Abstract
Calcium-dependent protein kinases (CDPKs) and CDPK-related kinases (CRKs) play multiple roles in plant. Nevertheless, genome-wide identification of these two families is limited to several plant species, and role of CRKs in disease resistance remains unclear. In this study, we identified the CDPK and CRK gene families in genome of the economically important crop tomato (Solanum lycopersicum L.) and analyzed their function in resistance to various pathogens. Twenty-nine CDPK and six CRK genes were identified in tomato genome. Both SlCDPK and SlCRK proteins harbored an STKc_CAMK type protein kinase domain, while only SlCDPKs contained EF-hand type Ca2+ binding domain(s). Phylogenetic analysis revealed that plant CRK family diverged early from CDPKs, and shared a common ancestor gene with subgroup IV CDPKs. Subgroup IV SlCDPK proteins were basic and their genes contained 11 introns, which were distinguished from other subgroups but similar to CRKs. Subgroup I SlCDPKs generally did not carry an N-terminal myristoylation motif while those of the remaining subgroups and SlCRKs universally did. SlCDPK and SlCRK genes were differently responsive to pathogenic stimuli. Furthermore, silencing analyses demonstrated that SlCDPK18 and SlCDPK10 positively regulated nonhost resistance to Xanthomonas oryzae pv. oryzae and host resistance to Pseudomonas syringae pv. tomato (Pst) DC3000, respectively, while SlCRK6 positively regulated resistance to both Pst DC3000 and Sclerotinia sclerotiorum in tomato. In conclusion, CRKs apparently evolved from CDPK lineage, SlCDPK and SlCRK genes regulate a wide range of resistance and SlCRK6 is the first CRK gene proved to function in plant disease resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Calcium-dependent protein kinases (CDPKs) are main receptors of Ca2+ signal (Sanders et al. 2002; Kudla et al. 2010; Reddy et al. 2011). They are multifunctional in plants, including regulation of plant growth, development as well as abiotic and biotic stress resistance (Boudsocq and Sheen 2013; Romeis and Herde 2014). CDPK-related kinase (CRK) is another type of protein kinase closely related to CDPKs (Harmon et al. 2000). Some CRKs are involved in plant development or abiotic stress tolerance (Leclercq et al. 2005; Li et al. 2006; Liu et al. 2008; Rigo et al. 2013; Tao and Lu 2013). Currently, genome-wide identification of these two families is mainly conducted in model plant species. Moreover, role of CRKs in plant disease resistance remains unclear. The aim of this study is to identify CDPK and CRK gene families in genome of the economically important crop tomato (Solanum lycopersicum L.), and analyze their function in resistance to various pathogens. The results provide new insights into composition, phylogeny and function of plant CDPK and CRK families.
CDPKs and CRKs are two structurally related families of protein kinases. CDPKs carry two kinds of key domains, Ser/Thr kinase domain and EF-hand type calcium-binding domain. In addition, they contain an N-terminal variable domain, an auto-inhibitory junction region and a C-terminus (Cheng et al. 2002; Harper et al. 2004; Ludwig et al. 2004; Harper and Harmon 2005). However, CRKs solely harbor a Ser/Thr kinase domain at their N-termini of protein sequences. Their C-termini only contain degenerated EF-hand-like sequences (Harmon et al. 2000). Therefore, CDPKs function in a calcium-dependent way, whereas CRKs are thought to act in a calcium-independent way (Harmon et al. 2000).
CDPK genes have been identified in plant as well as in green algae, oomycetes and protists, but are not found in fungi and animals (Hamel et al. 2014; Valmonte et al. 2014). Genome-wide identification of CDPK genes has been conducted individually in a number of plant species, such as Arabidopsis thaliana (Cheng et al. 2002), Oryza sativa (Asano et al. 2005; Ray et al. 2007; Boudsocq and Sheen 2013), Triticum aestivum (Li et al. 2008), Gossypium raimondii (Liu et al. 2014), Brassica napus (Zhang et al. 2014), Populus trichocarpa (Zuo et al. 2013), Selaginella moellendorffii, and Physcomitrella patens (Hamel et al. 2014), as well as comprehensively in many species (Valmonte et al. 2014). Results of these studies reveal that CDPK proteins in a plant species are typically encoded by a gene family, which is usually classified into four distinct subgroups (Hamel et al. 2014; Valmonte et al. 2014). There are 34 and 31 CDPK genes in Arabidopsis (Cheng et al. 2002) and rice genome (Ray et al. 2007; Boudsocq and Sheen 2013), respectively. Compared with CDPKs, CRKs are relatively less studied. Genome-wide analysis has identified eight CRKs in Arabidopsis (Hrabak et al. 2003), five CRKs in rice (Asano et al. 2005) and nine CRKs in P. trichocarpa (Zuo et al. 2013), respectively. Nevertheless, genome-wide identification of the CDPK and CRK gene families in many economically important crop plant species such as tomato (Solanum lycopersicum L.) has not yet been conducted.
CDPK substrates have increasingly been identified. CDPK may directly phosphorylate an MAPK to compromise MAPK signaling to regulate stress responses and disease resistance (Ludwig et al. 2005; Xie et al. 2014). LeCDPK2 directly phosphorylates LeACS2 to regulate ethylene biosynthesis in response to wound signaling (Kamiyoshihara et al. 2010), while AtCDPK4 and AtCDPK11 and StCDPK2 may target ABA-responsive transcription factors ABF1 and ABF4 to regulate ABA signaling (Zhu et al. 2007). StCDPK5 and AtCDPK1/2/4/5/11 phosphorylate and thereby activate NADPH oxidase to promote ROS production in response to abiotic and biotic stimuli (Kobayashi et al. 2007; Gao et al. 2013). AtCDPK32 interacts with AtCNGC18 to confer severe depolarization of pollen tube growth in tobacco (Zhou et al. 2013). OsCDPK4 plays a positive role in rice tolerance to salt and drought stress by protection of cellular membranes from lipid peroxidation (Campo et al. 2014). CDPKs are widely involved in the regulation of various types of disease resistance (Boudsocq et al. 2010; Boudsocq and Sheen 2013; Romeis and Herde 2014). AtCDPK28 phosphorylates BIK1 to attenuate PTI and antibacterial immunity (Monaghan et al. 2014), while AtCDPK1 plays a positive role in Arabidopsis resistance to various pathogens by promoting salicylic acid (SA) signaling pathway (Coca and San Segundo 2010). Six AtCDPKs are involved in the Arabidopsis NLR immune signaling via distinct functions, AtCDPK1/2 regulating the initiation of programmed cell death, AtCDPK4/5/6/11 phosphorylating specific WRKY transcription factors to regulate the immune gene expression, while AtCDPK1/2/4/11 phosphorylate NADPH oxidases to induce the production of ROS (Gao et al. 2013). NtCDPK2 is involved in Cf-4/Avr4 and Cf-9/Avr9 dependent hypersensitive response (HR) induction (Romeis et al. 2001). Additionally, CDPKs not only locally but also systemically regulate plant defense (Romeis and Herde 2014). Moreover, a CDPK gene may simultaneously regulate several biological processes. For example, OsCDPK12 positively regulates salt tolerance while negatively affects the blast resistance by affecting ABA signaling and suppressing ROS production (Asano et al. 2012).
