Abstract
Although human and gibbons are classified in the same primate superfamily (Hominoidae), their karyotypes differ by extensive chromosome reshuffling. To date, there is still limited understanding of the events that shaped extant gibbon karyotypes. Further, the phylogeny and evolution of the twelve or more extant gibbon species (lesser apes, Hylobatidae) is poorly understood, and conflicting phylogenies have been published. We present a comprehensive analysis of gibbon chromosome rearrangements and a phylogenetic reconstruction of the four recognized subgenera based on molecular cytogenetics data. We have used two different approaches to interpret our data: (1) a cladistic reconstruction based on the identification of ancestral versus derived chromosome forms observed in extant gibbon species; (2) an approach in which adjacent homologous segments that have been changed by translocations and intra-chromosomal rearrangements are treated as discrete characters in a parsimony analysis (PAUP). The orangutan serves as an "outgroup", since it has a karyotype that is supposed to be most similar to the ancestral form of all humans and apes. Both approaches place the subgenus Bunopithecus as the most basal group of the Hylobatidae, followed by Hylobates, with Symphalangus and Nomascus as the last to diverge. Since most chromosome rearrangements observed in gibbons are either ancestral to all four subgenera or specific for individual species and only a few common derived rearrangements at subsequent branching points have been recorded, all extant gibbons may have diverged within relatively short evolutionary time. In general, chromosomal rearrangements produce changes that should be considered as unique landmarks at the divergence nodes. Thus, molecular cytogenetics could be an important tool to elucidate phylogenies in other species in which speciation may have occurred over very short evolutionary time with not enough genetic (DNA sequence) and other biological divergence to be picked up.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Humans and great apes, together with the gibbons or lesser apes (Hylobatidae), form the primate subfamily of Hominoidae. With 12 or more extant species, gibbons are the most variable members of this taxon. Although gibbons represent our closest relatives, other than the great apes, there is still very limited consent concerning their taxonomy, phylogeny, and evolution. Early systematics (Napier and Napier 1967) divided gibbons into two distinct subgenera that included the siamang (subgenus Symphalangus, one species, Hylobates syndactylus) and Hylobates (all other lesser apes). More recently, it has become evident that four distinct major divisions in lesser apes should be recognized that should include the subgenera Bunopithecus, Hylobates, Symphalangus, and Nomascus. This division is also reflected by the four different karyomorphs found in the four subgenera with representatives such as the hoolock (Bunopithecus hoolock, 2n=38), the white-handed gibbon (Hylobates lar, 2n=44), the siamang (Symphalangus syndactylus, 2n=50), and the white-cheeked gibbon (Nomascus concolor, 2n=52); (Bender and Chu 1963; Prouty et al. 1983; Wurster and Benirschke 1969).
On the basis of various biological traits, different conflicting phylogenetic trees have been published for gibbons (Bruce and Ayala 1979; Garza and Woodruff 1992; Groves 1972; Haimoff et al. 1982; Hall et al. 1996, 1998; Roos and Geissmann 2001; Geissmann 2002). For example, Haimoff et al. (1982), Roos and Geissmann (2001), and Geissmann (2002) have suggested that the evolutionary branching sequence is Nomascus {Symphalangus {Bunopithecus and Hylobates}}, whereas Garza and Woodruff (1992) have identified Symphalangus as the most basal group of the Hylobatidae, followed by Nomascus, with Hylobates and Bunopithecus as the last to diverge. Figure 1 summarizes the taxonomic classification of gibbons into four (sub-) genera with 12 recognized species as is currently accepted by a majority of authors, together with a phylogenetic tree proposed by Geissmann and coworkers, based on vocal and molecular data (Geissmann 2002; Roos and Geissmann 2001).
Preliminary phylogenetic tree of the gibbons, combining trees based on vocal and molecular data (Geissmann 2002; Roos and Geissmann 2001). The vernacular name of each species and the diploid chromosome number for each (sub)-genus are given in brackets. Note: Bunopithecus hoolock, Symphalangus syndactylus, and the concolor group were formerly classified as Hylobates hoolock, Hylobates syndactylus, and Hylobates concolor ssp., i.e. Hylobates concolor leucogenys). Species highlighted bold were included in this study
These various conflicting interpretations of gibbon phylogeny indicate that extant gibbons may have diverged within short evolutionary time in which they have not accumulated enough new characters for a conclusive phylogenetic tree when based on quantitative traits. "Rare genomic events", however, have been found to be highly informative for elucidating phylogenies in species that diverged within relatively short evolutionary time (for a review, see Rokas and Holland 2000). They can provide cladistic landmarks with low levels of homoplasy that would link species phylogenetically. Changes in chromosome morphology represent one class of these rare events that may be especially helpful in the understanding of gibbon phylogeny.
