Abstract
Chinese cabbage (Brassica rapa subsp. pekinensis) is an economically important vegetable that has encountered four rounds of polyploidization. The fourth event, whole genome triplication (WGT), occurred after its divergence from Arabidopsis. Expansins (EXPs) are cell wall loosening proteins that participate in cell wall modification processes. In this study, the impacts of WGT on the B. rapa expansin (BrEXP) superfamily were evaluated. Whole genome screening of B. rapa identified 32 loci coding 53 expansin genes. Fifteen of the loci maintained a single gene copy, 15 maintained two gene copies and 2 maintained three gene copies. Six loci had no synteny to any Arabidopsis thaliana orthologs. Two loci were involved in tandem duplication. Segmental duplication and fragment recombination were dominant in accelerating BrEXP evolution. Three genes (BrEXPA7, BrEXLA1 and BrEXLA2) lost one of their ancestral introns, two genes (BrEXPA18 and BrEXPB6) gained new introns, and a domain tandem repeat (BrEXPA18) and domain recombination (Bra016981; not considered as expansin) were observed in one gene each. Further, domain deletion was observed in an additional five genes (Bra033068, Bra000142, Bra025800, Bra016473 and Bra004891, not considered as expansins) that lost one of their expansin-specific domains evolutionarily. These findings provide a basis for the evolution and modification of the BrEXP superfamily after a WGT event, which will help in determining the functional characteristics of BrEXPs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polyploidization is an evolutionary force that occurred in ~70 % of angiosperms and is recognized as a core factor in the evolution of the plant kingdom (Masterson 1994). Genome reorganization is a rapid and common process associated with polyploidization, in which the duplicated gene copies are either deleted or retained for new, complementary and/or extra function (Barker et al. 2012). Chinese cabbage (Brassica rapa ssp. pekinensis) is an important vegetable that has encountered four polyploidization events; three of them occurred before its divergence, and the fourth, whole genome triplication (WGT), occurred after its divergence from Arabidopsis (Bowers et al. 2003). The B. rapa genome sequence seems to show that >50 % of the duplicated genes were lost in Brassica plants after the fourth event, indicating that gene loss/retention is a vital process after polyploidization (Wang et al. 2011).
Expansins (EXPs) are non-enzymatic cell wall loosening proteins (Cosgrove 2000) that were first identified and characterized in the early 1990s (McQueen-Mason et al. 1992; McQueen-Mason and Cosgrove 1994). Since then, expansins have been identified in almost all plant genera, together forming the expansin superfamily. Based on a phylogenetic sequence analysis, the multigene expansin superfamily has been classified into four families, α-expansin (EXPA), β-expansin (EXPB), expansin-like A (EXLA) and expansin-like B (EXLB) (Kende et al. 2004). The largest and most diverse family is EXPA followed by EXPB. The EXLA and EXLB families are small and have been identified only by their conserved gene sequences. Despite not knowing the exact mechanism of action of the expansins, it has been widely acknowledged that, in all plant cell wall modification processes, expansins stimulate turgor-driven cell expansion by acid-induced disruption of the non-covalent bonds linking cellulose microfibrils and hemicellulose fibers (Sampedro and Cosgrove 2005). The influence of WGT on the expansin gene family of B. rapa has not been reported so far, despite the significance of this gene family in plant growth and development. Hence, it is appropriate to conduct a systematic analysis to understand and establish the impacts of WGT on the expansion of the B. rapa expansin gene family.
Expansins consist of ~250–275 amino acids that form two domains (domain 1 with unknown function and domain 2 that is hypothesized to bind cell wall polysaccharides) preceded by a signal peptide at the N-terminus. The signal peptide is presumably removed during processing in the endoplasmic reticulum (Cosgrove 2000). Glycoside hydrolase family 45 proteins (GS45; mostly fungal β-1,4-d-endoglucanases) and grass-2 pollen allergens (G2As family) shown distant homology to expansin domains 1 and 2, respectively (Sampedro and Cosgrove 2005). Expansins comprise a highly conserved cysteine-rich region at the N-terminus, four tryptophan residues at the C-terminus and an HFD (histidine–phenylalanine–aspartate) motif in the middle. Conserved cysteine residues help to stabilize the binding domain by forming disulfide bonds (Yu et al. 2011). Besides sharing many conserved amino acid residues and characteristic motifs, expansins in the different expansin families show some family-specific characteristics (Sampedro and Cosgrove 2005). For example, EXPAs differ from other expansins by the presence of a large α-insertion and a nearby deletion in domain 1, the conserved HFD motif is present in EXPA and EXPB but absent in EXLA and EXLB, and only the EXLAs have a conserved CDRC motif at domain 1 and a 17 amino acid extension at domain 2.
