Abstract
Lymphoedema-distichiasis (LD) is a syndromic form of primary lymphoedema, where mutations in the gene for the developmental transcription factor FOXC2 have been shown to be causative. The disorder has been considered very rare, but our group has now ascertained 34 families and 11 sporadic cases in the UK. Two families with LD have no mutation in the coding region of FOXC2, although both are consistent with linkage to the FOXC2 locus. A deletion has been ruled out as a possible cause of LD in these families, leaving promoter mutations as the most likely cause. Sixteen previously unpublished mutations are reported, plus an estimate of the frequency of new mutations in this disorder.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Primary lymphoedema shows an autosomal dominant pattern of inheritance with varying degrees of penetrance, and is characterised by chronic swelling of the limbs, most often of the legs (Mortimer 1995). There are three major forms of dominantly inherited primary lymphoedema: Milroy disease (MD) [MIM 153200], lymphoedema-distichiasis (LD) [MIM 153400] and Meige disease (MGD) [MIM 153200]. Of these, the causative genes are identified as VEGFR3 for at least a subset of MD patients (Ferrell et al. 1998, Irrthum et al. 2000), FOXC2 for LD (Mangion et al. 1999; Fang et al. 2000; Bell et al. 2001), but as yet no gene locus has been identified for MGD.
Patients with LD usually show signs of lymphoedema (late childhood or later onset) associated with distichiasis, which is the development of aberrant eyelashes arising from the meibomian glands of the eyelid (Dale et al. 1985; Robinow et al. 1970). There has been a report that some families with MGD disease have mutations in FOXC2 (Finegold et al. 2001). Our experience is that this is not the case, and that stringent phenotyping will show some of the syndromic features of LD in at least some family members. It is possible that in isolated cases lymphoedema could be the only manifestation of LD, as the expression of the clinical signs is highly variable. Any case where a mutation is found in FOXC2 should, in our opinion, be classed as LD, not MGD, lymphoedema with ptosis, or any other form of lymphoedema.
We have previously reported 18 families with LD, and six isolated cases. All bar one family had a mutation in FOXC2 (Bell et al. 2001; Brice et al. 2002). This family was consistent with linkage to a marker near the LD locus (D16S520), but was not tested at that time with markers in the FOXC2 gene itself. In this paper we show that markers 5′ and 3′ to FOXC2 support the linkage for the previously described family with no mutation, report 16 novel mutations in FOXC2 (including two missense changes), another family with no coding FOXC2 mutation, and provide an estimate of new mutations.
Materials and methods
DNA extraction
DNA was extracted from venous blood using the Nucleon extraction and purification kit (Amersham Life Sciences). The protocol supplied by the manufacturer was followed.
Mutation analysis
The FOXC2 gene was screened for mutations using sequence analysis. DNA sequencing was carried out on an ABI 3100 DNA analyser, using Bigdye chemistry (Applied Biosystems, Foster City, Calif., USA). The FOXC2 gene is composed of a 1.5-kb single exon, which was divided into four overlapping regions that were amplified and sequenced. This enabled the entire coding region to be analysed for mutations. In cases where no mutations were identified in the coding region of FOXC2, the 5′ and 3′ regions of the FOXC2 gene were also sequenced and analysed. The primers and their conditions are listed in Table 1. Those for the coding region of FOXC2 have been previously described (Bell et al. 2001), as have those in the 5′ region and 3′ region (Kovacs et al. 2003). A second set of primers for the 3′ region was designed for this study. Cycling involved an initial denaturation step of 95 °C for 5 min, followed by 35 cycles each of 95 °C for 1 min, various annealing temperatures (see Table 1) for 1 min and 72 °C for 1 min, and a final extension of 72 °C for 10 min.
Haploid cell line
Haploid cell lines were made of chromosome 16, using circulating lymphocytes from an affected LD patient showing no FOXC2 mutation (GMP Genetics, USA). DNA was provided by GMP Genetics from cell hybrids containing single copies of the human chromosome 16 s, which were confirmed by microsatellite analysis using D16S520, and the polymorphisms 5′ and 3′ to FOXC2 used in the current study. The entire FOXC2 gene of both haploid cell lines was amplified, including 639 bp upstream and 303 bp downstream of the coding region.
