Abstract
Previous studies have indicated that hormonal control is essential for plant root growth. The root of the carrot is an edible vegetable with a high nutritional value. However, molecular mechanisms underlying hormone-mediated root growth of carrot have not been illustrated. Therefore, the present study collected carrot root samples from four developmental stages, and performed transcriptome sequencing to understand the molecular functions of plant hormones in carrot root growth. A total of 160,227 transcripts were generated from our transcriptome, which were assembled into 32,716 unigenes with an average length of 1,453 bp. A total of 4,818 unigenes were found to be differentially expressed between the four developmental stages. In total, 87 hormone-related differentially expressed genes were identified, and the roles of the hormones are extensively discussed. Our results suggest that plant hormones may regulate carrot root growth in a phase-dependent manner, and these findings will provide valuable resources for future research on carrot root development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The carrot (Daucus carota L.), a plant of the Apiaceae family, is one of the most economically important vegetables worldwide known for its edible and nutritious root (Luby et al. 2014). The carrot root undergoes a notable size change during its growth process, but only limited information regarding carrot root development is available. In higher plants, the root is an essential organ that absorbs nutrients and water for plant growth (Petricka et al. 2012). It is also involved in plant anchorage, hormone synthesis, and storage (Schiefelbein and Benfey 1991). However, root development has not been as extensively investigated as that of other plant organs, although the root exhibits several unique characteristics.
Transcriptome sequencing has been a useful method of identifying novel transcripts and splice isoforms as well as performing expression analysis. Indeed, numerous sequences and plant molecular information have been obtained using transcriptome sequencing (Klaus et al. 2000; Lu et al. 2010; Torti et al. 2012; Duarte et al. 2014; Li et al. 2014), but such studies have not been conducted on carrot development.
Previous work has suggested that hormones are the major intrinsic regulators of plant growth (Durbak et al. 2012). Auxin derived from both shoots and roots contributes to normal root development (Reed et al. 1998; Saini et al. 2013), and several lines of evidence indicate that auxin promotes proliferation, elongation, and vascular tissue formation in the roots (Aloni 2013; Takatsuka and Umeda 2014). As previously reported, auxin transport toward the root tip seems to contribute largely to taproot elongation (Blilou et al. 2005). However, excess auxin results in the severe inhibition of root growth (López-Bucio et al. 2005). Cytokinins are critical regulators of root vascular development (Yokoyama et al. 2007), but may also act as growth inhibitors by affecting the root apical meristem (Werner et al. 2003; Ren et al. 2009). Gibberellins appear to control root growth through promoting cell elongation (Inada and Shimmen 2000; Shani et al. 2013). Recent studies report that gibberellins also regulate cell production to control root growth (Ubeda-Tomás et al. 2009). Ethylene and abscisic acid are believed to have similar effects on root growth, with low levels stimulating root growth and higher levels inhibiting it (Joshi-Saha et al. 2011; Luo et al. 2014). Furthermore, ethylene-induced inhibition of root growth appears to require the involvement of abscisic acid (Ghassemian et al. 2000; Luo et al. 2014). The newly identified hormones, brassinosteroids and jasmonates also play important roles in root growth (Müssig et al. 2003; Hacham et al. 2011).
As shown above, these hormones have overlapping functions and often interact with each other to form a complex regulatory network for root growth (Jung and McCouch 2013). Using advanced molecular genetic analysis, the crosstalk and cooperation between their biosynthesis and signaling pathways have been extensively observed, thus substantially improving our understanding of hormone-mediated root growth (Mouchel et al. 2006; Zhou et al. 2011). Plant hormones are clearly essential for root growth and development (Jung and McCouch 2013; Liu et al. 2014), and such regulation may be achieved in a developmental stage-dependent manner (Sharp et al. 2004; Joshi-Saha et al. 2011).
The present study investigated the growth characteristics and anatomical structure of the carrot root during root enlargement. Transcriptome analysis of the carrot root was also conducted, and genes involved in hormone biosynthesis and signaling pathways were extensively investigated to help elucidate the hormonal control of root growth based on digital gene expression.