Whole genome screening has been applied to explore several species-specific structural features of expansins in A. thaliana (Sampedro et al. 2005), rice (Oryza sativa; Sampedro et al. 2005), poplar (Populus trichocarpa; Sampedro et al. 2006), grapevine (Vitis vinifera; DeSanto et al. 2013) and soybean (Glycine max; Zhu et al. 2014). These genome-wide identifications and comparisons among different species have provided rich resources for the evolutionary analysis of expansins. In this study, we describe the identification, structural characteristics, phylogeny and evolutionary dynamics of B. rapa expansin (BrEXP) genes by comparative genomic analysis and suggest a systematic nomenclature for these genes. Our findings will be useful resources for future studies to unravel the functions of the BrEXP genes and will contribute to our understanding of the effects of polyploidy on the expansion of the BrEXP superfamily.
Materials and methods
Identification of expansin genes in the B. rapa genome
Nucleotide and amino acid sequences of 36 known A. thaliana expansins (AtEXPs) were obtained from The Arabidopsis Information Resource database (TAIR; http://www.arabidopsis.org/browse/genefamily/expansin.jsp) and the NCBI database (http://www.ncbi.nlm.nih.gov/). The downloaded sequences were used for homology search against the genome sequences of B. rapa line Chiifu-401-42 (version 1.5 and 1.2) in the Brassica (BRAD; http://brassicadb.org/brad/) and Phytozome (http://www.phytozome.net) databases. The identified BrEXP gene sequences were then used as queries in BLAST searches against BRAD, Phytozome, Superfamily (http://supfam.org/SUPERFAMILY/index.html) and UniProt (http://www.uniprot.org/) databases to ensure that no expansin and/or expansin-like sequences were missed. All the obtained sequences were scanned using ScanProsite (http://prosite.expasy.org/prosite.html) to confirm the presence of the expansin-specific domains (domains 1 and 2). The signal peptide was predicted using the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/). Isoelectric points (pIs) and molecular weights (MWs) of the BrEXPs were retrieved from the ExPASy Proteomics Server (http://web.expasy.org/compute_pi/). All tools/programs used in this study were run with default settings; otherwise the changes are mentioned.
Synteny and gene duplication analyses
The synteny relationship between the BrEXPs and AtEXPs was evaluated using the search syntenic genes tool in BRAD (http://brassicadb.org/brad/searchSynteny.php). The IDs of the BrEXP and AtEXP genes were individually used to identify synteny blocks in the two genomes that contain expansin-coding genes. Genes on syntenic loci in the same genome were defined as paralogous groups. Tandem duplications were defined if two EXP genes were separated by four or fewer other genes. Fifty-three BrEXP genes were aligned and analyzed using the Recombination Detection Program (RDP4; Martin et al. 2010) to check the possibility of conversion events. The synonymous (K s) and nonsynonymous (K a) nucleotide substitution rates per site between orthologous gene pairs were retrieved from the Plant Genome Duplication Database (PGDD; http://chibba.agtec.uga.edu/duplication/). PGDD uses ClustalW and PAL2NAL programs to align the protein and coding sequences, respectively, of the homologous gene pairs inferred from syntenic alignments and then uses the Nei-Gojobori method implemented in the PAML package to calculate the K s rates (Lee et al. 2012).
Gene structure and chromosomal mapping
The chromosomal coordinates and loci of the identified BrEXP genes were inferred from the Brassica Genome Browser (http://brassicadb.org/cgi-bin/gbrowse/Brassica_v1.5/). Chromosomal loci were reconfirmed using the genomic block arrangement that was obtained from the synteny search. We used the Exon–Intron Graphic Maker (http://wormweb.org/exonintron) online tool to draw the structure of the expansin gene sequences that comprised both the exons and introns. This tool was run with default settings and the ranges of the conserved domains were input manually.