Unlike CDPKs, whose functions have been widely studied, functional analysis of CRKs has been mainly conducted in Arabidopsis. AtCRK1 binds CaM in a Ca2+-dependent manner but phosphorylates itself and substrates such as histone IIIS and syntide-2 in a Ca2+-independent manner (Wang et al. 2004). It positively regulates plant tolerance to salt and heat stresses (Liu et al. 2008; Tao and Lu 2013). AtCRK3 interacts with a cytosolic glutamine synthetase AtGLN1;1 to regulate nitrogen remobilization during leaf senescence (Li et al. 2006), while AtCRK5 functions in primary root elongation and gravitropic bending of shoots and roots in Arabidopsis (Rigo et al. 2013). Additionally, a tomato CRK (LeCRK1) was found to play a role in the fruit ripening process (Leclercq et al. 2005). The role of CRKs in plant disease resistance remains unclear.
In this study, we conducted a genome-wide identification of the CDPK and CRK families in tomato and analyzed the function of a set of these genes in disease resistance. Our data demonstrated that the structural and biochemical features of SlCDPK (previously LeCDPK) family are obviously subgroup dependent. The SlCRK family shared the same ancestor with subgroup IV SlCDPKs. Additionally, our results revealed that SlCDPK genes regulate a wide range of resistance in tomato but effectiveness against individual pathogen is CDPK gene dependent. SlCRK6 was proved to function in tomato resistance to both Sclerotinia sclerotiorum and Pseudomonas syringae pv. tomato DC3000. This is the first report that demonstrates a role of a CRK gene in plant disease resistance.
Materials and methods
Identification of CDPK and CRK genes in tomato genome and CRK genes in Selaginella moellendorffii and Physcomitrella patens
To identify CDPK and CRK genes in tomato (Solanum lycopersicum L.), all 34 Arabidopsis CDPK protein sequences were collected through searching the genome sequence databases TAIR (The Arabidopsis Information Resource, http://www.arabidopsis.org/). All retrieved AtCDPK protein sequences were used to BLASTp search the tomato genome database (http://solgenomics.net/). All non-redundant sequences were collected, and subjected to domain analysis using the Pfam (http://pfam.sanger.ac.uk/), SMART (http://smart.embl-heidelberg.de/), COG and Conserved Domain Database (CDD) (http://www.ncbi.nlm.nih.gov/cdd) programs. The proteins containing both an STKc_CAMK kinase domain and two EF-hand type domains (four EF-hand motifs) were considered as prototypical CDPK proteins. The remaining full length proteins that possessed an STKc_CAMK kinase domain were further subjected to phylogenetic tree construction together with the Arabidopsis and rice CRKs. Those clustered with Arabidopsis and rice CRKs were recognized as tomato CRKs. Similar approaches were used to identify CRKs in Selaginella moellendorffii, Physcomitrella patens and algal species (Chlamydomonas reinhardtii, Volvox carteri, Coccomyxa subellipsoidea C-169, Micromonas pusilla CCMP1545, Micromonas sp. RCC299, Ostreococcus lucimarinus) whose genomes are deposited in Phytozome database version 10 (http://phytozome.jgi.doe.gov/pz/portal.html). The pI value and molecular weight of CDPK and CRK proteins were predicted by DNAStar software.
Sequence comparison, gene structure and phylogenetic analyses of SlCDPK and SlCRK genes
CDPK protein sequences from Arabidopsis and tomato and CRK protein sequences from Arabidopsis, tomato, rice, Populus trichocarpa, Selaginella moellendorffii, Physomitrella patens and representatives of algal CDPKs were aligned using MUSCLE program (Edgar 2004). The phylogenetic trees were constructed based on the alignments using MEGA 5.0 by maximum likelihood (ML) with the JTT model (Jones et al. 1992; Tamura et al. 2011). One thousand bootstrap replicates were performed to evaluate the support of clusters and nodes. Three apicomplexan CDPKs, TgCDPK1 (ToxoDB ID TGME49_301440), PfCDPK3 (PlasmoDB ID PF3D7_0310100) and CpCDPK1 (CryptoDB ID cgd3_920), were included as outgroup for rooted tree construction (Valmonte et al. 2014). The exon/intron structure of SlCDPK and SlCRK genes was analyzed online using the Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn/) with default settings (Guo et al. 2007). For comparison of EF-hands of tomato CDPKs with the corresponding region of tomato CRKs, the sequences were aligned using MUSCLE program (Edgar 2004) and visualized using GeneDoc 2.6 software.
N-terminal myristoylation prediction
The N-terminal myristoylation of all SlCDPK, SlCRK, SmCRK and PpCRK proteins was predicted using the Myristoylator program in ExPASy (http://web.expasy.org/myristoylator/) with default settings (Bologna et al. 2004).
Plant materials for expression analysis
Tomato plants were grown in growth chambers at 28 °C with a 16 h/8 h light/dark daily cycle. For fungi pathogen inoculation, Sclerotinia sclerotiorum was grown at 25 °C on potato dextrose agar (PDA) plates for 2 days. PDA plugs of 5 mm in diameter containing actively growing S. sclerotiorum mycelia were placed on the fully developed leaves of the 7- to 8-week-old tomato plants. For bacterial inoculation, Pseudomonas syringae pv. tomato (Pst) DC3000 and Xanthomonas oryzae pv. oryzae (Xoo) were incubated overnight at 28 °C on King’s B plates containing rifampicin (50 μg/ml) and NA medium plates containing carbenicillin (50 μg/ml), respectively. After overnight shaking, the bacterial cells were collected by centrifugation, resuspended in 10 mM MgCl2 buffer or sterilized ddH2O and diluted to an OD600 of 0.002 and 0.5, respectively. The prepared bacterial suspensions (with 10 mM MgCl2 buffer or sterilized ddH2O as controls) were infiltrated into leaves of tomato plants (Zhao et al. 2013). For oxalic acid (OA) treatment, tomato leaves were infiltrated with 500 μM of OA (Kim et al. 2011). Samples were collected for gene expression analysis at two time points after inoculation or treatment; 0 and 12 h for S. sclerotiorum, 0 and 8 h for Xoo, 0 and 4 h for Pst DC3000 as well as 0 and 4 h for OA treatment.
Gene expression analyses by RT-qPCR
Real-time quantitative RT-PCR (RT-qPCR) analyses and subsequent statistical analyses of the gene expression data were conducted as described (Zhao et al. 2013). The primers used in RT-qPCR analyses are listed in Table S1.