Although very closely related to humans and great apes, the classical banding pattern comparison of gibbon chromosomes have rarely identified homologies between lesser apes and great apes. Except for the X chromosome, which is well conserved, only a few gibbon autosomes show a banding pattern reminiscent of any of those found in humans and the great apes. In contrast, Old World monkeys, such as macaques and baboons, which are much more distantly related to human and great apes than gibbons, have most of their chromosomal syntenies in common with the great apes (Wienberg et al. 1992). Unfortunately, the limited gene mapping studies available for gibbons are mostly restricted to one species (white-cheeked gibbon) and thus have not been helpful in elucidating chromosome homologies in these species either (Créau-Goldberg 1993; Turleau et al. 1983; Van Tuinen and Ledbetter 1989). A comparison of chromosome morphology between gibbon species has revealed additional extensive differences in chromosome banding patterns suggesting various translocations and other rearrangements. Only a few chromosome homologies have been proposed on the basis of banding patterns. Thus, gibbons have clearly experienced a dramatic change in chromosome morphology not found in other primates (Dutrillaux et al. 1975; Marks 1982; Stanyon and Chiarelli 1983; Van Tuinen and Ledbetter 1983). Examinations within distinct gibbon species have further revealed polymorphisms for inversions and translocations (Couturier et al. 1982; Couturier and Lernould 1991; Stanyon et al. 1987). Since only a few individuals had previously been analyzed, it was unclear whether these rearrangements were polymorphisms or defined karyological differences of sub-species or of not yet recognized species. More recently, however, more than 60 individuals of the subgenus Hylobates have been analyzed by chromosome banding and extensive inversion/translocation chromosome polymorphisms have been described (Van Tuinen et al. 1999). No such chromosome polymorphism is known in other primates.
Molecular cytogenetic techniques, such as chromosome painting and fine mapping with defined DNA probes by fluorescence in situ hybridization, allowed, for the first time, a detailed description of such complex chromosome changes during evolution as found in gibbons (Arnold et al. 1996; Wienberg et al. 1990). When applied to entire gibbon karyotypes, all four karyomorphs were analyzed in detail with human chromosome-specific painting probes: three species of the subgenus Hylobates (H.lar, H. agilis, H. klossii; Jauch et al. 1992), the white-cheeked gibbon (Koehler et al. 1995b; Schröck et al. 1996), the siamang, (Koehler et al. 1995a), and more recently, the hoolock (Yu et al. 1997). In their analysis of the chromosome rearrangements in three of the four genera that had been studied at that time, Koehler et al. (1995a) suggested that many of the rearrangements were shared by all three subgenera and probably occurred in their common ancestor. Common derived rearrangements could not be defined with certainty, since in most cases, rearrangements were still too complex. Thus, a firm phylogenetic interpretation of gibbon chromosome evolution could not be proposed by using human chromosome-specific painting probes alone.
To obtain more information about chromosome homologies, we introduced "reciprocal chromosome painting" between human and gibbon species (Arnold et al. 1996). This approach employs both human paint probes and those of at least one representative of the species group to be investigated. It allows the identification of homologous chromosome sub-regions and therefore a more precise interpretation of the origin of complex chromosome rearrangements. This strategy has been helpful in establishing detailed homology maps between human and three gibbon species (N. leucogenys, B. hoolock, and H. lar; Müller et al. 2002, 1998; Nie et al. 2001) representing species of three of the four subgenera.