Sequence alignment and phylogenetic analysis
The translated amino acid sequences of 53 BrEXPs were aligned by ClustalW using the MEGA6 program (Tamura et al. 2013). The resulting alignment file was used to construct phylogenetic trees using MEGA6. Trees were generated using the neighbor-joining (NJ) method with a number of differences and complete delete options. Tree reliability was assessed using 1,000 bootstrap replicates. The numbers generated for each clade represent the bootstrap support values expressed as percentages. For the phylogenetic clade assignment analysis, a multiple sequence alignment file consisting of the protein sequences of 53 BrEXPs, 35 AtEXPs (representing 15 clades) and 2 O. sativa expansins (OsEXPs; representing clades EXPA-XI and EXLB-II) was used in MEGA6 with the above-mentioned parameters.
Results
Genome-wide identification, selection and classification of the expansin gene superfamily in Chinese cabbage
A total of 59 expansins were identified. Based on the proposed nomenclature for expansin superfamily members (Kende et al. 2004), five of the expansins were excluded because of the absence of either domain 1 or domain 2 in their amino acid sequences: one protein (Bra004891) did not contain domain 1, while four proteins (Bra000142, Bra016473, Bra025800, and Bra033068) did not contain domain 2. Surprisingly, Bra016981 contained domains 1 and 2 plus two different macro domains (Pfam: PF01661) and a ribosomal protein S26e domain (Pfam: PF01283). This structure is unusual for standard expansins; therefore, Bra016981 was removed from the list of BrEXPs. Another protein (Bra017059: later named BrEXPA18) contained one domain 1 and two domain 2s that shared 100 % nucleotide and amino acid sequence similarity. This feature is known commonly as a tandem domain repeat (Bjorklund et al. 2006). Bra017059 was included in the list of BrEXPs because, unlike Bra016981, it contained only the expansin-specific domains. Thus, the 53 remaining B. rapa expansins were used in this study.
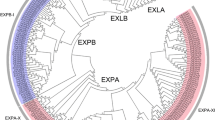
The s tructures of the 53 selected BrEXPs are shown in Fig. 1b. Domains 1 and 2 were preceded by a signal peptide of 15–29 amino acids in most of the BrEXPs, while 13 BrEXPs lacked a signal peptide (Online Resource 1). Multiple sequence alignments and conserved amino acid residues of the BrEXPs are given in Online Resource 2. In domain 1 in the BrEXPA proteins, seven conserved cysteine (Cys) residues were identified (an exception was BrEXPA39 in which only three Cys were conserved), and in the BrEXPB, BrEXLA, and BrEXLB proteins five Cys were conserved. In domain 2 in BrEXPA, three tryptophan (Try) residues were conserved and in BrEXPB, BrEXLA, and BrEXLB two, five, and four Try were conserved, respectively. Overall, excluding the first methionine (Met), five Cys, three glycine (Gly), and one alanine (Ala) in domain 1, and one Try in domain 2 were highly conserved in all the B. rapa expansins. Forty-five BrEXPs contained an HFD motif in the middle of the sequence, and 36 of these included the characteristic large insertion and deletion in domain 1 (Table S2). One expansin (Bra011901) contained an HLE motif and two expansins (Bra021365 and Bra032006) contained an HFV motif instead of the HFD motif, but all three carried a large insertion and deletion in domain 1. These results suggest that the B. rapa genome contains 39 EXPA and 9 EXPB genes. Among the remaining BrEXPs, two were classified as EXLA based on the CDRC motif and EXLA-specific extension contained at the C-terminus, and the other three were classified as EXLB members. This classification was supported by the results of the phylogenetic analysis (Fig. 1a). We compared the numbers of BrEXPs with the numbers of EXPs in related species (Table 1). In the soybean genome, the maximum numbers of expansins belonged to the EXPA and EXLB families, while in the rice genome the majority of expansins belonged to the EXPB and EXLB families. The BrEXP superfamily (n = 53) comprised more members than the AtEXP superfamily (n = 36), but the number of BrEXPs was not triple that of the AtEXPs as might have been expected. This finding indicates that >50 % of BrEXP genes were deleted during evolution of Brassica after the WGT event. The numbers of EXPs in each expansin family varied greatly among the three legume species, soybean, common bean, and Medicago truncatula. The number of expansins was highest in soybean, medium in common bean, and lowest in M. truncatula. This result suggests the expansion of the expansin families was species specific.
