dependent on TMPRSS3 catalytic capacity. This suggests that TMPRSS3 cleavage occurs as the result of autocatalytic cleavage. Similar results have been obtained for TMPRSS2: Western blot on cells expressing a TMPRSS2 protein harboring a mutation of the catalytic serine S441 to alanine showed that the proteolytic cleavage is dependent on catalytic activity (Afar et al. 2001). The non-cleavage of the R216L mutant protein is consistent with our hypothesis of TMPRSS3 as the protease responsible for this cleavage.
Pathogenic mutations in the R/K-IVGG consensus have already been identified in several serine proteases, including factor VII (Chaing et al. 1994; Wulff and Herrmann 2000) and factor IX (Sommer et al. 1992), both implicated in coagulation, and protein C (Grundy et al. 1989). The mutation can affect any amino acid of the consensus site and in all cases impairs the activating cleavage. In contrast, the PRSS1 (protease serine 1) pathogenic mutation N21I, which maps six amino acids after the cleavage site, results in an increased propensity to autoactivation, thus causing pancreatitis (Sahin-Toth 2000; Sahin-Toth and Toth 2000). All these observations underscore the biological importance of activation cleavage. Zymogen protease activation by proteolytic cleavage is a regulatory mechanism to prevent undesired proteolysis. The current accepted view is that all proteases are synthesized as inactive zymogens. In the majority of the cases, catalytic cleavage of the zymogen induces a conformational change of the protease domain, allowing the formation of the active site and thus an active protease (Lazure 2002). By analogy with other proteases (Hedstrom 2002), we can surmise that in the case of TMPRSS3, I217 would form a salt bridge with R400, allowing the formation of the active site and the oxyanion hole. The identification and characterization of the R216L mutation demonstrates the importance of the activation cleavage for TMPRSS3 function and emphasizes the currently accepted view of activation sequences as major players in protease function. The level of TMPRSS3 R216L mutant in our Xenopus model system does not seem lower than TMPRSS3 wild type. Thus, our data do not argue in favor of an obvious altered turnover of the mutant protein compared with the wild type. The possibility that decreased ENaC activation activity is due to increased degradation by the ER protein quality control system in collaboration with the proteosome is thus unlikely. Nevertheless, we cannot exclude the possibility that the R216L mutation induces a drastic conformational change thus masking the cleavage activation site.
In the frame of this study we identified the c.1221C→T transition that leads to the P404L missense, previously identified in a Tunisian family (Masmoudi et al. 2001). Although the Turkish and the Tunisian families harbor the same mutation, the deafness phenotype is different: in the Tunisian family deafness was congenital (Masmoudi et al. 2001), whereas in the Turkish family the onset of deafness was at age 6–7 years. Consistently, the haplotypes of polymorphic sites located in a region of 500 kb encompassing the TMRSS3 mutation are different in the two families, indicating an independent origin of the mutation (Fig. 4). We postulate that besides the c.1221C→T mutation, other genetic factors influence the severity of the disease, leading to the phenotypic variability. A similar situation has already been observed in the case of sickle cell disease (Wood et al. 1980; Kulozik et al. 1987; Serjeant 1989). Phenotypic variability among patients harboring the same CFTR mutation has also been observed in families with cystic fibrosis (Mekus et al. 2000). It becomes evident that phenotypic variability of monogenic disorders is modified by complex genetic and environmental factors. The hearing phenotype associated with the TMPRSS3 P404L mutation is consistent with the proposed idea that the current classification of disorders into monogenic vs multifactorial diseases is an oversimplification (Scriver and Waters 1999).