Materials and methods
Plant material and tissue preparation
‘Kurodagosun’ carrot seeds were sown at Nanjing Agricultural University (32°02′N, 118°50′E) in December 2013. The plants were cultivated in a container with a mixture of vermiculite and organic soil. The artificial weather was maintained at 25 °C for 14 h in the daytime with 320 μmol m−2s−1 light intensity and at 18 °C for 10 h during the night. Plants were divided into four stages of 22 (Stage 1), 40 (Stage 2), 56 (Stage 3), and 95 days (Stage 4) after the seeds were planted. The roots from these four stages were harvested and stored at −80 °C until analysis.
Anatomical structure analysis
To investigate the changes in anatomical structure of the carrot root, fresh root samples were cut into small pieces and stored in phosphate buffer (pH 7.2) with 2.5 % glutaraldehyde. The slices were dehydrated with ethanol and treated with epoxy propane the soaked and embedded with Spurr resin (Spurr 1969). Using a Leica ultramicrotome (Germany), we cut the samples into thin sections (~1 μm) and stained them with methyl violet for 3 min. The slices were observed and then photographed using a Leica DMLS microscope (Germany).
RNA extraction and mRNA-seq library construction
Total RNA from roots of all four stages was extracted using an RNAprep pure plant kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. An Illumina HiSeq™ library was constructed according to the manufacturer’s instructions. Briefly, mRNA was purified by attaching beads containing oligo-dT to the poly-A. mRNAs were then cut into short fragments using a chemical reagent and applying a high temperature. Subsequently, these short fragments were used as templates to synthesize first-strand cDNA. Buffer, dNTPs, RNase H, and DNA polymerase I were used to synthesize second-strand cDNA from first-strand cDNA. The overhangs from fragmentation were converted into blunt ends using an end-repair mix (Illumina). Indexing adapters were ligated to the ends of double stranded cDNA when single A nucleotides were added to the 3′ ends of blunt fragments to prepare these adapters for hybridization in a flow cell. The short fragments with adapters ligated on both ends were enriched by PCR amplification and establishment of a cDNA library. After checking and quantifying, we mixed the multiplexed DNA libraries with normalized 10 nM concentration in equal volumes. The library was then sequenced using an Illumina HiSeq™ 2000 platform.
Data filtering and de novo assembly
Raw reads from four samples were collected to remove unsuitable reads and to perform subsequent de novo assembly. Data filtering was performed according to standard procedures. In brief, the adapter sequences and reads with a low quality score or a final length <50 were removed. High quality reads were assembled into contigs, transcripts, and unigenes using Trinity (http://trinityrnaseq.sourceforge.net/), non-redundant (NR) protein database (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and NCBI Blast software (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Unigenes were submitted to BLASTX search to identify homology against the NR database with a cut-off E value of 1e−5, and top-hit species were indicated.
Gene annotation and analysis
We used the evolutionary genealogy of genes: nonsupervised orthologous groups (eggNOG), a database of orthologous groups of genes, to identify and annotate unigene distribution. Gene ontology (GO) was also introduced to analyze the complicated biological behaviors of these unigenes during root enlargement. A Venn plot was constructed in which numbers in a circle denoted phase-specific genes, while numbers in two or more intersecting circles indicated phase overlapping genes.
Identification of differentially expressed genes (DEGs)
Reads per kilobase per million mapped reads (RPKM) were used to quantify gene expression. RPKM values were normalized, and genes with expression levels exhibiting more than twofold changes between two phases were clustered into DEGs. GO enrichment was adopted to further investigate the DEGs (significant difference at the 0.05 and 0.01 level). Relative RPKM was log2 (RPKM + 1) transformed to perform cluster analysis, and a heat map was then generated from log2 ‘relative RPKM’ using average linkage analysis.
Results
Plant growth analysis
We harvested carrots at all four stages of growth: 22 days old (Stage 1), 40 days old (Stage 2), 56 days old (Stage 3), and 95 days old (Stage 4) (Fig. 1). In Stage 1, the root was white, but had become orange in color and increased in size by Stage 2. Fresh root and shoot weights increased over the different stages (Fig. 2), while the middle diameter of the root gradually increased during root development.