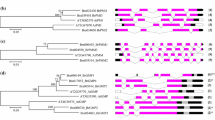
Phylogenetic tree and exon–intron organization of BrEXP genes. a Phylogenetic tree of the deduced amino acid sequences of 53 BrEXPs. ClustalW and the neighbor-joining method in the MEGA6 program were used to construct the tree. The reliability of the tree was tested by bootstrapping with 1,000 replicates. Bootstrap values below 75 are not shown. Colored diamonds indicate the sequence similarity between sister gene pairs: green 95–99, blue 90–94, red 80–89, black 70–79, and empty 60–60 %. Phylogenetic clades and their ancestral intron patterns are shown to the right of the gene names. b Exon–intron structures of the BrEXP genes. Exons are represented as boxes (drawn to scale) and introns are represented as lines (not drawn to scale). Domains 1 and 2 are shown in gray and yellow, respectively. Previously defined intron positions are represented at the top (Sampedro and Cosgrove 2005). The intron pattern for each BrEXP gene is indicated on the right
All the BrEXP members were named based on the accepted nomenclature and chromosomal position within each family (Online Resource 1). The average pI, MW, and number of amino acids for the BrEXPs were 8.6 ± 1.5 pI, 28 ± 3 kDa, and 261 ± 22 residues, respectively. No correlations were found between the pI, MW, gene length, ORF length, and amino acid length. The lengths and MWs of the BrEXPs ranged from 230 to 275 residues and 25 to 30 kDa, respectively; exceptions were BrEXPA18 (aa = 348; MW = 38 kDa), BrEXPB4 (360 aa; 41 kDa), and BrEXPB6 (318 aa; 33 kDa). The majority of BrEXPAs and BrEXLAs, and all the BrEXPBs had pI values from 8 to 10. The three BrEXLBs all had pI values less than 7.
Evolutionary relationship among BrEXPs
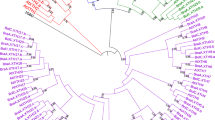
To determine the evolutionary relationship among the BrEXPs, the 53 aligned translated BrEXP amino acid sequences were used to construct an NJ-oriented unrooted tree in MEGA6. The 53 BrEXPs clustered into four major clades as expected: EXPA, EXPB, EXLA, and EXLB (Fig. 1a). Twenty-one sister pair genes were found with very strong bootstrap support (>90 %). Nine of the gene pairs shared 95–99 % sequence similarity, six pairs shared 90–94 %, two pairs shared 80–89 %, three pairs shared 70–79 %, and one gene pair shared 60–60 % similarity. Most of the gene pairs (n = 15) had very short branch lengths indicating that they may have evolved recently.
The common ancestor of monocots and eudicots was predicted to contain 15–17 expansin genes (Table 1). With this consideration, the four expansin gene families in angiosperms were subdivided into 17 clades (Sampedro and Cosgrove 2005). Therefore, all the genes that descended from the same ancestral gene should be included in the same clade. The expansin superfamily in O. sativa was found to contain all 17 clades, while the Arabidopsis expansin superfamily contained only 15 clades (Sampedro et al. 2005). Hence, to determine the number of clades in B. rapa, we constructed a phylogenetic tree with the translated amino acid sequences of the 53 BrEXPs, 35 AtEXPs representing 15 clades, and 2 OsEXPs representing the two clades (EXPA-XI and EXLB-II) that were missing in Arabidopsis (Online Resource 3). The near-identical ortholog sequences from B. rapa and A. thaliana clustered together with very short branch lengths. We found that the same two monocot/grass-specific ancestral clades (EXPA-XI and EXLB-II) that were absent in the AtEXPs were also absent in the BrEXPs (Fig. 1a). The highest number of expansins was in clade EXPA-IV (n = 10), followed by clades EXPA-I (n = 7) and EXPB-I (n = 6). Three clades (EXPA-II, EXPA-V, and EXPA-VIII) contained one expansin each and all the remaining clades contained from two to five expansins.