A significant part of recessive non-syndromic deafness can be attributed to mutations in the Connexin26 (GJB2) gene. However, this contribution varies between populations. Forty-two percent of Americans (Green et al. 1999), 42–46% of European Mediterraneans (Estivill et al. 1998; Rabionet et al. 2000; Pampanos et al. 2002), 32% of Turks (Uyguner et al. 2003), 27% of Japanese (Fuse et al. 1999), and 8% of Koreans (Park et al. 2000) affected by non-syndromic recessive deafness show GJB2 mutations, raising the possibility that other gene(s) may contribute significantly to childhood non-syndromic recessive deafness in the Turkish, Japanese, and Korean populations. We collected 25 unrelated Turkish families in schools for deaf. As we studied non-syndromic deafness, we can consider that having collected only families with children in deaf schools did not include a bias in the enrolling procedure. Thus, the 25 families we collected can be considered as a representative sample of childhood non-syndromic onset deafness in Turkey. In this manuscript, we present evidence suggesting that in the Turkish population studied, TMPRSS3 mutations significantly contribute to non-syndromic recessive deafness. Hence, TMPRSS3 appears to be an important locus, in addition to GJB2, involved in a substantial fraction of non-syndromic deafness cases in a defined population. In the Turkish population, we observe a prevalence of 11% of TMPRSS3 mutations among patients with childhood hearing loss and negative for GJB2 mutations, which enables us to estimate that this locus plays a role in about 8% of the total childhood deaf Turkish population. Since we did not identify a recurrent mutation, we can consider that there is no obvious founder effect. Similar to GJB2, the contribution of TMPRSS3 differs in different populations. Previously reported TMPRSS3 mutations have been identified in one Palestinian, five Pakistani, and two Tunisian families, as well as in two sporadic Caucasian patients from Spain and Greece (Ben-Yosef et al. 2001; Masmoudi et al. 2001; Scott et al. 2001; Wattenhofer et al. 2002). All these analyses reported very low percentages of TMPRSS3 mutations in the studied deaf populations, i.e. 2.5% (4/159) in Pakistani, 0.4% (2/448) in European Mediterranean, and even 0% (0/64) in North American populations.
References
Afar DE, Vivanco I, Hubert RS, Kuo J, Chen E, Saffran DC, Raitano AB, Jakobovits A (2001) Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res 61:1686–1692
Ahmed ZM, Li XC, Powell SD, Riazuddin S, Young TL, Ramzan K, Ahmad Z, Luscombe S, Dhillon K, MacLaren L, Ploplis B, Shotland LI, Ives E, Friedman TB, Morell RJ, Wilcox ER (2004) Characterization of a new full length TMPRSS3 isoform and identification of mutant alleles responsible for nonsyndromic recessive deafness in Newfoundland and Pakistan. BMC Med Genet 5:24
Ben-Yosef T, Wattenhofer M, Riazuddin S, Ahmed ZM, Scott HS, Kudoh J, Shibuya K, Antonarakis SE, Bonne-Tamir B, Radhakrishna U, Naz S, Ahmed Z, Pandya A, Nance WE, Wilcox ER, Friedman TB, Morell RJ (2001) Novel mutations of TMPRSS3 in four DFNB8/B10 families segregating congenital autosomal recessive deafness. J Med Genet 38:396–400
Berry A, Scott HS, Kudoh J, Talior I, Korostishevsky M, Wattenhofer M, Guipponi M, Barras C, Rossier C, Shibuya K, Wang J, Kawasaki K, Asakawa S, Minoshima S, Shimizu N, Antonarakis S, Bonne-Tamir B (2000) Refined localization of autosomal recessive nonsyndromic deafness DFNB10 locus using 34 novel microsatellite markers, genomic structure, and exclusion of six known genes in the region. Genomics 68:22–29
Bonne-Tamir B, DeStefano AL, Briggs CE, Adair R, Franklyn B, Weiss S, Korostishevsky M, Frydman M, Baldwin CT, Farrer LA (1996) Linkage of congenital recessive deafness (gene DFNB10) to chromosome 21q22.3. Am J Hum Genet 58:1254–1259
Chaing S, Clarke B, Sridhara S, Chu K, Friedman P, VanDusen W, Roberts HR, Blajchman M, Monroe DM, High KA (1994) Severe factor VII deficiency caused by mutations abolishing the cleavage site for activation and altering binding to tissue factor. Blood 83:3524–3535