Anatomical observation of carrot roots
Leica DMLS microscopy revealed the internal structure of the roots at different developmental stages. In Stage 1, the protoxylem and the primary phloem were relatively thin (Fig. 3a), but the number and size of cells in these parts had increased by Stage 2, resulting in thickening of the carrot roots. This thickening could also be attributed almost exclusively to the constant division of the vascular cambium (VC), located between the xylem and the phloem (Fig. 3b). The VC differentiated inwards to form the secondary xylem, and outwards to form the secondary phloem (SP). After this time point, expansion of the outer vascular parenchyma cells was visible (Fig. 3b, c, f), and starch granules (SGs) were also seen during the later stages of growth (Fig. 3b–d, f, g).
Anatomical structure of carrot roots from a Stage 1, b Stage 2, (c–e) Stage 3, and (f–h) Stage 4. c and f show the outer vascular parenchyma cells close to the epidermis; d and g represent the inner vascular parenchyma cells; e and h represent the central part of carrot roots. BC border cells, Ep epidermis, PC parenchymal cell, Ph phellogen, PP primary phloem, Px protoxylem, SG starch granule, SP secondary phloem, VC vascular cambium, Ve vessels. Scale bars in a and b, 100 μm; scale bars in c–h, 50 μm
Assembly, functional annotation, and classification
A total of 22,940,679 clean reads were obtained for Stage 1, 48,885,725 for Stage 2, 36,758,033 for Stage 3, and 35,189,581 for Stage 4. The Trinity software generated 160,227 transcripts by de novo assembly, and each transcript was aligned with the NR database (Fig. 4). Based on the results of the top-hits from BLASTX, we assembled the transcripts into 32,716 unigenes with an average length of 1,453 bp and a 41.03 % GC content. Among these unigenes, 13,832 (42.28 %) were in the size range 200–1,000 bp, and only 419 (1.28 %) were >2 kb (Fig. 5).
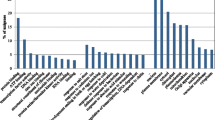
All unigenes were predicted and classified on the basis of eggNOG database, which is a database that can explore the ancestry of a protein. A total of 26,188 unigenes were found to be similar with eggNOG, and were then clustered into 26 categories (Fig. 6). The largest category was “function unknown”, followed by “general function prediction only” (4,564, 17.43 %), and “signal transduction mechanisms” (2,329, 8.89 %).
Function classification of unigenes in evolutionary genealogy of genes: non-supervised orthologous groups (eggNOG). A total of 26, 188 unigenes showed significant similarity to sequences in eggNOG database and were classified into 26 groups. Letters on x-axis indicates groups (shown on right of histogram), and y-axis represents unigene number
Based on GO annotations, the unigenes were classified into different functional categories (Fig. 7). “Molecular process” and “cellular process” were the most abundant GO slims within the biological process category, while “cell” and “binding” were the most highly represented groups within the cellular component category and molecular function category, respectively. Genes involved in other important biological processes such as stress response, anatomical structure, and growth were also identified.
Figure 8 shows the Venn diagrams presenting the commonly or uniquely expressed genes detected at each developmental stage. A total of 26,730 genes were expressed in all four stages, with approximately 237, 311, 439, and 62 genes exclusively expressed in Stages 1, 2, 3, and 4, respectively.
Analysis of differentially expressed genes
Compared with Stage 1, there were 408 up-regulated and 462 down-regulated genes in Stage 2. For each comparison, the number of down-regulated genes exceeded that of up-regulated genes (Fig. 9). To gain more insights into the functions of differentially expressed genes (DEGs), we searched for markedly enriched GO terms compared with the reference gene background (Fig. 10). According to molecular function, the DEGs mapped to “binding” were enriched during the later stages of development (Stages 3 and 4). According to cellular component, the GO term “extracellular region” was enriched in all developmental stages. Considering biological processes, the DEGs that mapped to “sequence-specific DNA binding transcription factor activity” and “response to stress” were enriched in all stages. The DEGs in later developmental stages (Stages 2, 3, and 4) constituted a high proportion of “metabolic process” genes, suggesting that a substantial number of this type of gene may be involved in metabolic processes.