Here, we present the first attempt to reconstruct the chromosomal phylogeny of all four gibbon subgenera based on "multi-directional" painting data. For this purpose, the siamang, representing the only gibbon not yet studied with reciprocal chromosome painting, has been analyzed both with human and N. leucogenys paint probes. Further, the orangutan has been included in this study as an "outgroup" for lesser apes, since it has a karyotype that is supposed to be most similar to the ancestral form of humans and great apes (Müller and Wienberg 2001). Comparisons of gibbons with the orangutan instead of humans should eliminate the "noise" that stems from various additional derived chromosome rearrangements that has occurred in human but not orangutan genome evolution and that may obscure the identification of rearrangements that happened in the phylogenies leading to extant gibbons. Thus, for the first time, a comprehensive phylogenetic analysis of gibbons by using chromosome rearrangements as traits should be possible.
We present two approaches for the reconstruction of phylogenies: a cladistic approach distinguishing between ancestral and derived chromosome forms in gibbons by a stepwise reconstruction of chromosome rearrangements starting from an inferred ancestral karyotype for all hominoids (Müller and Wienberg 2001). Further, we use an approach in which adjacent homologous segments derived from translocations and intra-chromosomal rearrangements are treated as discrete characters in a parsimony analysis by using the PAUP software package (phylogenetic analysis using parsimony; Swofford 1998).
Materials and methods
Cell samples and chromosome preparation
Chromosome preparations for in situ hybridization experiments were obtained from lymphoblastoid cell lines of a female siamang previously described (Koehler et al. 1995a) and from a female Pongo pygmaeus (orangutan, purchased from ECACC (EB(JC)185, ECACC No. 89072705). Chromosome preparation followed standard cytogenetic protocols.
Probe composition, labeling, in situ hybridization, and microscopy
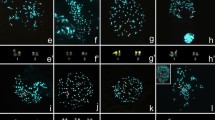
White-cheeked gibbon (N. leucogenys, formerly classified as H. concolor leucogenys or H. leucogenys) and human chromosome-specific painting probes were as described by Müller et al. (1998). Human 24-color karyotyping followed the protocol as described by Müller et al. (2002). All white-cheeked gibbon probes were hybridized in four sets of six to seven combinatorially labeled paint probes as illustrated in Fig. 2. Probe labeling was performed by degenerate-oligonucleotide-primed PCR (Telenius et al. 1992) in the presence of biotin-dUTP, digoxigenin-dUTP (Roche), and TAMRA-dUTP (Applied Biosystems/PE) respectively. In situ hybridization, probe detection, and microscopy was as previously described (Müller et al. 2002). For each hybridization experiment, at least ten metaphases were analyzed.
Multi-color fluorescence in situ hybridization experiments with all 26 white-handed gibbon paint probes on (A–D) orangutan and (E–H) siamang metaphases. To visualize the hybridization pattern and chromosomal counter-stain simultaneously, an overlay of both images is shown. The respective probe composition and false color assignment is given with each metaphase (left). I Hybridization of a human 24-color painting probe set to siamang metaphases
Quantification of homologous chromosome segments
The multi-directional chromosome painting approach was the basis for the generation of an operationally defined chromosomal painting unit termed "chromosomal segment homolog" (CSH). The idea behind the approach is the use of painting probes from highly rearranged karyotypes to define sub-regional chromosome homologies. A CSH for any group of species is that set of chromosome segments that all hybridize to the homologous region of a reference species. The size and numbers of CSHs depend on the number of chromosome rearrangements that have occurred in evolution and on the difference in chromosome numbers of the two or more species that are going to be compared. The advantage of this approach is that it allows a more precise description of chromosome rearrangements and a better quantification of changes by using CSHs as discrete phylogenetic characters (see below). In the present experiments, we used the white-cheeked gibbon as the reference species and defined the CSHs on the basis of the hybridization pattern of its chromosome-specific probes relative to the p-terminus of homologous orangutan chromosomes, which served as the "outgroup" species. For example, the white-cheeked gibbon chromosome 9 probe defines one CSH on orangutan chromosome 1, which is CSH 1d (Fig. 3A). In those cases in which chromosome segments not only differ by translocations, but also by inversions, one white-cheeked gibbon probe may define more than one CSH on the orangutan homolog. For example, the gibbon chromosome 5 probe defines two CSHs 1c on orangutan chromosome 1.