VIGS manipulation procedure and plant disease resistance analysis
SlCDPK10/12/13/18 and SlCRK4/6 were selected for virus-induced gene silencing (VIGS) analysis. VIGS analysis in tomato was conducted as described (Wang et al. 2006; Cai et al. 2007; Zhao et al. 2013) except using the recombinant pTRV2 with insertion of an eGFP fragment instead of empty pTRV2 as a negative control vector to repress the viral symptom efficiently (Cheng et al. 2012). Gene-specific VIGS-targeted fragments from CDS regions of SlCDPK10, SlCDPK12 and SlCDPK18 and 5′ UTR regions of SlCDPK13 as well as SlCRK4 and SlCRK6 were cloned and ligated into the VIGS vector PYL156 (pTRV2), which were immediately electroporated into Agrobacterium tumefaciens strain GV3101 for VIGS analyses (Saand et al. 2015). Primers used in the VIGS experiments are listed in Table S1. VIGS analyses were conducted with the vacuum-infiltration delivery approach. The agro-inoculated plants were grown in a plant growth chamber at 21 °C with a 16 h/8 h light/dark regime. Three weeks later, the plants were subjected to disease resistance analyses (Zhao et al. 2013; Saand et al. 2015). They were inoculated with nonhost pathogen Xoo and host pathogens S. sclerotiorum and Pst DC3000 as described above. For each pathogen, at least six silenced plants were examined and the experiments were conducted three times independently. Data were analyzed using SPSS (verson19.0) by Student’s t test and Duncan’s multiple range test (DMRT) (P value ≤0.05).
Results
Identification of CDPK and CRK genes in tomato (Solanum lycopersicum L.) genome and CRK genes in spikemoss (Selaginella moellendorffii) and moss (Physcomitrella patens) genomes
BLASTp searches of the tomato genome using all 34 Arabidopsis CDPK protein sequences as templates retrieved 197 non-redundant sequences. After domain composition analysis of these sequences using Pfam, SMART, COG and CDD programs (Table S2), 29 of them were found to possess both STKc_CAMK protein kinase and EF-hand domains and were thus recognized as tomato CDPKs. To identify the tomato CRKs, a phylogenetic tree was constructed for the remaining 15 full length sequences containing solely an STKc_CAMK kinase domain together with the known Arabidopsis and rice CRKs. Six of them clustered with the known Arabidopsis and rice CRKs into a distinct clade, separated from the remaining tomato sequences, and were thus identified as tomato CRKs (Fig. S1). Following similar BLASTp searching, domain composition and phylogenetic analyses, two and five CRKs were identified in spikemoss (Selaginella moellendorffii) and moss (Physcomitrella patens) genomes, respectively (Table 1). However, no CRK was identified from algal species (data not shown). We assigned names of all individual SlCDPK and CRK members in ascending order in accordance with group numbers on the basis of phylogenetic tree for easy recognition.
Phylogeny of CDPK and CRK families
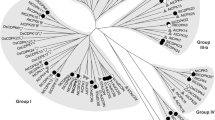
To examine the phylogenetic relationship among the CDPKs and CRKs, kinase domains of CDPKs from Arabidopsis and tomato, and CRKs from four higher flowering plant species, one primitive lycophyte and one moss were subjected to multiple sequence alignment along with representatives of algal CDPKs. Subsequently, a rooted maximum likelihood (ML) phylogenetic tree was constructed using three apicomplexan CDPKs as outgroup (Fig. 1). The ML tree demonstrated that the CDPK family of both tomato and Arabidopsis and all CRKs clustered distinctly from those of algal CDPKs. CDPKs formed four subgroups, while the CRK family of all six plant species including both higher and lower plant species was divided into two subgroups. All CRKs of lower plant species clustered in subgroup I along with a subclade of CRKs of higher plant species, while the remaining CRKs of higher plant species formed subgroup II which was split into two clades (Fig. 1). Together with the finding that CRK family does not exist in algal species (data not shown), it is obvious that CRK family may have emerged very early in land plant species before the divergence of nonvascular and vascular plant species, and the diversification of CRKs in land plant species apparently resulted from gene duplications and rearrangement. Remarkably, the CRK family clustered with the subgroup IV CDPKs, which clearly separated from the other three subgroups of CDPK family with a strong bootstrap support (Fig. 1). This result reveals that the CRK family shares ancestral CDPK gene with subgroup IV CDPKs, which is distinct from the remaining three subgroups of CDPK family. Collectively, CRK gene family is most likely to have arisen from a CDPK ancestor and its appearance possibly predated diversification of CDPKs and separation of land plant into nonvascular and vascular plants, but after the split of algae and the ancestor of plant lineage.
Phylogenetic tree of plant CDPK and CRK proteins. The tree was created based on the alignment of kinase domain of protein sequences using MUSCLE program. Maximum likelihood (ML) method was used with bootstrap of 1000 in MEGA 5.0. CDPK sequences from Arabidopsis, tomato and algae as well as CRK sequences from Arabidopsis, tomato, rice, Populus trichocarpa, Selaginella moellendorffii and Physcomitrella patens were subjected to tree construction. Three apicomplexan CDPKs, TgCDPK1 (ToxoDB ID TGME49_301440), PfCDPK3 (PlasmoDB ID PF3D7_0310100) and CpCDPK1 (CryptoDB ID cgd3_920), were also included as outgroup. Tomato CDPKs and CRKs are indicated as a solid circle
In the SlCDPK family, 13 members, SlCDPK1 to SlCDPK13, clustered into subgroup I; eight members, SlCDPK14 to SlCDPK21, comprised subgroup II; six members, SlCDPK22 to SlCDPK27, constituted subgroup III, while the remaining two members SlCDPK28 and SlCDPK29 formed subgroup IV (Fig. 1). Two members of the SlCRK family, SlCRK1 and SlCRK2, belong to subgroup I, while four members, SlCR K3 to SlCRK6, represent subgroup II (Figs. 1, 2, 3). Regarding the CRK family in the lower plants S. moellendorffii and P. patens, all two and five members, respectively, converged in subgroup I (Fig. 1).