A total of 4,818 DEGs were identified in root enlargement, which were then clustered using hierarchical cluster analysis (Fig. 11). DEGs in Stage 1 were located close to those of Stage 2, while a similar pattern was observed between DEGs of Stages 3 and 4.
Hormonal control of carrot root growth
Figure 12 shows the number of DEGs involved in plant hormone metabolism and signaling during carrot root development. A total of 87 DEGs were hormone-related, involving auxin, cytokinin, abscisic acid, gibberellins, ethylene, brassinosteroids, and jasmonic acid pathways (Table 1).
In the auxin metabolism and signaling pathway, 10 of 15 DEGs showed down-regulated trends, whereas only one was up-regulated. Similar results were observed in the cytokinin, gibberellin, jasmonic acid, and brassinosteroid pathways. In the abscisic acid biosynthesis pathway, where some unigenes also regulate the accumulation of lycopene, carotenes, and lutein, six of 11 DEGs showed up-regulated trends, while five of nine DEGs in the abscisic acid signaling pathway were down-regulated during root growth. In the ethylene pathway, ACS1 and ACS3 encoding 1-aminocyclopropane-1-carboxylic acid (ACC) synthase showed a similar pattern, indicating redundant roles for ACS1 and ACS3. However, two unigenes, ACO1 and ACO5, encoding ACC oxidase showed opposite trends (Fig. 12).
Discussion
Molecular mechanisms play vital roles in plant growth and development (Wu and Cheng 2014), and it is important to identify the genes responsible for controlling plant development to fully comprehend these mechanisms. To date, studies based on increasingly available sequence data have detected numerous genes and expression profiles that occur during the development of plant tissues such as flowers, leaves, and fruits (Ando et al. 2012; Sweetman et al. 2012; Singh et al. 2013; Lou et al. 2014). The root provides the plant growth with water, nutrients, and anchorage (Rich and Watt 2013), and an understanding of how root growth is controlled by hormones is essential for crop production, especially for fleshy root plants such as the carrot (Herder et al. 2010; Remans et al. 2012). However, the dynamics of the genes involved in carrot root development remain unclear.
‘Kurodagosun’ is a carrot variety widely cultivated for its high and stable production, which is also used as a model for genetic and breeding research (Huang et al. 2014; Xu et al. 2014a, b). Its root undergoes a significant size change during plant growth, providing useful material for our present work. As previously described, root growth is a consequence of an increase in both the number of cells in the root and their size (Perilli et al. 2012). Our results show that carrot root enlargement can be attributed to the differentiation of VC, a cylindrical meristematic tissue that gives rise to the secondary xylem toward the inner part, and to SP toward the outer part. We also observed SGs deposition in the carrot root; its distribution is similar to that of sugar and carotene, which are located in the cortex (Vilaine et al. 2013).
To gain a better understanding of root growth and development, carrot roots from four stages were analyzed by high-throughput Illumina sequencing. A total of 32,716 unigenes were assembled, of which 26,730 were co-expressed during all four developmental stages, indicating that they are essential for root development. The genes that were expressed in one to three developmental stages may instead correlate with phase-specific processes. In total, 4,818 DEGs were identified in four developmental stages, which can provide novel genetic material for further studies aimed to measure and characterize root development.
Plant hormones are intrinsic regulators of plant growth in response to environmental cues (Malamy 2005), and have been shown to be necessary for carrot root growth and development (Michalczuk et al. 1992; Mitsuhashi et al. 2003; Kulka 2008). We identified 87 DEGs involved in hormone biosynthesis, deactivation, and signaling pathways, indicating that plant hormones regulate carrot root growth in a stage-dependent manner (Michalczuk et al. 1992). Most DEGs showed down-regulated trends, leading us to hypothesize that hormones play vital roles in root formation and initiation (Aloni et al. 2006). Alternatively, they may work actively in metabolism regulation and pigmentation (Woeste et al. 1999; Titapiwatanakun and Murphy 2009), which is supported by the observed roles of hormones in sugar and anthocyanin metabolism (Shan et al. 2009; LeClere et al. 2010). On the other hand, jasmonic acid and cytokinin inhibit root growth (Creelman 1998; Kuderová et al. 2008), which may explain the observed down-regulated trends of the unigenes associated with these two hormones. Furthermore, the similar expression profiles in different hormones reveal that extensive hormonal crosstalk occurs during the regulation of root development (Nemhauser et al. 2006).