An idiogram of orangutan G-banded chromosomes, together with the assignment of human and white-handed gibbon chromosome-specific painting probes. Chromosome numbering is given below each chromosome, numbers beside chromosomes indicate white-handed gibbon (left) and human (right) homologous chromosome regions, and horizontal bars indicate the borders of homologous regions. Human homologous regions were subdivided into CSHs. Each CSH was defined by the homology of both the human and the corresponding white-handed gibbon homologous chromosome segments and was coded according to its mapping order in relation to the p-terminus of the corresponding orangutan chromosome. B The idiogram of Siamang G-banded chromosomes (adapted from Koehler et al. 1995a), together with the assignment of human and white-handed gibbon chromosome-specific painting probes. Chromosome numbering is given below each chromosome, numbers beside chromosomes indicate white-handed gibbon (left) and human (right) homologous chromosome regions, and horizontal bars indicate the borders of homologous regions
The white-cheeked gibbon was used as the reference species, since it has the highest chromosome number of all gibbon species and thus would probably provide the largest number of CSHs. Further, including the present experiments on the siamang (see below) and published results comparing karyotypes within gibbon species (Müller et al. 1998, 2002; Nie et al. 2001), the white-cheeked gibbon has the most complete hybridization results with other gibbons. The numbering of the individual CSHs defined by white-cheeked gibbon probes follows human chromosome nomenclature, i.e., orangutan chromosome 2, which is homologous to human chromosome 3, shows CSHs 3a–3d (see Fig. 2A).
PAUP analysis
To define discrete characters for a phylogenetic analysis, a binary data matrix based on the presence or absence of each two adjacent CSHs in each species was established (electronic supplementary material 1 and 2). These data were subjected to a maximum parsimony analysis (PAUP 4.0 software; Swofford 1998), by using the exhaustive search option. All characters had the same weight, based on the premise that all chromosome rearrangements occurred by equal chance. The relative stability of nodes was assessed by bootstrap estimates based on 1000 iterations. Each bootstrap replicate involved a heuristic parsimony search with 10 random taxon additions and tree-bisection-reconnection branch swapping.
Results
Multi-directional painting between white-cheeked gibbon, orangutan, and human
To define an ancestral karyotype for all gibbons and CSHs between white-cheeked gibbon and the orangutan, white-cheeked gibbon chromosome-specific probes were painted on orangutan chromosomes. All probes were hybridized in four sets of multicolor probes (see above, Fig. 2A–H), since these probe sets were simpler to analyze in cross species chromosome painting than when using all probes in a single hybridization (i.e. M-FISH or spectral karyotyping, Schröck et al. 1996; Speicher et al. 1996). Since homologies between white-cheeked gibbon and human when using human probes are well documented (Koehler et al. 1995b; Schröck et al. 1996), these experiments were also helpful in the re-evaluation of previously published assignments to human chromosomes. Compared with these previous assignments, novel homologous regions on orangutan chromosomes 4q and 9q (white-cheeked gibbon chromosomes 18 and 12, respectively) were evident that had not previously been identified on homologous human chromosomes. Further, previously unresolved homologies between some white-cheeked gibbon and human chromosomes when using gibbon probes (Müller and Wienberg 2001) and that were attributable to the color redundancy of the probe set could be assigned in the hybridization to the orangutan: orangutan chromosomes 4 (white-cheeked gibbon chromosomes 17 and 20), 5p (white-cheeked gibbon chromosomes 3, 8, 17, and 22), 7 (white-cheeked gibbon chromosomes 9 and 18), and 11 (white-cheeked gibbon chromosomes 17 and 20).
White-cheeked gibbon chromosome-specific probes divided the orangutan karyotype into 76 CSHs (Fig. 3A). Except for homologs to human chromosomes 15, 18, 21, 22, and the sex chromosomes, all other autosomes exhibited at least two CSHs with the maximum number of six different CSHs on orangutan chromosome 5.
Multi-directional painting between white-cheeked gibbon, siamang, and human
The hybridization of white-cheeked gibbon probes on siamang chromosomes revealed a maximum of 83 CSHs. Only siamang chromosome 21 and the sex chromosomes had a single CSH, whereas all other autosomes showed up to six CSHs (Fig. 3B). To clarify inconsistencies with a previous study (Koehler et al. 1995a), the siamang was reinvestigated by using all 24 different human chromosome specific probes simultaneously employing the hybridization protocol described in Müller et al. (2002; Fig. 2I). Additional hybridization signals for human chromosomes 8 and 12 were found that hybridized to siamang chromosomes 1qter, and to 6q and 10q, respectively (Fig. 3B).