Multiple sequence alignment of EF-hands of tomato CDPKs with the corresponding region of tomato CRKs. Sequence alignment was conducted using MUSCLE program and visualized using GeneDoc 2.6 software. Conserved residues are shaded gray and black, while predicted functional Ca2+ binding residues are highlighted in red
Schematic diagram indicating the exon/intron structure of CDPK and CRK genes. Exon/intron configuration was analyzed with tomato CDPK genes and CRK genes from tomato, Selaginella moellendorffii and Physcomitrella patens. The locus number of these CDPK and CRK genes is listed in Table 1. Exons and introns are indicated as green boxes and black lines, respectively. The intron phase numbers 0, 1 and 2 are labeled at the beginning of each intron. The diagram is drawn to scale. An unrooted tree for their protein sequences is also shown in the left side
Prediction of biochemical characteristics of SlCDPKs and plant CRKs
All SlCDPK proteins were composed of approximately 500–600 amino acids (Table 1). As a result, the molecular weight of the SlCDPK proteins was around 60 kDa. Additionally, the predicted pI value of the SlCDPKs was from 4.94 (SlCDPK7) to 9.28 (SlCDPK29) and apparently varied among the different subgroups of the SlCDPKs. It seems to increase from subgroup I to IV as shown by both the averaged pI and the range of pI of the subgroups. The predicted averaged pI of the subgroups I–IV was 5.40, 6.01, 6.33 and 9.12, respectively, while the range of pI of the subgroups I–IV was 4.94–5.92, 5.24–6.87, 5.84–6.84 and 8.95–9.28, respectively (Table 1). The predicted protein pI data demonstrated that the two SlCDPKs of subgroup IV are basic proteins while all the remaining 27 SlCDPKs are either acidic or neutral proteins. The biochemical features of CRKs from three species with different evolutionary positions were compared. The six SlCRK proteins contained 574–607 amino acids and their corresponding predicted molecular weight was 64.32–67.92 kDa. They were all basic proteins with a predicted pI value of 7.77–9.07 (Table 1). All the five CRKs from the nonvascular plant P. patens (PpCRKs) were found to have around 600 amino acids with predicted molecular weight of about 67 kDa. They also appeared to be basic proteins with a pI value of 7.93–9.28 (Table 1), while the two CRK genes from the vascular nonflowering plant S. moellendorffii (SmCRKs) encode proteins with around 580 amino acids with a molecular weight of about 65 kDa. These were basic proteins as well as their pI value was estimated to be 9.17 and 9.33, respectively (Table 1). These data reveal that plant CRKs are highly conserved with respect to their biochemical characteristics, which are highly similar to those of subgroup IV CDPKs. This supports the clustering of CRKs and CDPKs in the phylogenetic tree (Figs. 1, 3).
Domain and motif composition of SlCDPKs and plant CRKs
CDPKs should contain at least a protein kinase domain and a calcium-binding domain. As expected, all SlCDPKs harbored an STKc_CAMK type protein kinase domain and a calcium-binding domain, which consisted of two pairs of EF-hands (Table 1; Fig. S2). CRKs from tomato, S. moellendorffii and P. patens also contained an STKc_CAMK type protein kinase domain at a similar position of CDPK proteins. However, they did not contain a canonical complete EF-hand domain. Instead, they possessed degenerated EF-hand-like sequences. For example, SlCRK4, SlCRK5, PpCRK4 and PpCRK5 carried a FRQ1-like domain, while SlCRK6 possessed an incomplete EH-like domain (Table 1; Fig. S3). FRQ1 and EH are two members of the EF-hand superfamily (Confalonieri and Di Fiore 2002; Huttner et al. 2003). To further clarify the differences in the C terminal region of the SlCDPK and SlCRK protein sequences, pairwise comparison of this region in the two protein families was performed. Results revealed low similarities among SlCDPK and SlCRK protein sequences in this region. Importantly, most of the calcium-binding sites in EF-hand motifs of the SlCRKs were substituted by physico-chemically distinct amino acids except the third EF-hand. SlCRK4 displayed a complete third EF-hand motif, while for the other SlCRKs, three of the four calcium-binding residues of this EF-hand were conserved, whereas the remaining one was a D–E substitution. Hence, this EF-hand of SlCRKs might be still functional. In addition, this sequence analysis also further reflected closeness between subgroup IV SlCDPKs and SlCRKs. For example, SlCRK4 exhibited up to 39 % similarity to SlCDPK28 and SlCDPK29 each, while the SlCRK protein sequences generally showed only less than 28 % similarity to SlCDPKs of other subgroups. These results unveiled that degeneration of EF-hand motifs of SlCRKs during the emergence of this family from CDPK involved mutational substitution of residues required for Ca2+ binding with eventual activity reduction or even loss of the EF-hand domains (Fig. 2).
Additionally, the N-terminal myristoylation motif of the SlCDPK and CRK proteins was predicted by a myristoylator program in ExPASy (http://web.expasy.org/myristoylator/) (Bologna et al. 2004). Results showed that 14 SlCDPKs were predicted to bear an N-terminal myristoylation motif for membrane association (Table 1). The presence of an N-terminal myristoylation motif in SlCDPKs was subgroup-dependent. All members of subgroup II, the majority of subgroups III and one of the two subgroup IV SlCDPKs contained an N-terminal myristoylation motif whereas all but one member of subgroup I did not (Table 1). All CRKs from tomato, S. moellendorffii and P. patens were predicted to have the N-terminal myristoylation motif (Table 1), suggesting that the N-terminal myristoylation was highly conserved during the evolution of the plant CRKs.
Gene structure and chromosome location of tomato CDPK and plant CRK genes
To further understand the relationship among the members of the SlCDPK and plant CRK gene families, we examined the exon/intron gene structure of all SlCDPK genes and CRK genes from tomato, S. moellendorffii and P. patens. Results of comparison of SlCDPK genomic coding sequences showed that the intron number and phase pattern of the SlCDPK genes varied obviously in a subgroup-dependent manner (Table 1; Fig. 3). The majority (9 out of 13) of the subgroup I SlCDPK genes contained six introns with a phase pattern of 111000. The remaining four genes of this subgroup bore seven introns with an additional one in the 5′ or 3′ end. All subgroup II SlCDPK genes carried seven introns with a phase pattern of 1110020 except SlCDPK15, which had eight introns with an extra intron gain in the 5′ end of the gene. Subgroup III SlCDPK genes possessed 6–8 introns. Among them, three (SlCDPK25, SlCDPK26 and SlCDPK27) were constituted of seven introns with a phase pattern of 0111000; two (SlCDPK22 and SlCDPK23) contained six introns with a phase pattern of 111000, while the remaining one (SlCDPK24) carried eight introns with a phase pattern of 20111000. Strikingly, the subgroup IV SlCDPK genes were composed of 11 introns, which were significantly more than those found in SlCDPK genes of any other subgroups. Their phase pattern was 02201010000 (Table 1; Fig. 3). The exon/intron structure of the CRK genes from tomato, S. moellendorffii and P. patens was similar to each other. All of them carried ten introns with a phase pattern of 0220110000 except the PpCRK4 gene, which contained 11 introns with an extra phase 1 intron in the 3′ end. Thus, except for loss of one intron of phase 0 in the middle of the genes, the exon/intron structure of the SlCRK genes was highly similar to that of the subgroup IV SlCDPK genes (Table 1; Fig. 3). Collectively, these gene structural data fit the classification of the SlCDPK and SlCRK families based on the phylogenetic tree of protein sequences, and indicate the possible diversification in gene expression and functions among the different subgroups.