Results
Eleven of the mutations are deletions, two are duplications, one is nonsense, and two are missense (Table 2). This brings to four the number of missense mutations reported by our group, S125L and R121H being those previously identified (Bell et al. 2001; Brice et al. 2002). We are not aware of any other missense mutations in FOXC2 being reported as causative for LD, with those previously published being either frame shift or nonsense mutations (Fang et al. 2000; Finegold et al. 2001; Erickson et al. 2001; Bell et al. 2001; Brice et al. 2002; Traboulsi et al. 2002; Brooks et al. 2003; Yildirim-Toruner et al. 2004).
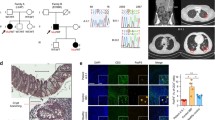
We previously reported a pedigree with LD but no FOXC2 mutation (Brice et al. 2002), which showed linkage to the microsatellite D16S520, which is about 100 kb from the FOXC2 gene. We have used polymorphisms in the FOXC2 gene to confirm that this linkage applies to the gene itself (Fig.1a, lodscore of one using the haplotype as a single marker). A second pedigree (Fig.1b) has now been ascertained, where the clinical signs are completely consistent with LD, but there is no mutation in the coding region of FOXC2. Polymorphisms in FOXC2 were used to investigate the possibility of a gene deletion in either family A or B. The 5′ polymorphism (−512C > T) has previously been reported (Ridderstrale et al. 2002), while that in the 3′ region (1761G > A) is newly identified in this study. This 1761G>A polymorphism has a frequency of 18.8% in our control population (Caucasians from the UK). The heterozygosity for both polymorphisms is shown in Fig.1. The same polymorphisms have been utilised to show that family B is also consistent with linkage to FOXC2, and also to demonstrate that there is no large-scale deletion of the FOXC2 gene in either family. To ensure that there was no small deletion between the polymorphisms, we had haploid cell lines made from lymphocytes from an affected individual in family A. The PCR of a 2,445-bp fragment using primers that were 5′ and 3′ to the two polymorphisms showed that the same sized fragment was produced for both chromosomes (data not shown). Amplification of DNA from an affected individual in each family with the same primers also produced only a single fragment. There were no deletions identified in either case. Sequencing of the 2,445-bp fragment in the haploid lines and the DNA from the individual from whom the lines originated showed that there were no small deletions outside the coding region, and that the −512 and 1761 polymorphisms had the C and A, and G and T alleles in cis, as predicted from the linkage studies (see Fig.1).
From our families and sporadic cases, we were able to calculate an approximate value for the frequency of new mutations in LD. Excluding the two families where we have not found a mutation, we have 32 families and 11 sporadic cases with LD. Of these, one apparently sporadic case was adopted and the parental phenotype was not available. Seven others were demonstrated to be new mutations by sequencing DNA from the probands and both parents, but in the other three cases it was not possible to get DNA from both parents. There were eight examples of only a parent and child apparently being affected, and of these one was definitely a new mutation and one probably so, as the affected father had four unaffected siblings and there was no clinical suggestion that the deceased grandparents had been affected. Only the definite new mutation was scored as such. The remaining six affected parent/child pairs had insufficient information to determine whether the parent was a new mutation. In none of the more extended familial cases was a new mutation suggested, based on DNA analysis coupled with clinical family history. The seven definite de novo mutations we identified are given in Table 3.
Discussion
We have shown, along with other groups, that LD is mainly produced as a result of nonsense mutations, and insertions or deletions that produce frameshifts (Bell et al. 2001; Finegold et al. 2001; Erickson et al. 2001; Brice et al. 2002). Although the mutations are scattered throughout the gene, there is a hotspot for recombination in the region 893–930 bp, where we have found seven mutations, none of them identical. There are three reports of the 914-921delACGCCGCC mutations in the literature (Bell et al. 2001; Erickson et al. 2001; Bahau et al. 2002). The presence of a GCCGCCGC element either side of a 14-bp sequence, plus an additional GCCGCC in this region, might explain how misalignment and recombination could produce deletions and insertions.
We have also now found four missense mutations that co-segregate with the condition. Of these, there is compelling evidence that those previously reported, S125L and R121H, are definitely causative, since analogous changes in FOXC1 (S131L and R127H) have been shown by computer modelling and cell studies to have severe effects on protein function (Saleem et al. 2003). S131L does not transactivate at all in a luciferase reporter assay (Saleem et al. 2003), and is considered by that research group as a completely inactive protein in this respect (M. Walter, personal communication). R127H shows greatly reduced nuclear localisation compared to wild type, and transactivation in the luciferase reporter assay was 16% of wild-type levels. Computer modelling for R127H shows that the mutation alters the electrostatic charge on the DNA binding surface of the fork head domain, which presumably affects the ability of the mutant protein to attach to its DNA target.