Couloigner V, Fay M, Djelidi S, Farman N, Escoubet B, Runembert I, Sterkers O, Friedlander G, Ferrary E (2001) Location and function of the epithelial Na channel in the cochlea. Am J Physiol Renal Physiol 280:F214–F222
Estivill X, Fortina P, Surrey S, Rabionet R, Melchionda S, D’Agruma L, Mansfield E, Rappaport E, Govea N, Mila M, Zelante L, Gasparini P (1998) Connexin-26 mutations in sporadic and inherited sensorineural deafness. Lancet 351:394–398
Fuse Y, Doi K, Hasegawa T, Sugii A, Hibino H, Kubo T (1999) Three novel connexin26 gene mutations in autosomal recessive non-syndromic deafness. Neuroreport 10:1853–1857
Geering K, Theulaz I, Verrey F, Hauptle MT, Rossier BC (1989) A role for the beta-subunit in the expression of functional Na+–K+-ATPase in Xenopus oocytes. Am J Physiol 257:C851–C858
Green GE, Scott DA, McDonald JM, Woodworth GG, Sheffield VC, Smith RJ (1999) Carrier rates in the midwestern United States for GJB2 mutations causing inherited deafness. JAMA 281:2211–2216
Grimberg J, Nawoschik S, Belluscio L, McKee R, Turck A, Eisenberg A (1989) A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res 17:8390
Grundy C, Chitolie A, Talbot S, Bevan D, Kakkar V, Cooper DN (1989) Protein C London 1: recurrent mutation at Arg 169 (CGG–TGG) in the protein C gene causing thrombosis. Nucleic Acids Res 17:10513
Guipponi M, Vuagniaux G, Wattenhofer M, Shibuya K, Vazquez M, Dougherty L, Scamuffa N, Guida E, Okui M, Rossier C, Hancock M, Buchet K, Reymond A, Hummler E, Marzella PL, Kudoh J, Shimizu N, Scott HS, Antonarakis SE, Rossier BC (2002) The transmembrane serine protease (TMPRSS3) mutated in deafness DFNB8/10 activates the epithelial sodium channel (ENaC) in vitro. Hum Mol Genet 11:2829–2836
Hedstrom L (2002) Serine protease mechanism and specificity. Chem Rev 102: 4501–4524
Kulozik AE, Thein SL, Kar BC, Wainscoat JS, Serjeant GR, Weatherall DJ (1987) Raised Hb F levels in sickle cell disease are caused by a determinant linked to the beta globin gene cluster. Prog Clin Biol Res 251:427–439
Lazure C (2002) The peptidase zymogen proregions: nature’s way of preventing undesired activation and proteolysis. Curr Pharm Des 8:511–531
Lee YJ, Park D, Kim SY, Park WJ (2003) Pathogenic mutations but not polymorphisms in congenital and childhood onset autosomal recessive deafness disrupt the proteolytic activity of TMPRSS3. J Med Genet 40:629–631
Masmoudi S, Antonarakis SE, Schwede T, Ghorbel AM, Gratri M, Pappasavas MP, Drira M, Elgaied-Boulila A, Wattenhofer M, Rossier C, Scott HS, Ayadi H, Guipponi M (2001) Novel missense mutations of TMPRSS3 in two consanguineous Tunisian families with non-syndromic autosomal recessive deafness. Hum Mutat 18:101–108
Mekus F, Ballmann M, Bronsveld I, Bijman J, Veeze H, Tummler B (2000) Categories of deltaF508 homozygous cystic fibrosis twin and sibling pairs with distinct phenotypic characteristics. Twin Res 3:277–293
Nance WE, Kearsey MJ (2004) Relevance of connexin deafness (DFNB1) to human evolution. Am J Hum Genet 74:1081–1087
Pampanos A, Economides J, Iliadou V, Neou P, Leotsakos P, Voyiatzis N, Eleftheriades N, Tsakanikos M, Antoniadi T, Hatzaki A, Konstantopoulou I, Yannoukakos D, Gronskov K, Brondum-Nielsen K, Grigoriadou M, Gyftodimou J, Iliades T, Skevas A, Petersen MB (2002) Prevalence of GJB2 mutations in prelingual deafness in the Greek population. Int J Pediatr Otorhinolaryngol 65:101–108
Park HH, Chun YM, Park K, Kim HN (2000) Connexin 26 mutations associated with nonsyndromic hearing loss. Laryngoscope 110:1535–1538
Petit C (1996) Genes responsible for human hereditary deafness: symphony of a thousand. Nat Genet 14:385–391
Rabionet R, Zelante L, Lopez-Bigas N, D’Agruma L, Melchionda S, Restagno G, Arbones ML, Gasparini P, Estivill X (2000) Molecular basis of childhood deafness resulting from mutations in the GJB2 (connexin 26) gene. Hum Genet 106:40–44