Defining chromosomal segment homologs in H. lar and B. hoolock karyotypes
Multi-directional painting experiments have been published for representatives of the two remaining subgenera (Bunopithecus and Hylobates) including humans and white-cheeked gibbon chromosome-specific probes (Jauch et al. 1992; Müller et al. 2002; Nie et al. 2001; Yu et al. 1997). These data were used here to establish CSHs in both species based on homologous chromosome regions defined by the white-cheeked gibbon probes. The published data and the results presented in this paper were consistent for all CSHs, except for two small bands that were found in the hoolock (Nie et al. 2001) and that included bands on hoolock chromosome 8 and 13qter for which no homologous segments were reported in other gibbons. The complete set of CSHs and the observed syntenic associations thereof for all gibbons and for the orangutan karyotypes is given in electronic supplementary material 1.
Ancestral karyotype of all gibbons
The first step in this analysis was to identify the ancestral chromosome forms for all gibbons that would then allow a detailed stepwise examination of derived traits at all nodes of gibbon phylogeny. To determine the ancestral karyotype of all gibbons, individual CSHs and associations of different CSHs were inspected to ascertain whether they were conserved in a putative common ancestor of all gibbon species. Compared with the outgroup, the ancestral gibbon karyotype should incorporate the following chromosome forms: (1) conserved synteny of entire chromosomes, i.e., chromosomes homologous to human 7, 9, 13, 14, 15, 18, 20, 21, 22, and the sex chromosomes, which all showed conserved synteny in at least one extant gibbon species, (2) chromosome regions represented by more than one CSH that showed a conserved segment order between the outgroup and at least one extant gibbon, viz., 1c/1d, 2a/2b, 2d/2e, 4c/4d, 6a/6b/6c/6d, 6e/6f/6e, 10b/10c, and 17a/17b, (3) individual CSHs 1a, 1b, 3a, 8c, 11b, and 12a, which were found as separate entities in one or more gibbon karyotypes, (4) individual CSHs 4a and 5a that were involved in different rearrangements in each phylogenetic lineage. For both (3) and (4), the most parsimonious interpretation is that the entire CSH was fissioned in the common ancestor and then experienced different rearrangements or remained unchanged. (5) We additionally included syntenic associations of CSHs 19b/12d/19b/12d, 10a/4b, 18/11a, 4c/10b, 3d/12b, 12c/3c/8b, 16b/5c/16b/5c, 5b/16a, 3b/8a, and 2b/17b, which are derived translocation products and are present in all four gibbon species (at least as a part of further rearrangements). Putative ancestral chromosome forms 10c/10b/4c/4d and 17b/2b/17b/2b/2a represent compound products of (2) and (5). Association of 12b/19a was only present in three gibbon karyotypes but was nevertheless included since it clearly represented the product of a reciprocal translocation of chromosomes homologous to human 3d/12b/12d and 19a/19b, resulting in ancestral gibbon chromosome forms 19b/12d/19b/12d and 3d/12b/19a. In the white-cheeked gibbon, the 19a fragment was further translocated and was determined to be a species-specific rearrangement.
In conclusion, the suggested ancestral karyotype common to all extant gibbons would have had 2n=66 chromosomes and differed from the putative ancestral hominoid by at least 24 rearrangements: five reciprocal translocations, eight inversions, ten fissions, and one fusion.
Ancestral and derived chromosome rearrangements in the individual gibbon subgenera
Starting from the proposed ancestral gibbon karyotype, we were able to identify common derived rearrangements for the four different subgenera and to distinguish them from those that were species-specific. Except for those already present in the putative gibbon ancestor, the hoolock does not share any chromosome change found in any other gibbon species. However, it has a number of derived rearranged chromosomes; 16 out of 18 autosomes are the product of species-specific rearrangements that include four reciprocal translocations, 19 fusions, and five fissions.
In contrast to the hoolock, the white-cheeked gibbon, siamang, and white-handed gibbon share at least five chromosome forms that should be considered as common derived forms: fusion products 2e/2d/7a/7b/7c, 8a/3b/11a/18, 4a/5a, and 22/5b/16a, and the 6e/6f/6e to 6e/6f inversion. From this assumed ancestral karyotype of these three species (2n=60 chromosomes), the karyotype observed in extant white-handed gibbons can be derived by six fusions, three reciprocal translocations, and one inversion.