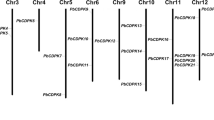
Chromosomal localization analysis showed that the SlCDPKs were distributed in all 12 chromosomes of the tomato genome. However, this distribution in each chromosome was unequal. Chromosomes 5, 7, 8, 9 and 12 each contained only a single SlCDPK gene copy, while chromosomes 1, 10 and 11 carried 5, 5 and 4 SlCDPK genes, respectively. Moreover, many SlCDPK genes were found to form clusters on their respective chromosomes (Fig. 4), suggesting repeated gene duplication events and thus the expansion of this gene family. The six SlCRKs are scattered on four chromosomes. Interestingly, SlCRK3 is located in the middle of a cluster of five SlCDPKs on chromosome 10 (Fig. 4), implying that these SlCDPKs and SlCRK3 might have similar functions.
Expression of SlCDPK and SlCRK genes was highly responsive to diverse stimuli
To gain information about potential gene function in plant disease resistance, expression patterns of a set of six SlCDPK genes (SlCDPK9/10/11/12/13/18) and two SlCRK (SlCRK4/6) genes in response to host pathogens [Sclerotinia sclerotiorum, Pseudomonas syringae pv. tomato (Pst) DC3000] and a non-host pathogen (Xanthomonas oryzae pv. oryzae, Xoo) as well as a pathogenicity factor of the pathogen S. sclerotiorum, oxalic acid (OA) in tomato were investigated. These SlCDPK genes were selected because they might be the orthologs of Arabidopsis CDPK4/5/6/11 and rice CDPK12 (Fig. 1, this study; Boudsocq and Sheen 2013), which have been reported to function in plant disease resistance (Boudsocq et al. 2010; Asano et al. 2012).
At 12 h post-inoculation (hpi) of S. sclerotiorum, expression of the majority (5 out of 6) of the SlCDPK genes was dramatically down-regulated. However, the expression of SlCDPK9 of subgroup I was slightly up-regulated (Fig. 5a). The expression pattern of the SlCDPK genes in response to OA treatment was very similar to that in response to inoculation with S. sclerotiorum, which produces OA during plant infection, except SlCDPK18, a gene of subgroup II, which was in contrast in response to S. sclerotiorum inoculation and OA treatment (Fig. 5b). Upon inoculation with the bacterial host pathogen Pst DC3000, the expression pattern of the SlCDPK genes was similar to that of inoculation with S. sclerotiorum (Fig. 5c). When inoculated with a non-host pathogen Xoo, expression of SlCDPK10, SlCDPK12 and SlCDPK13 was induced at 8 hpi, while that of SlCDPK9 was reduced significantly, which was opposite to its expression after inoculation with S. sclerotiorum. Expression of all the remaining SlCDPK genes was reduced as observed for their expression after inoculation with S. sclerotiorum (Fig. 5d). These data indicate that expression of the SlCDPK genes is diverse in a gene- and stimulus-dependent manner. However, unlike SlCDPK genes, the two SlCRK genes SlCRK4 and SlCRK6 exhibited similar response to all stimuli in this study. They were all down-regulated by all pathogen inoculations and OA treatment. This indicates that these SlCRK genes might be involved in plant resistance to a wide range of pathogens.
The expression patterns of selected SlCDPK and SlCRK genes in response to pathogen inoculation and pathogenecity factor treatment. Gene expression was analyzed at 12 h after S. sclerotiorum inoculation (a), 4 h after OA treatment (b), 4 h after Pst DC3000 infiltration (c) and 8 h after Xoo infiltration (d). The small letters indicate the significance in expression of SlCDPK and SlCRK genes under each stimulus (P ≤ 0.05, by Student’s t test)
Knock-down of a set of SlCDPK and SlCRK genes altered the resistance to S. scelrotiorum, Pst DC3000 and Xoo in tomato
To understand the function of SlCDPKs and SlCRKs in plant disease resistance, virus-induced gene silencing (VIGS) was performed for the SlCDPK10/12/13/18 and SlCRK4/6 genes. These genes were selected for VIGS analysis because they are highly responsive to various pathogen inoculations (Fig. 5). Moreover, orthologs of these SlCDPK genes are involved in plant disease resistance (Boudsocq et al. 2010; Asano et al. 2012). A vector containing a fragment of eGFP was used as the control in agro-infiltrated plants (Zhao et al. 2013). Three weeks post-agro-infiltration, the VIGS-treated (VT) tomato plants were inoculated with the host pathogens S. sclerotiorum and Pst DC3000 and the nonhost pathogen Xoo, and thereafter the resistance was evaluated.
When inoculated with S. sclerotiorum, the SlCRK6-VT plants displayed more severe disease symptom than the eGFP-control plants. The lesion diameter of these plants was 9.4 mm at 36 hpi, which was significantly larger than that of eGFP-control plants (6.6 mm) (P ≤ 0.05). However, the SlCDPK10-VT and SlCDPK18-VT plants did not show significant difference from the control plants (Fig. 6a). This result indicated that SlCRK6 plays a positive role in basal resistance to S. sclerotiorum. In case of inoculation with Pst DC3000, the SlCDPK10-VT and SlCRK6-VT plants exhibited highly and weakly stronger necrosis disease symptom, respectively, than the eGFP-control plants. Meanwhile, the bacterial number on these plants was 1.0 and 0.5 orders of magnitude, respectively, higher than the control plants. However, the SlCDPK18-VT plants did not show obvious difference from the control plants (Fig. 6b). This result showed that SlCDPK10 and SlCRK6 are positively involved in resistance to Pst DC3000. When inoculated with Xoo, the HR necrosis was significantly weaker in the Xoo-infiltrated areas of the SlCDPK18-VT plants at 9 hpi when compared with the eGFP-control plants. Coincidently, the bacterial number on these plants was 1.8 orders of magnitude higher than the control plants. However, the SlCDPK10-VT and SlCRK6-VT plants did not show obvious difference in either HR symptom or bacterial number in the Xoo-infiltrated areas in comparison with the control plants (Fig. 6c). This result demonstrated that SlCDPK18 plays a positive role in tomato nonhost resistance to Xoo. In addition, VIGS treatment of the SlCDPK12, SlCDPK13 and SlCRK4 showed no influence on tomato resistance to these three pathogens (data not shown), suggesting that these genes are not involved in these resistances or that other members of the SlCDPK and SlCRK families act redundantly.