The number of introns in the BrEXPs varied from two to five (Fig. 1b). In general, members of same family shared similar exon–intron structures. BrEXPA18 contained one new intron (H) and BrEXPB6 contained two new introns (H and I). BrEXPB4 contained a very long exon after domain 2; this feature was not found in the other BrEXPBs. In all phylogenetic clades, the clustered expansins followed the same ancient exon–intron pattern with some exceptions. BrEXPA7, which was expected to cluster in clade EXPA-IX, contained only the B intron; however, it clustered with BrEXPA29 in clade EXPA-I. The genes that encode EXLA proteins usually contain four introns (ACBF) (Sampedro et al. 2005), but all three BrEXLAs contained only the A and B introns.
Synteny analysis and chromosomal mapping of BrEXPs
We used the term ‘locus’ instead of ‘gene’ to evaluate the synteny relationship of the expansin-coding loci between A. thaliana and B. rapa because ‘locus’ is evolutionarily more accurate. The ancestral genomes, such as ancestral crucifer karyotype (n = 8), proto-calepineae karyotype (n = 7), and translocation proto-calepineae karyotype (n = 7), each comprises 24 ancestral genomic blocks (Cheng et al. 2013). These ancient blocks were present in triplicate in 10 B. rapa chromosomes as a result of the WGT event. The triplicate blocks (71 blocks in total, because one copy of block G was lost; Cheng et al. 2013) in B. rapa were classified into three sub-genomes; i.e., least fractionated blocks (LF), medium fractionated blocks (MF1), and most fractionated blocks (MF2) that retained 23, 18, and 12 expansin genes, respectively (Fig. 2). Expansin genes were distributed on the ten chromosomes (A) of B. rapa with different densities. Two unmapped genes BrEXPA39 and BrEXPB9 were located on Scaffold000317 (gene position: 22,499–24,350) and Scaffold000111 (gene position: 290,653–291,849), respectively. The maximum number of expansin genes was found on A03 (n = 8), followed by A02 (n = 7) and A05 (n = 7). The A10 (n = 2) and A08 (n = 2) had the lowest density of expansin genes.
Position of the BrEXP genes on the Brassica rapa chromosomes. Chromosome (A) size is indicated by their relative lengths. The genome structure is taken from Cheng et al. (2013). LF least fractionated blocks, MF1 medium fractionated blocks, MF2 most fractionated blocks represent the three defined sub-genomes of B. rapa. Chromosomal coordinates of all the BrEXPs (except BrEXPA33) coincided with the positions of the genomic blocks. BrEXPA39 and BrEXPB9 were located on Scaffold000317 (gene position: 22,499–24,350) and Scaffold000111 (gene position: 290,653–291,849), respectively
The synteny analysis revealed that the 53 BrEXP genes were encoded in 32 loci: 25 loci encoded 39 BrEXPA genes, 5 loci encoded nine BrEXPB genes, 1 locus encoded two BrEXLA genes, and 1 locus encoded three BrEXLB genes (Table 2). Among the 32 loci, five EXPA and one EXPB loci were not in synteny with any of the A. thaliana homologs. On the other hand, of the 29 expansin loci in A. thaliana, 3 loci [one EXPA (gene id: AT3G29365), 1 EXPB (AT1G65680), and 1 EXLA (AT3G45970)] showed no syntenic relationship with any of the B. rapa homologs. Only two BrEXP loci maintained three copies, while most of the BrEXP loci maintained either a single copy (15 loci) or two copies (15 loci). Seventeen gene loci (53.1 %) were found to be involved in segmental duplication and two gene loci (6.3 %) were found to be involved in tandem duplication events. Each pair of segmental duplicates was distributed on different chromosomes. Among the tandem duplicates, BrEXPB5:BrEXPB6 shared 75.9 % similarity, whereas BrEXLB2:BrEXLB3 shared 97.6 % similarity. Recombination analysis showed that 14 BrEXPA and 3 BrEXLA genes may have undergone recombination events (Online Resource 4). No recombination was found among the BrEXPB and BrEXLB genes.