The two-missense changes reported in this paper do not have an analogy with any known FOXC1 mutations, and it is therefore difficult to be sure of their effect on the protein. Both are absent from 200 control chromosomes. W116R is in the fork head domain and exhibits an R group change in both size and charge. The tryptophan is part of a motif (including R121) that is conserved across all mouse Fox genes (Carlson et al. 2002) and other species (Fig.2). The likelihood is that this is a causative mutation. For S235I the position is less certain. It is outside the fork head domain of FOXC2, and the function of this region of the protein is unclear. The residue is either conserved in homologues from a BLAST search (Fig.2), or is replaced by a glycine (e.g. in mouse Foxc2). Clearly a change to isoleucine could produce more profound effects, but with so little information on the function of this part of the protein it is impossible to be definite as to the effect in this case. If it is causative, future studies using this in a construct could help elucidate the function of the protein C-terminal to the fork head domain in FOXC2.
FOXC2 protein alignment of nine different species and one homologous protein to human FOXC2. The letters in bold are the equivalent amino acids to those altered by the SNPs. The alignments were obtained using the computer program ClustalW (http://www.ebi.ac.uk/clustalw/)
Our data on the number of new mutations in LD is not robust, but it does give some idea of the frequency of new mutations in LD. Of the 43 probands in our analysis, 11 were apparently sporadic, but one was adopted and so removed from the analysis. All the other seven cases were de novo FOXC2 mutations, as was one from families where only a parent and child were affected. From our 42 cases, we therefore have 19% new mutations at the lowest possible estimate, with a top limit of 43% in the unlikely event of all six parent/child couplets being new mutations. With the one highly probable case included plus the three other sporadics, and one from the six parent/child pedigrees assumed to be a new mutation, a figure of 30% emerges as a reasonable estimate of new mutations in LD.
The seven definite new mutations are a mixture of insertions deletions and a missense, with no obvious clustering (Table 3).
The two families with no mutation in the coding region of FOXC2 are clinically indistinguishable from those where mutations have been identified. In view of haploinsufficiency as a postulated mechanism underlying LD (Fang et al. 2000; Bell et al. 2001; Erickson et al. 2001; Finegold et al. 2001; Brice et al. 2002), we had expected that gene deletions might be involved in these cases, but polymorphism studies, and haploid cell line analysis showed that this is not the case. Both pedigrees are consistent with linkage to the FOXC2 gene using 5′ and 3′ polymorphic markers, but the lod scores are only 1 and 0.75 due to the sizes of the families. The disorder in these families could be due to mutations in another gene, but the linkage data are more in favour of FOXC2 being the gene affected than an alternative locus. Promoter mutations could well be involved in these cases, but there is very little information on the promoter sites of FOXC2. Analysis of possible sites, and upstream sequencing, needs to be conducted to see if any changes can be identified.
Our present data suggest that LD is more common than was supposed in the pubertal onset lymphoedema population, is most likely to be genetically homogeneous, and has a new mutation rate of at least 20%. The nature of the promoter region of FOXC2, and the downstream targets of the gene, are currently unknown, and will be the subject of further work.