Knock-down of a set of SlCDPK and SlCRK genes by VIGS decreased the tomato disease resistance. a The necrosis symptoms caused by S. sclerotiorum inoculation and statistical analysis of lesion diameter at 36 hpi. b The necrosis symptoms and bacterial numbers in the areas infiltrated with Pst DC3000 at 36 hpi. c The HR symptoms and bacterial numbers in the areas infiltrated with Xoo at 9 hpi. Significant differences of bacterial numbers and lesion diameter are indicated as different lowercase letters (P ≤ 0.05, by Student’s t test and DMRT)
To ensure the silencing efficiency of the SlCDPK and SlCRK genes, the expression of the target genes in the VIGS-treated and non-silenced eGFP control plants was compared. Results of RT-qPCR analysis showed that transcript of all the SlCDPK and SlCRK genes in the VIGS-treated plants accumulated to less than 30 % of that of control plants (Fig. 7), indicating that they were effectively knocked down, and the observed alteration in disease resistance is attributed to the SlCDPK and SlCRK genes.
Taken together, these results revealed that SlCDPK and SlCRK genes play roles in a wide range of resistance in tomato but their effectiveness against individual pathogen is gene-dependent. SlCDPK18 is required for nonhost resistance to Xoo, SlCDPK10 for basal resistance to Pst DC3000, while SlCRK6 for basal resistance to both S. sclerotiorum and Pst DC3000.
Discussion
The phylogeny of CDPK and CRK gene families and their function in plant disease resistance are not well understood. In the present study, this issue was addressed through different approaches. Following identification of the CDPK family in tomato and the CRK family in tomato, a primitive lycophyte (Selaginella moellendorffii) and a moss (Physcomitrella patens), we performed various bioinformatics analyses including prediction of protein domain composition and physico-chemical characteristics, dissection of gene exon/intron structure, pairwise comparison of protein sequences and construction of rooted phylogenetic tree. As a result, we clarify that the plant CRK family evolved from the CDPK family and emerged very early in land plant species before the divergence of nonvascular and vascular plant species, sharing the same ancestor gene with the subgroup IV CDPKs. Moreover, we report for the first time the role of a CRK (SlCRK6) in plant disease resistance and the function of plant CDPK genes in nonhost resistance.
Phylogenetic relationship between the CDPK and CRK families
CDPK and CRK are two types of closely related protein kinases. They differ in presence or absence of EF-hand motifs. CRKs are thought to lack canonical EF-hand motifs and thus can not bind calcium (Harmon et al. 2000). The evolution of these two protein kinase families has been studied, but their phylogenetic relationship has not yet been well understood. Phylogenetic trees for CDPK and CRK families have been constructed previously based on the alignment of kinase catalytic domains (Harmon et al. 2000; Hrabak et al. 2003). It has been suggested that the plant CDPK and CRK genes shared a single common origin (Harmon et al. 2000) and protist and plant CDPKs have a monophyletic origin (Zhang and Choi 2001). In another study, it has been proposed that CRKs have arisen relatively recently in evolution from a distinct subgroup of CDPKs and had been only identified in angiosperm until that moment (Hrabak et al. 2003). In this study, we identified the complete CDPK and CRK gene families in tomato genome and the CRK genes in genomes of a primitive lycophyte (S. moellendorffii) and a moss (P. patens) (Table 1) and their existence in algal species was searched. We found no CRK gene in genomes of algal species, but did identify two and five CRKs in S. moellendorffii and P. patens genomes, respectively (Table 1), which is different from CDPKs as they have been previously identified in all these ancient green plant species (Hamel et al. 2014). Furthermore, we constructed the rooted phylogenetic tree for CDPK family of both tomato and Arabidopsis and CRK family from six plant species including both higher and lower plant species using apicomplexan CDPKs as outgroup based on the alignment of kinase domains of protein sequences. In this tree, all CRKs clustered along with mainstream CDPK sequences and were found to be the closest relatives of subgroup IV CDPKs as they shared node with 100 % bootstrap support (Fig. 1). Since plant CRKs did not cluster with any algal CRK, it is suggestive of their early emergence from the ancestral CDPK which is common to that of group IV CDPKs. In addition, it is highly probable that CRK and group IV ancestors separated after the split of green algae and the last common ancestor of land plant lineage, which is in agreement with the absence of CRKs in green algae. However, the CRK expansion and diversification seem to be lineage independent like those of CDPKs that are estimated to have taken place after the split of land plants into vascular and non-vascular plants as recently revealed (Valmonte et al. 2014). Here, we reveal that the plant CRK family evolved from the CDPK family and emerged very early in land plant species before the divergence of nonvascular and vascular plant species, sharing the same ancestor with the subgroup IV CDPKs.
In addition to the phylogenetic tree, data on sequence similarity, prediction of biochemical characteristics and exon/intron structure of the CDPKs and CRKs also support our conclusion. Protein sequences of the CRKs are much more similar to those of the subgroup IV CDPKs than to those of any other CDPK subgroup (Fig. 2). Additionally, all identified CRKs are basic proteins, which are highly similar to the subgroup IV SlCDPKs, but distinguished from SlCDPKs of the remaining subgroups (Table 1). Moreover, all but one of the identified CRK genes possessed ten introns, which was similar to the subgroup IV CDPKs (11 introns), but distinct to CDPK genes of the other subgroups (6–8 introns) (Table 1; Fig. 3, this study; Hamel et al. 2014; Valmonte et al. 2014).
Collectively, these results clarify that the CRK lineage appeared very early from the last common ancestor and shares the immediate ancestral gene with subgroup IV CDPKs. Besides, it is discernible that the degeneration of EF-hand motifs of CRKs involved events such as mutational substitution of residues required for Ca2+ binding that occurred during the emergence of this family from CDPK gene of the last common ancestor of all land plant species.