Further, the putative common ancestor of the white-cheeked gibbon and the siamang may have acquired seven chromosome forms not found in the white-handed gibbon: the products of one fusion and four fissions resulting in chromosome forms 10c/6e/6f, 10b/4c/4d, 2d/7a, 2e, 7b, and 7c, and an inversion that changed 6a/6b/6c/6d to 6a/6b/6d/6c. Again, both species show a number of derived rearrangements not found in any other gibbon: a minimum of six fusions, seven reciprocal translocations, and one inversion in the white-cheeked gibbon and seven fusions, three reciprocal translocations, and one inversion in the siamang.
In conclusion, this analysis suggests a phylogeny in which the hoolock split first from the ancestral gibbon line, then the white-handed gibbon, and finally, the white-cheeked gibbon and siamang (Fig. 4).
Reconstruction of chromosomal changes in gibbons leads to one most parsimonious phylogenetic tree (PPY Pongo pygmaeus, BHO B. hoolock, HLA H. lar, SSY S. syndactylus, NCO N. leucogenys, black circles fusion, rectangles inversions, × reciprocal translocations). Bold numbers at the two nodes indicate bootstrap values obtained in the phylogenetic analysis by using the PAUP software package. NCO/SSY/HLA share derived fusion products 2e/2d/7a/7b/7c, 8a/3b/11a/18, 4a/5a, and 22/5b/16a, and the 6e/6f/6e to 6e/6f inversion. NCO/SSY share the products of one fusion and four fissions resulting in chromosome forms 10c/6e/6f, 10b/4c/4d, 2d/7a, 2e, 7b, and 7c, and an inversion that changed 6a/6b/6c/6d to 6a/6b/6d/6c. Chromosome nomenclature refers to homology with human chromosomes coded as CSHs
PAUP analysis of gibbon chromosome reshuffling
We employed adjacent CSHs as discrete characters in a PAUP analysis for all four gibbon species by using the white-cheeked gibbon chromosome-specific hybridizations on the siamang presented here and published data for the other two subgenera (Jauch et al. 1992; Müller et al. 2002; Nie et al. 2001; Yu et al. 1997). These data resulted in 130 different associations of CSHs and thus in different numerical characters (electronic supplementary material 1). From these, 32 characters were parsimony informative. The resulting binary data matrix with all species included in this study is given in electronic supplementary material 2. Maximum parsimony analysis resulted in one most parsimonious tree (consistency index=0.95; retention index=0.56; homoplasy index=0.05) with the branching sequence {hoolock {white-handed gibbon {white-cheeked gibbon and siamang}}}. This maximum parsimony tree is illustrated in Fig. 4.
Discussion
Both the cladistic reconstruction based on the identification of ancestral versus derived chromosome forms and the PAUP analysis resulted in the same most parsimonious phylogenetic tree, placing the representatives of the subgenus Bunopithecus as the most basal group of the Hylobatidae, followed by the representative of Hylobates, with Symphalangus and Nomascus as the last to diverge. These results contrast with the findings based on other biological traits (see above). Various published phylogenies, however, agree with the close relationship of Bunopithecus and Hylobates and that of Symphalangus and Nomascus (Haimoff et al. 1982; Roos and Geissmann 2001; Geissmann 2002). Since the hoolock does not share a single derived chromosome rearrangement with other gibbons, whereas other nodes show several common derived traits, this is strong support for the basal position of Bunopithecus.
Most quantitative approaches for the reconstruction of phylogenies from molecular data presume constant change over time (a "molecular clock"). This is certainly not valid for chromosome rearrangements in general (Wienberg et al. 2000), and gibbons are the most outstanding known exception from this presumption as compared with other primates. Nevertheless, since many chromosome rearrangements are either ancestral to all four subgenera or species-specific, and just five and six common derived rearrangements at the two subsequent branching points have been observed, this may argue for a rapid evolutionary divergence of all extant gibbon subgenera.
Whether chromosome rearrangements play a specific role in species divergence has been disputed over the decades. In any case, the majority of chromosomal rearrangements result in changes that should be considered as unique landmarks at the divergence nodes. Various other mammalian taxa in which speciation may have occurred over very short evolutionary time, but with not enough genetic and other biological divergence, may be picked up by other methods. In these cases, the approach presented here may be an important tool to elucidate phylogenies.