References
Bahuau M, Houdayer C, Tredano M, Soupre V, Couderc R, Vazquez MP (2002) FOXC2 truncating mutation in distichiasis, lymphedema, and cleft palate. Clin Genet 62:470–473
Bell R, Brice G, Child AH, Murday VA, Mansour S, Sandy CJ, Collin JRO, Brady AF, Callen DF, Burnand K, Mortimer P, Jeffery S (2001) Analysis of lymphoedema-distichiasis families for FOXC2 mutations reveals small insertions and deletions throughout the gene. Hum Genet 108:546–551
Brice G, Mansour S, Bell R, Collin JRO, Child AH, Brady AF, Sarfarazi M, Burnand KG, Jeffery S, Mortimer P, Murday VA (2002) Analysis of the phenotypic abnormalities in lymphoedema-distichiasis syndrome in 74 patients with FOXC2 mutations or linkage to 16q24. J Med Genet 39:478–483
Brooks BP, Dagenais SL, Nelson CC, Glynn MW, Caulder MS, Downs CA, Glover TW (2003) Mutation of the FOXC2 gene in familial distichiasis. J AAPOS 7:354–357
Carlsson P, Mahlapuu M (2002) Forkhead transcription factors: key players in development and metabolism. Dev Biol 250:1–23
Dale RF (1985) The inheritance of primary lymphoedema. J Med Genet 22:274–278
Erickson R, Dagenais S, Caulder M, Downs C, Herman G, Jones M, Kerstjens-Frederikse W, Lidral A, McDonald M, Nelson C, Witte M, Glover T (2001) Clinical heterogeneity in lymphoedema-distichiasis with FOXC2 truncating mutations. J Med Genet 38:761–766
Fang J, Dagenais SL, Erickson RP, Arlt MF, Glynn MW, Gorski JL, Seaver LH, Glover TW (2000) Mutations in FOXC2 (MFH-1), a forkhead family transcription factor, are responsible for the hereditary lymphedema-distichiasis syndrome. Am J Hum Genet 67:1382–1388
Ferrell RE, Levinson KL, Esman JH, Kimak MA, Lawrence EC, Barmada MM, Finegold DN (1998) Hereditary lymphoedema: evidence for linkage and genetic heterogeneity. Hum Mol Genet 7:2073–2078
Finegold DN, Kimak MA, Lawrence EC, Levinson KL, Cherniske EM, Pober BR, Dunlap JW, Ferrell RE (2001) Truncating mutations in FOXC2 cause multiple lymphedema syndromes. Hum Mol Genet 10:1185–1189
Irrthum A, Karkkainen MJ, Devriendt K, Alitalo K, Vikkula M (2000) Congenital hereditary lymphedema caused by a mutation that inactivates VEGFR3 tyrosine kinase. Am J Hum Genet 67:295–301
Kovacs P, Lehn-Stefan A, Stumvoll M, Bogardus C, Baier LJ (2003) Genetic variation in the human winged helix/forkhead transcription factor gene FOXC2 in Pima Indians. Diabetes 52:1292–1295
Mangion J, Rahman N, Mansour S, Brice G, Rosbotham J, Child AH, Murday VA, Mortimer PS, Barfoot R, Sigurdsson A, Edkins S, Sarfarazi M, Burnand K, Evans AL, Nunan TO, Stratton MR, Jeffery S (1999) A gene for lymphedema-distichiasis maps to 16q24.3. Am J Hum Genet 65:427–432
Mortimer PS (1995) Managing lymphoedema. Clin Dermatol 12:499–505
Ridderstrale M, Carlsson E, Klannemark M, Cederberg A, Kosters C, Tornqvist H, Storgaard H, Vaag A, Enerback S, Groop L (2002) FOXC2 mRNA expression and a 5′ untranslated region polymorphism of the gene are associated with insulin resistance. Diabetes 51:3554–3560
Robinow M, Johnson GF, Verhagen AD (1970) Distichiasis-lymphedema: a hereditary syndrome of multiple congenital defects. Am Dis Child 119:343–347
Saleem RA, Banerjee-Basu S, Berry FB, Baxevanis AD, Walter MA (2003) Structural and functional analyses of disease-causing missense mutations in the forkhead domain of FOXC1. Hum Mol Genet 12:2993–3005
Traboulsi EI, Al-Khayer K, Matsumoto M, Kimak MA, Crowe S, Wilson SE, Finegold DN, Ferrell RE, Meisler DM (2002) Lymphedema-distichiasis syndrome and FOXC2 gene mutation. Am J Ophthalmol 134:592–596
Yildirim-Toruner C, Subramanian K, El Manjra L, Chen E, Goldstein S, Vitale E (2004) A novel frameshift mutation of FOXC2 gene in a family with hereditary lymphedema-distichiasis syndrome associated with renal disease and diabetes mellitus. Am J Med Genet 131A:281–286
Acknowledgments
C.S.D.V. is supported by the British Heart Foundation. The research was carried out within the network of the London IDEAS Genetic Knowledge Park, and used the resources of the Biomics Unit at SGHMS.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sholto-Douglas-Vernon, C., Bell, R., Brice, G. et al. Lymphoedema-distichiasis and FOXC2: unreported mutations, de novo mutation estimate, families without coding mutations. Hum Genet 117, 238–242 (2005). https://doi.org/10.1007/s00439-005-1275-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-005-1275-2