In conclusion, root development appears to be a complex process involving several changes. Our current work has provided a novel view of carrot root development, indicating that 4,818 DEGs are involved in root enlargement, and that 87 unigenes are hormone-related. As a result, plant hormones may regulate root growth in a developmental stage-dependent manner. These transcriptome sequencing data will increase the genetic resources available for scientists working on root enlargement.
References
Aloni R (2013) Role of hormones in controlling vascular differentiation and the mechanism of lateral root initiation. Planta 238:819–830
Aloni R, Aloni E, Langhans M, Ullrich CI (2006) Role of cytokinin and auxin in shaping root architecture: regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann Bot 97:883–893
Ando K, Carr K, Grumet R (2012) Transcriptome analyses of early cucumber fruit growth identifies distinct gene modules associated with phases of development. BMC Genomics 13:518
Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433:39–44
Creelman RA (1998) Jasmonate perception: characterization of COI1 mutants provides the first clues. Trends Plant Sci 3:367–368
Duarte J, Rivière N, Baranger A, Aubert G, Burstin J, Cornet L, Lavaud C, Lejeune-Hénaut I, Martinant JP, Pichon JP, Pilet-Nayel ML, Boutet G (2014) Transcriptome sequencing for high throughput SNP development and genetic mapping in Pea. BMC Genomics 15:126
Durbak A, Yao H, McSteen P (2012) Hormone signaling in plant development. Curr Opin Plant Biol 15:92–96
Ghassemian M, Nambara E, Cutler S, Kawaide H, Kamiya Y, McCourt P (2000) Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12:1117–1126
Hacham Y, Holland N, Butterfield C, Ubeda-Tomas S, Bennett MJ, Chory J, Savaldi-Goldstein S (2011) Brassinosteroid perception in the epidermis controls root meristem size. Development 138:839–848
Herder GD, Van Isterdael G, Beeckman T, De Smet I (2010) The roots of a new green revolution. Trends Plant Sci 15:600–607
Huang Y, Li MY, Wang F, Xu ZS, Huang W, Wang GL, Ma J, Xiong AS (2014) Heat shock factors in carrot: genome-wide identification, classification, and expression profiles response to abiotic stress. Mol Biol Rep. doi:10.1007/s11033-11014-13826-x
Inada S, Shimmen T (2000) Regulation of elongation growth by gibberellin in root segments of Lemna minor. Plant Cell Physiol 41:932–939
Joshi-Saha A, Valon C, Leung J (2011) Abscisic acid signal off the STARTing block. Mol Plant 4:562–580
Jung JKH, McCouch S (2013) Getting to the roots of it: genetic and hormonal control of root architecture. Front Plant Sci 4:186
Klaus M, Aaron L, Thomas E, Allen M, Jürg S, Kay AL, Jeffery LD, Robert AD (2000) The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet 26:403–410
Kuderová A, Urbánková I, Válková M, Malbeck J, Brzobohatý B, Némethová D, Hejátko J (2008) Effects of conditional IPT-dependent cytokinin overproduction on root architecture of Arabidopsis seedlings. Plant Cell Physiol 49:570–582
Kulka RG (2008) Hormonal control of root development on epiphyllous plantlets of Bryophyllum (Kalanchoë) marnierianum: role of auxin and ethylene. J Exp Bot 59:2361–2370
LeClere S, Schmelz EA, Chourey PS (2010) Sugar levels regulate tryptophan-dependent auxin biosynthesis in developing maize kernels. Plant Physiol 153:306–318
Li MY, Wang F, Jiang Q, Ma J, Xiong AS (2014) Identification of SSRs and differentially expressed genes in two cultivars of celery (Apium graveolens L.) by deep transcriptome sequencing. Hort Res 1:10
Liu J, Rowe J, Lindsey K (2014) Hormonal crosstalk for root development: a combined experimental and modeling perspective. Front Plant Sci 5:116