References
Arnold N, Stanyon R, Jauch A, O'Brien P, Wienberg J (1996) Identification of complex chromosome rearrangements in the gibbon by fluorescent in situ hybridization (FISH) of a human chromosome 2q specific microlibrary, yeast artificial chromosomes, and reciprocal chromosome painting. Cytogenet Cell Genet 74:80–85
Bender MA, Chu EHY (1963) The chromosomes of primates. In: Buettner-Janusch J (ed) Evolutionary and genetic biology of primates, vol 1. Academic Press, New York, pp 261–310
Bruce EJ, Ayala FJ (1979) Phylogenetic relationships between man and the apes: electrophoretic evidence. Evolution 33:1040–1056
Couturier J, Lernould J-M (1991) Karyotypic study of four gibbon forms provisionally considered as subspecies of Hylobates (Nomascus) concolor (Primates, Hylobatidae). Folia Primatol (Basel) 56:95–104
Couturier J, Dutrillaux B, Turleau C, Grouchy J de (1982) Comparative karyotyping of our gibbon species or subspecies. Ann Génét (Paris) 25:5–10
Créau-Goldberg N (1993) Primate genetic maps. In: O´Brien SJ (ed) Genetic maps, locus maps of complex genomes; nonhuman vertebrates, vol 4. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Dutrillaux B, Rethore MO, Aurias A, Goustard M (1975) Karyotype analysis of 2 species of gibbons (Hylobates lar and H. concolor) with different banding species. Cytogenet Cell Genet 15:81–91
Garza JC, Woodruff DS (1992) A phylogenetic study of the gibbons (Hylobates) using DNA obtained noninvasively from hair. Mol Phylogenet Evol 1:202–210
Geissmann T (2002) Taxonomy and evolution of gibbons. In: Soligo C, Anzenberger G, Martin RD (eds).Anthropology and primatology into the third millennium: the centenary congress of the Zürich Anthropological Institute. Evolutionary anthropology, vol 11, supplement 1. Wiley-Liss, New York, pp 28–31
Groves CP (1972) Systematics and phylogeny of gibbons. In: Rumbaugh DM (ed) Gibbon and siamang, vol 1. Karger, Basel, pp 1–89
Haimoff EH, Chivers DJ, Gittins SP, Whitten T (1982) A phylogeny of gibbons (Hylobates spp) based on morphological and behavioural characters. Folia Primatol (Basel) 39: 213–237
Hall LM, Jones D, Wood B (1996) Evolutionary relationships between gibbon subgenera inferred from DNA sequence data. Biochem Soc Trans 24:416S
Hall LM, Jones DS, Wood BA (1998) Evolution of the gibbon subgenera inferred from cytochrome b DNA sequence data. Mol Phylogenet Evol 10:281–286
Jauch A, Wienberg J, Stanyon R, Arnold N, Tofanelli S, Ishida T, Cremer T (1992) Reconstruction of genomic rearrangements in great apes and gibbons by chromosome painting. Proc Natl Acad Sci USA 89:8611–8615
Koehler U, Arnold N, Wienberg J, Tofanelli S, Stanyon R (1995a) Genomic reorganization and disrupted chromosomal synteny in the siamang (Hylobates syndactylus) revealed by fluorescence in situ hybridization. Am J Phys Anthropol 97:37–47
Koehler U, Bigoni F, Wienberg J, Stanyon R (1995b) Genomic reorganization in the concolor gibbon (Hylobates concolor) revealed by chromosome painting. Genomics 30:287–292
Marks J (1982) Evolutionary tempo and phylogenetic inference based on primate karyotypes. Cytogenet Cell Genet 34:261–264
Müller S, Wienberg J (2001) "Bar-coding" primate chromosomes: molecular cytogenetic screening for the ancestral hominoid karyotype. Hum Genet 109:85–94
Müller S, O'Brien PC, Ferguson-Smith MA, Wienberg J (1998) Cross-species colour segmenting: a novel tool in human karyotype analysis. Cytometry 33:445–452
Müller S, Neusser M, Wienberg J (2002) Towards unlimited colors for fluorescence in-situ hybridization (FISH). Chromosome Res 10:223–232