Functions of SlCDPKs and SlCRKs in plant disease resistance
There has been increasing evidences supporting the involvement of CDPKs in plant disease resistance (Boudsocq and Sheen 2013). Nevertheless, the role of CDPKs in plant disease resistance has been studied only in the limited phytopathosystems. Additionally, whether CRKs play a role in plant disease resistance remains unknown. To gain more information about potential gene function in plant disease resistance, we checked expression patterns of a set of SlCDPK and SlCRK genes and performed their VIGS functional analyses in three pathosystems. These include tomato-Sclerotinia sclerotiorum, tomato-Pseudomonas syringae pv. tomato (Pst) DC3000 and tomato-Xanthomonas oryzae pv. oryzae (Xoo), representing three different types of resistance: host basal resistance to necrotrophic fungal pathogen, host basal resistance to biotrophic bacterial pathogen and nonhost resistance to bacterial pathogen, respectively. Expression data demonstrated that different SlCDPK genes display diverse expression in response to the same pathogen, and the same SlCDPK gene exhibits various expression patterns in response to different pathogens such as host and nonhost pathogens (Fig. 5), indicating that expression of the SlCDPK genes is diverse in a gene- and stimulus-dependent manner. This is similar to what have been reported in other systems (Valmonte et al. 2014). Furthermore, our VIGS functional analyses revealed that different SlCDPK and SlCRK genes are involved in different resistance to various pathogens. For example, SlCDPK18 is required for tomato nonhost resistance to the rice pathogen Xoo, SlCDPK10 is involved in tomato basal resistance to Pst DC3000, while SlCRK6 affects basal resistance to both S. sclerotiorum and Pst DC3000 (Fig. 6). According to the phylogenetic tree, SlCDPK18 is the ortholog of AtCDPK29 and OsCDPK12. These genes have identical intron phase pattern of 1110020 (Figs. 1, 3 of this study; Fig. S13 of Valmonte et al. 2014). OsCDPK12 was found to negatively regulate the blast resistance in rice (Asano et al. 2012). Similarly, SlCDPK10 is phylogenetically very close to AtCDPK4/11 (Fig. 1) which were found to play a positive role in Arabidopsis resistance to Pst DC3000 (Boudsocq et al. 2010). These observations indicate that the function in disease resistance is conserved in othologs of SlCDPK10/AtCDPK4/11 and SlCDPK18/AtCDPK29/OsCDPK12 in various plant species. In addition, to our knowledge, this is the first report on the role of CRK in plant disease resistance, and SlCRK6 is the first plant CRK gene that is proved to function in disease resistance. This is also the first report on function of tomato CDPK genes in disease resistance and is the first finding that plant CDPK genes are involved in nonhost resistance. Our results extended the spectrum of resistance and pathogens that are regulated by the plant CDPK genes, and revealed that plant CDPK genes play roles in a wide range of resistance with effectiveness against individual pathogen being CDPK gene dependent.
The functional mechanism of these SlCDPKs to regulate plant resistance remains unclear. Some CDPKs target NADPH oxidase to regulate reactive oxygen species (ROS) production. For example, the orthologs of SlCDPK10, AtCDPK4/11 were found to play a positive role in Arabidopsis resistance to Pst DC3000 through promoting ROS production, potentially by directly phosphorylating NADPH oxidase RBOHB (Boudsocq et al. 2010). On the contrary, the ortholog of SlCDPK18, OsCDPK12, negatively modulates blast resistance through reducing ROS accumulation (Asano et al. 2012). We wondered whether SlCDPK10 and SlCDPK18 function similarly. However, DAB staining analysis shows that the ROS accumulation level of the SlCDPK-knock-down plants both before and after pathogen inoculation does not alter significantly when compared with non-silenced control plants (data not shown). This result suggests that SlCDPK10 and SlCDPK18 might be not involved in ROS production. Alternatively, their function is likely overlapped by other functionally redundant SlCDPK gene(s) such as the phylogenetically closest paralog SlCDPK11 (Fig. 1).
In addition to alter ROS accumulation, CDPKs may target MAPKs (Xie et al. 2014), BIK1 (Monaghan et al. 2014), WRKY transcription factors (Gao et al. 2013), and/or affect defense hormones (Coca and San Segundo 2010) locally or systemically (Romeis and Herde 2014) in response to pathogen infection. Whether SlCDPK10 and SlCDPK18 target similar substrates awaits further analyses. Unlike CDPKs, no target of CRKs has yet been identified under pathogenic conditions. Therefore, identification of the targets of SlCRK6 will provide new insights into the molecular mechanism of SlCRK6 to regulate plant resistance. Additionally, whether the function of SlCRK6 depends on Ca2+ is worth clarifying, considering that this protein only carries degenerated EF-hand motifs (Fig. 2).
References
Asano T, Tanaka N, Yang G, Hayashi N, Komatsu S (2005) Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol 46:356–366
Asano T, Hayashi N, Kobayashi M, Aoki N, Miyao A, Mitsuhara I, Ichikawa H, Komatsu S, Hirochika H, Kikuchi S (2012) A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J 69:26–36
Bologna G, Yvon C, Duvaud S, Veuthey AL (2004) N-terminal myristoylation predictions by ensembles of neural networks. Proteomics 4:1626–1632
Boudsocq M, Sheen J (2013) CDPKs in immune and stress signaling. Trends Plant Sci 18:30–40
Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J (2010) Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464:418–422
Cai X, Wang C, Xu Y, Xu Q, Zheng Z, Zhou X (2007) Efficient gene silencing induction in tomato by a viral satellite DNA vector. Virus Res 125:169–175
Campo S, Baldrich P, Messeguer J, Lalanne E, Coca M, San Segundo B (2014) Overexpression of a Calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol 165:688–704
Cheng SH, Willmann MR, Chen HC, Sheen J (2002) Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol 129:469–485
Cheng WS, Xu QF, Li F, Xu YP, Cai XZ (2012) Establishment of a suitable control vector for Tobacco rattle virus-induced gene silencing analysis in Nicotiana benthamiana. J Zhejiang Univ (Agric Life Sci) 38:10–20
Coca M, San Segundo B (2010) AtCPK1 calcium-dependent protein kinase mediates pathogen resistance in Arabidopsis. Plant J 63:526–540
Confalonieri S, Di Fiore PP (2002) The Eps15 homology (EH) domain. FEBS Lett 513:24–29
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Gao X, Chen X, Lin W, Chen S, Lu D, Niu Y, Li L, Cheng C, McCormack M, Sheen J (2013) Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog 9:e1003127
Guo A, Zhu Q, Chen X, Luo J (2007) GSDS: a gene structure display server. Yi chuan 29:1023–1026
Hamel LP, Sheen J, Séguin A (2014) Ancient signals: comparative genomics of green plant CDPKs. Trends Plant Sci 19:79–89
Harmon AC, Gribskov M, Harper JF (2000) CDPKs–a kinase for every Ca2+ signal? Trends Plant Sci 5:154–159