López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Pérez-Torres A, Rampey RA, Bartel B, Herrera-Estrella L (2005) An auxin transport independent pathway is involved in phosphate stress-induced root architectural alterations in Arabidopsis. Identification of BIG as a mediator of auxin in pericycle cell activation. Plant Physiol 137:681–691
Lou Q, Liu Y, Qi Y, Jiao S, Tian F, Jiang L, Wang Y (2014) Transcriptome sequencing and metabolite analysis reveals the role of delphinidin metabolism in flower colour in grape hyacinth. J Exp Bot 65:3157–3164
Lu T, Lu G, Fan D, Zhu C, Li W, Zhao Q, Feng Q, Zhao Y, Guo Y, Li W, Huang X, Han B (2010) Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Res 20:1238–1249
Luby CH, Maeda HA, Goldman IL (2014) Genetic and phenological variation of tocochromanol (vitamin E) content in wild (Daucus carota L. var. carota) and domesticated carrot (D. carota L. var. sativa). Hort Res 1:15
Luo X, Chen Z, Gao J, Gong Z (2014) Abscisic acid inhibits root growth in Arabidopsis through ethylene biosynthesis. Plant J 79:44–55
Malamy JE (2005) Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ 28:67–77
Michalczuk L, Cooke TJ, Cohen JD (1992) Auxin levels at different stages of carrot somatic embryogenesis. Phytochemistry 31:1097–1103
Mitsuhashi W, Toyomasu T, Masui H, Katho T, Nakaminami K, Kashiwagi Y, Akutsu M, Kenmoku H, Sassa T, Yamaguchi S, Kamiya Y, Kamada H (2003) Gibberellin is essentially required for carrot (Daucus carota L.) somatic embryogenesis: dynamic regulation of gibberellin 3-oxidase gene expressions. Biosci Biotechnol Biochem 67:2438–2447
Mouchel CF, Osmont KS, Hardtke CS (2006) BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature 443:458–461
Müssig C, Shin G-H, Altmann T (2003) Brassinosteroids promote root growth in Arabidopsis. Plant Physiol 133:1261–1271
Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126:467–475
Perilli S, Di Mambro R, Sabatini S (2012) Growth and development of the root apical meristem. Curr Opin Plant Biol 15:17–23
Petricka JJ, Winter CM, Benfey PN (2012) Control of Arabidopsis root development. Annu Rev Plant Biol 63:563–590
Reed RC, Brady SR, Muday GK (1998) Inhibition of auxin movement from the shoot into the root Inhibits lateral root development in Arabidopsis. Plant Physiol 118:1369–1378
Remans T, Thijs S, Truyens S, Weyens N, Schellingen K, Keunen E, Gielen H, Cuypers A, Vangronsveld J (2012) Understanding the development of roots exposed to contaminants and the potential of plant-associated bacteria for optimization of growth. Ann Bot 110:239–252
Ren B, Liang Y, Deng Y, Chen Q, Zhang J, Yang X, Zuo J (2009) Genome-wide comparative analysis of type-A Arabidopsis response regulator genes by overexpression studies reveals their diverse roles and regulatory mechanisms in cytokinin signaling. Cell Res 19:1178–1190
Rich SM, Watt M (2013) Soil conditions and cereal root system architecture: review and considerations for linking Darwin and Weaver. J Exp Bot 64:1193–1208
Saini S, Sharma I, Kaur N, Pati P (2013) Auxin: a master regulator in plant root development. Plant Cell Rep 32:741–757
Schiefelbein JW, Benfey PN (1991) The development of plant roots: new approaches to underground problems. Plant Cell 3:1147–1154
Shan X, Zhang Y, Peng W, Wang Z, Xie D (2009) Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J Exp Bot 60:3849–3860
Shani E, Weinstain R, Zhang Y, Castillejo C, Kaiserli E, Chory J, Tsien RY, Estelle M (2013) Gibberellins accumulate in the elongating endodermal cells of Arabidopsis root. Proc Natl Acad Sci USA 110:4834–4839