Sahin-Toth M (2000) Human cationic trypsinogen. Role of Asn-21 in zymogen activation and implications in hereditary pancreatitis. J Biol Chem 275:22750–22755
Sahin-Toth M, Toth M (2000) Gain-of-function mutations associated with hereditary pancreatitis enhance autoactivation of human cationic trypsinogen. Biochem Biophys Res Commun 278:286–289
Scott HS, Kudoh J, Wattenhofer M, Shibuya K, Berry A, Chrast R, Guipponi M, Wang J, Kawasaki K, Asakawa S, Minoshima S, Younus F, Mehdi SQ, Radhakrishna U, Papasavvas MP, Gehrig C, Rossier C, Korostishevsky M, Gal A, Shimizu N, Bonne-Tamir B, Antonarakis SE (2001) Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat Genet 27:59–63
Scriver CR, Waters PJ (1999) Monogenic traits are not simple: lessons from phenylketonuria. Trends Genet 15:267–272
Serjeant GR (1989) Geography and the clinical picture of sickle cell disease. An overview. Ann N Y Acad Sci 565:109–119
Sommer SS, Bowie EJ, Ketterling RP, Bottema CD (1992) Missense mutations and the magnitude of functional deficit: the example of factor IX. Hum Genet 89:295–297
Uyguner O, Emiroglu M, Uzumcu A, Hafiz G, Ghanbari A, Baserer N, Yuksel-Apak M, Wollnik B (2003) Frequencies of gap- and tight-junction mutations in Turkish families with autosomal-recessive non-syndromic hearing loss. Clin Genet 64:65–69
Vallet V, Chraibi A, Gaeggeler HP, Horisberger JD, Rossier BC (1997) An epithelial serine protease activates the amiloride-sensitive sodium channel. Nature 389:607–610
Veske A, Oehlmann R, Younus F, Mohyuddin A, Muller-Myhsok B, Mehdi SQ, Gal A (1996) Autosomal recessive non-syndromic deafness locus (DFNB8) maps on chromosome 21q22 in a large consanguineous kindred from Pakistan. Hum Mol Genet 5:165–168
Wattenhofer M, Di Iorio MV, Rabionet R, Dougherty L, Pampanos A, Schwede T, Montserrat-Sentis B, Arbones ML, Iliades T, Pasquadibisceglie A, D’Amelio M, Alwan S, Rossier C, Dahl HH, Petersen MB, Estivill X, Gasparini P, Scott HS, Antonarakis SE (2002) Mutations in the TMPRSS3 gene are a rare cause of childhood nonsyndromic deafness in Caucasian patients. J Mol Med 80:124–131
Wood WG, Pembrey ME, Serjeant GR, Perrine RP, Weatherall DJ (1980) Hb F synthesis in sickle cell anaemia: a comparison of Saudi Arab cases with those of African origin. Br J Haematol 45:431–445
Wulff K, Herrmann FH (2000) Twenty two novel mutations of the factor VII gene in factor VII deficiency. Hum Mutat 15:489–496
Acknowledgements
We thank the families and the administrative bodies of the schools for the deaf for their kind participation in this study. We also thank N. Akarsu, J.-L. Blouin, I. Bouchardy, C. Borel, Ö. Refik Çaylan, L. Excoffier, M. Friedli, M. Gagnebin, J.-D. Horisberger, E. Kalay, N. Lin-Marq, M. Morris, L. Pignat, C. Rossier and J.D. Vassali for critical comments, helpful discussions, or technical assistance. This work was supported by grants from the Swiss National Science Foundation (31.57149.99), the European Union (QLRT-2001-00816), the NCCR frontiers in Genetics, the Jérôme Lejeune Foundation, the Child Care Foundation to AR and SEA, from the Swiss National Science Foundation (31-061966.00) and the Danish Research Council (22-05-0535) to BR and DA. BB was supported by the Geneva Research Programme for Medical Students (PREM). This study was supported by the Karadeniz Technical University Research Fund (Project no: 2002.114.001.3), Stichting Irene Kinderziekenhuis and Stichting Vrienden van Effatha.
Author information
Authors and Affiliations
Corresponding author
Additional information
Marie Wattenhofer and Nilüfer Sahin-Calapoglu contributed equally to this work
Rights and permissions
About this article
Cite this article
Wattenhofer, M., Sahin-Calapoglu, N., Andreasen, D. et al. A novel TMPRSS3 missense mutation in a DFNB8/10 family prevents proteolytic activation of the protein. Hum Genet 117, 528–535 (2005). https://doi.org/10.1007/s00439-005-1332-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-005-1332-x