Harper JF, Harmon A (2005) Plants, symbiosis and parasites: a calcium signalling connection. Nat Rev Mol Cell Biol 6:555–566
Harper JF, Breton G, Harmon A (2004) Decoding Ca2+ signals through plant protein kinases. Annu Rev Plant Biol 55:263–288
Hrabak EM, Chan CWM, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, Thomas M, Walker-Simmons K, Zhu JK, Harmon AC (2003) The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132:666–680
Huttner IG, Strahl T, Osawa M, King DS, Ames JB, Thorner J (2003) Molecular interactions of yeast frequenin (Frq1) with the phosphatidylinositol 4-kinase isoform, Pik1. J Biol Chem 278:4862–4874
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282
Kamiyoshihara Y, Iwata M, Fukaya T, Tatsuki M, Mori H (2010) Turnover of LeACS2, a wound-inducible 1-aminocyclopropane-1-carboxylic acid synthase in tomato, is regulated by phosphorylation/dephosphorylation. Plant J 64:140–150
Kim H-J, Chen C, Kabbage M, Dickman MB (2011) Identification and characterization of Sclerotinia sclerotiorum NADPH oxidases. Appl Environ Microbiol 77(21):7721–7729
Kobayashi M, Ohura I, Kawakita K, Yokota N, Fujiwara M, Shimamoto K, Doke N, Yoshioka H (2007) Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19:1065–1080
Kudla J, Batistic O, Hashimoto K (2010) Calcium signals: the lead currency of plant information processing. Plant Cell 22:541–563
Leclercq J, Ranty B, Sanchez-Ballesta MT, Li ZG, Jones B, Jauneau A, Pech JC, Latche A, Ranjeva R, Bouzayen M (2005) Molecular and biochemical characterization of LeCRK1, a ripening-associated tomato CDPK-related kinase. J Exp Bot 56:25–35
Li RJ, Hua W, Lu YT (2006) Arabidopsis cytosolic glutamine synthetase AtGLN1;1 is a potential substrate of AtCRK3 involved in leaf senescence. Biochem Biophys Res Commun 342:119–126
Li AL, Zhu YF, Tan XM, Wang X, Wei B, Guo HZ, Zhang ZL, Chen XB, Zhao GY, Kong XY (2008) Evolutionary and functional study of the CDPK gene family in wheat (Triticum aestivum L.). Plant Mol Biol 66:429–443
Liu HT, Gao F, Li GL, Han JL, Liu DL, Sun DY, Zhou RG (2008) The calmodulin-binding protein kinase 3 is part of heat-shock signal transduction in Arabidopsis thaliana. Plant J 55:760–773
Liu W, Li W, He Q, Daud MK, Chen J, Zhu S (2014) Genome-wide survey and expression analysis of calcium-dependent protein kinase in Gossypium raimondii. PLoS One 9:e98189
Ludwig AA, Romeis T, Jones JD (2004) CDPK-mediated signalling pathways: specificity and cross-talk. J Exp Bot 55:181–188
Ludwig AA, Saitoh H, Felix G, Freymark G, Miersch O, Wasternack C, Boller T, Jones JD, Romeis T (2005) Ethylene-mediated cross-talk between calcium-dependent protein kinase and MAPK signaling controls stress responses in plants. Proc Natl Acad Sci USA 102:10736–10741
Monaghan J, Matschi S, Shorinola O, Rovenich H, Matei A, Segonzac C, Malinovsky FG, Rathjen JP, MacLean D, Romeis T (2014) The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 16:605–615
Ray S, Agarwal P, Arora R, Kapoor S, Tyagi AK (2007) Expression analysis of calcium-dependent protein kinase gene family during reproductive development and abiotic stress conditions in rice (Oryza sativa L. ssp. indica). Mol Genet Genomics 278:493–505
Reddy ASN, Ali GS, Celesnik H, Day IS (2011) Coping with stresses: roles of calcium-and calcium/calmodulin-regulated gene expression. Plant Cell 23:2010–2032
Rigo G, Ayaydin F, Tietz O, Zsigmond L, Kovacs H, Pay A, Salchert K, Darula Z, Medzihradszky KF, Szabados L, Palme K, Koncz C, Cseplo A (2013) Inactivation of plasma membrane-localized CDPK-RELATED KINASE5 decelerates PIN2 exocytosis and root gravitropic response in Arabidopsis. Plant Cell 25:1592–1608
Romeis T, Herde M (2014) From local to global: CDPKs in systemic defense signaling upon microbial and herbivore attack. Curr Opin Plant Biol 20:1–10
Romeis T, Ludwig AA, Martin R, Jones JD (2001) Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J 20:5556–5567
Saand MA, Xu YP, Li W, Wang JP, Cai XZ (2015) Cyclic nucleotide gated channel gene family in tomato:genome-wide identification and functional analyses in disease resistance. Front Plant Sci 6:303
Sanders D, Pelloux J, Brownlee C, Harper JF (2002) Calcium at the crossroads of signaling. Plant Cell 14(suppl.):S401–S417
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tao XC, Lu YT (2013) Loss of AtCRK1 gene function in Arabidopsis thaliana decreases tolerance to salt. J Plant Biol 56:306–314
Valmonte GR, Arthur K, Higgins CM, MacDiarmid RM (2014) Calcium-dependent protein kinases in plants: evolution, expression and function. Plant Cell Physiol 55:551–569
Wang Y, Liang SP, Xie QG, Lu YT (2004) Characterization of a calmodulin-regulated Ca2+-dependent-protein-kinase-related protein kinase, AtCRK1, from Arabidopsis. Biochem J 383:73–81
Wang C, Cai X, Wang X, Zheng Z (2006) Optimisation of tobacco rattle virus-induced gene silencing in Arabidopsis. Funct Plant Biol 33:347–355
Xie K, Chen J, Wang Q, Yang Y (2014) Direct phosphorylation and activation of a mitogen-activated protein kinase by a calcium-dependent protein kinase in rice. Plant Cell 26:3077–3089
Zhang XS, Choi JH (2001) Molecular evolution of calmodulin-like domain protein kinases (CDPKs) in plants and protists. J Mol Evol 53:214–224
Zhang H, Liu WZ, Zhang Y, Deng M, Niu F, Yang B, Wang X, Wang B, Liang W, Deyholos MK (2014) Identification, expression and interaction analyses of calcium-dependent protein kinase (CPK) genes in canola (Brassica napus L.). BMC Genom 15:211
Zhao Y, Liu W, Xu YP, Cao JY, Braam J, Cai XZ (2013) Genome-wide identification and functional analyses of calmodulin genes in Solanaceous species. BMC Plant Biol 13:70
Zhou L, Lan W, Jiang Y, Fang W, Luan S (2013) A calcium-dependent protein kinase interacts with and activates a calcium channel to regulate pollen tube growth. Mol Plant 7:369–376
Zhu SY, Yu XC, Wang XJ, Zhao R, Li Y, Fan RC, Shang Y, Du SY, Wang XF, Wu FQ (2007) Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19:3019–3036
Zuo R, Hu R, Chai G, Xu M, Qi G, Kong Y, Zhou G (2013) Genome-wide identification, classification, and expression analysis of CDPK and its closely related gene families in poplar (Populus trichocarpa). Mol Biol Rep 40:2645–2662
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was funded by the Genetically Modified Organisms Breeding Major Projects (no. 2014ZX0800905B), the Special Fund for Agro-scientific Research in the Public Interest (no. 201103016), the Program for Changjiang Scholars and Innovative Research Team in University (no. IRT0943) and the SRFDP (no. 20110101110092).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by C. Gebhardt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, JP., Xu, YP., Munyampundu, JP. et al. Calcium-dependent protein kinase (CDPK) and CDPK-related kinase (CRK) gene families in tomato: genome-wide identification and functional analyses in disease resistance. Mol Genet Genomics 291, 661–676 (2016). https://doi.org/10.1007/s00438-015-1137-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-015-1137-0