Sharp RE, Poroyko V, Hejlek LG, Spollen WG, Springer GK, Bohnert HJ, Nguyen HT (2004) Root growth maintenance during water deficits: physiology to functional genomics. J Exp Bot 55:2343–2351
Singh VK, Garg R, Jain M (2013) A global view of transcriptome dynamics during flower development in chickpea by deep sequencing. Plant Biotechnol J 11:691–701
Spurr AR (1969) A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res 26:31–43
Sweetman C, Wong D, Ford C, Drew D (2012) Transcriptome analysis at four developmental stages of grape berry (Vitis vinifera cv. Shiraz) provides insights into regulated and coordinated gene expression. BMC Genom 13:691
Takatsuka H, Umeda M (2014) Hormonal control of cell division and elongation along differentiation trajectories in roots. J Exp Bot 65:2633–2643
Titapiwatanakun B, Murphy AS (2009) Post-transcriptional regulation of auxin transport proteins: cellular trafficking, protein phosphorylation, protein maturation, ubiquitination, and membrane composition. J Exp Bot 60:1093–1107
Torti S, Fornara F, Vincent C, Andrés F, Nordström K, Göbel U, Knoll D, Schoof H, Coupland G (2012) Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell 24:444–462
Ubeda-Tomás S, Federici F, Casimiro I, Beemster GTS, Bhalerao R, Swarup R, Doerner P, Haseloff J, Bennett MJ (2009) Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr Biol 19:1194–1199
Vilaine F, Kerchev P, Clément G, Batailler B, Cayla T, Bill L, Gissot L, Dinant S (2013) Increased expression of a phloem membrane protein encoded by NHL26 alters phloem export and sugar partitioning in Arabidopsis. Plant Cell 25:1689–1708
Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15:2532–2550
Woeste KE, Ye C, Kieber JJ (1999) Two Arabidopsis mutants that overproduce ethylene are affected in the posttranscriptional regulation of 1-aminocyclopropane-1-carboxylic acid synthase. Plant Physiol 119:521–530
Wu W, Cheng S (2014) Root genetic research, an opportunity and challenge to rice improvement. Field Crop Res 165:111–124
Xu ZS, Huang Y, Wang F, Song X, Wang GL, Xiong AS (2014a) Transcript profiling of structural genes involved in cyanidin-based anthocyanin biosynthesis between purple and non-purple carrot (Daucus carota L.) cultivars reveals distinct patterns. BMC Plant Biol 14:262
Xu ZS, Tan HW, Wang F, Hou XL, Xiong AS (2014) CarrotDB: a genomic and transcriptomic database for carrot. Database 2014:bau096
Yokoyama A, Yamashino T, Amano Y-I, Tajima Y, Imamura A, Sakakibara H, Mizuno T (2007) Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant Cell Physiol 48:84–96
Zhou ZY, Zhang CG, Wu L, Zhang CG, Chai J, Wang M, Jha A, Jia PF, Cui SJ, Yang M, Chen R, Guo GQ (2011) Functional characterization of the CKRC1/TAA1 gene and dissection of hormonal actions in the Arabidopsis root. Plant J 66:516–527
Acknowledgments
The research was supported by the New Century Excellent Talents in University (NCET-11-0670); Jiangsu Natural Science Foundation (BK20130027); China Postdoctoral Science Foundation (2014M551609), Priority Academic Program Development of Jiangsu Higher Education Institutions Project.
Conflict of interest
The authors declare that there are no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S.Hohmann.
Rights and permissions
About this article
Cite this article
Wang, GL., Jia, XL., Xu, ZS. et al. Sequencing, assembly, annotation, and gene expression: novel insights into the hormonal control of carrot root development revealed by a high-throughput transcriptome. Mol Genet Genomics 290, 1379–1391 (2015). https://doi.org/10.1007/s00438-015-0999-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-015-0999-5