Napier JR, Napier PH (1967) A handbook of living primates. Academic Press, London
Nie W, Rens W, Wang J, Yang F (2001) Conserved chromosome segments in Hylobates hoolock revealed by human and H. leucogenys paint probes. Cytogenet Cell Genet 92:248–253
Prouty LA, Buchanan PD, Pollitzer WS, Mootnick AR (1983) A presumptive new hylobatid subgenus with 38 chromosomes. Cytogenet Cell Genet 35:141–142
Rokas A, Holland WH (2000) Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol 15:454–459
Roos C, Geissmann T (2001) Molecular phylogeny of the major hylobatid divisions. Mol Phylogenet Evol 19:486–494
Schröck E, du Manoir S, Veldman T, Schoell B, Wienberg J, Ferguson-Smith MA, Ning Y, Ledbetter DH, Bar-Am I, Soenksen D, Garini Y, Ried T (1996) Multicolor spectral karyotyping of human chromosomes. Science 273:494–497
Speicher MR, Gwyn Ballard S, Ward DC (1996) Karyotyping human chromosomes by combinatorial multi-fluor FISH. Nat Genet 12:368–375
Stanyon R, Chiarelli B (1983) Mode and tempo in primate chromosomal evolution: implications for hylobatid phylogeny. J Hum Evol 10:305–315
Stanyon R, Sineo L, Chiarelli B, Camperio-Ciani A, Haimoff EH, Mootnick AR, Suturman DR (1987) Banded karyotypes of the 44-chromosome gibbons. Folia Primatol (Basel) 48:56–64
Swofford DL (1998) PAUP*. Phylogenetic analysis using parsimony (*and other methods), 4th edn. Sinauer Associates, Sunderland, Mass.
Telenius H, Pelmear AHP, Tunnacliffe A, Carter NP, Behmel A, Ferguson-Smith MA, Nordenskjold M, Pfragner R, Ponder BAJ (1992) Cytogenetic analysis by chromosome painting using DOP-PCR amplified flow-sorted chromosomes. Genes Chromosome Cancer 4:257–263
Turleau C, Creau-Goldberg N, Cochet C, Grouchy J de (1983) Gene mapping of the gibbon. Its position in primate evolution. Hum Genet 64:65–72
Van Tuinen P, Ledbetter DH (1983) Cytogenetic comparison and phylogeny of three species of Hylobatidae. Am J Phys Anthropol 61:453–466
Van Tuinen P, Ledbetter DH (1989) New confirmatory and regional gene assignments in the white-cheeked gibbon Hylobates concolor. Cytogenet Cell Genet 51:1094–1095
Van Tuinen P, Mootnick AR, Kingswood SC, Hale DW, Kumamoto AT (1999) Complex, compound inversion/translocation polymorphism in an ape: presumptive intermediate stage in the karyotypic evolution of the agile gibbon Hylobates agilis. Am J Phys Anthropol 110:129–142
Wienberg J, Jauch A, Stanyon R, Cremer T (1990) Molecular cytotaxonomy of primates by chromosomal in situ suppression hybridization. Genomics 8:347–350
Wienberg J, Stanyon R, Jauch A, Cremer T (1992) Homologies in human and Macaca fuscata chromosomes revealed by in situ suppression hybridization with human chromosome specific DNA libraries. Chromosoma 101:265–270
Wienberg J, Frönicke L, Stanyon R (2000) Insights into mammalian genome organization and evolution by molecular cytogenetics. In: Clark MS (ed) Comparative genomics. Kluver, Dordrecht, pp 207–244
Wurster DH, Benirschke K (1969) Chromosomes of some primates. Mamm Chromosome Newsletter 10:3
Yu D, Yang F, Liu R (1997) A comparative chromosome map between human and Hylobates hoolock built by chromosome painting. Yi Chuan Xue Bao 24:417–423
Acknowledgments
This work was funded by the Deutsche Forschungsgemeinschaft (DFG Wi 970/6–1).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Müller, S., Hollatz, M. & Wienberg, J. Chromosomal phylogeny and evolution of gibbons (Hylobatidae). Hum Genet 113, 493–501 (2003). https://doi.org/10.1007/s00439-003-0997-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-003-0997-2