Synonymous (K s) and nonsynonymous (K a) nucleotide substitution rates
The K a and K s values can be used to predict the selective pressure on duplicated genes. A K a/K s ratio >1 indicates positive selection, K a/K s = 1 indicates neutral selection, and K a/K s < 1 indicates purifying (negative) selection. The orthologous expansin gene pairs between the B. rapa and A. thaliana genomes were used to estimate K a, K s, and K a /K s (Table 3). All the gene loci had K a values <0.10 except the loci of AT3G60570 (K a = 0.14). No K a/K s values were found for non-synteny genes, except for BrEXPA5 (Bra033099) and BrEXPA25 (Bra025397), which showed high Ks values (>2) and low Ka/Ks values (<0.2) from AtEXPA21 (gene id: AT5G39260). The single-copy gene loci had Ks values between 0.37 and 0.56, the two-copy loci had Ks values between 0.38 and 0.59, and the three-copy loci had K s values between 0.38 and 0.47. Most of the BrEXP genes had Ka/Ks ratios <0.2. The highest K a/K s ratio was found in the loci of AT5G39260 (K a /K s = 0.46; Bra028232 and Bra030353).
Discussion
Evolution of expansin-coding genes after whole genome triplication in B. rapa
Genome duplication expands genome content and diversifies gene function to ensure the best adaptability and evolution of plants. Gene loss/retention, fragmentation, and recombination are core processes that occur after genome duplication events. Gene gain/loss has interested researchers for decades and several hypotheses have been proposed (reviewed in Barker et al. 2012). B. rapa is a mesopolyploid, which can be used to study gene loss/retention in triplicated genomes. The Arabidopsis genome contain 36 expansin genes (Sampedro et al. 2005); therefore, the WGT event could be expected to produce >100 expansin genes in the B. rapa genome. However, we found that only 53 BrEXP genes were retained in the B. rapa genome. This finding implies that more than 50 % of duplicated expansin genes were lost after WGT. Besides gene lost, the WGT event has indeed expanded the BrEXP superfamily. Of the three principal evolutionary patterns, segmental duplication, tandem duplication, and transposition (retroposition and replicative transposition), the expansion of the BrEXP superfamily seems to have been achieved mainly by segmental duplication (68 %; 36 of 53 genes), while tandem duplication seems to have played only a minor role (6.3 %; four of 53 genes). This result is similar to the findings reported for expansion of expansin families in soybean and Arabidopsis where segmental duplication had a major influence (68 and 50 % of the genes, respectively); in rice, tandem duplication had a major influence (55.2 % of the genes). These findings coincide with the results of Cannon et al. (2004), who reported that segmental and tandem duplication events were the main causes of gene family expansion in plants. We observed recombination signatures in 35.9 % (19 of 53) of the BrEXP genes and, of the 19 genes, 16 (30.2 %) exhibited fragment recombination and 3 (5.7 %) exhibited large-scale recombination. Based on all of these findings, we propose that segmental duplication and fragment recombination were the major drivers of the expansion of the BrEXP superfamily.
One gene of a duplicated pair can generally follow one of three functional outcomes: nonfunctionalization (duplicated genes are silenced), neofunctionalization (duplicated genes gain new functions), and subfunctionalization (function is partitioned between the new paralogs) (Barker et al. 2012). In gene dosage balance model, duplicated genes that have pleiotropic effects are mostly over-retained after a whole genome duplication event than a small-scale duplication event (Veitia 2005). Further, because duplication increases the mutational targets and mutations in one of the gene copies may produce dominant-negative phenotypes, some genes are convergently restored to single-copy status (DeSmet et al. 2013). In the present study, we identified 32 loci encoding 53 genes in B. rapa; 15 loci encoded single copies, 15 loci encoded two copies, and 2 loci encoded three copies. It is unclear why some loci retained one gene copy, while other loci retained two or three gene copies. The K a /K s ratios showed no significant differences among the one-, two-, and three-copy loci (Table 3). Only one two-copy loci (Bra028232 and Bra030353) had a Ka/Ks ratio of 0.46, while all other loci had Ka/Ks ratios <0.2, indicating that all the BrEXP loci underwent strong purifying selection. We believe that comparing the comprehensive tissue-specific, temporal, and spatial expression profile analyses between A thaliana and B. rapa will help to reveal whether the single-copy gene loci follow the gene dosage balance and/or the dominant-negative hypotheses, and whether the two- and three-copy loci follow the neo- and/or sub-functionalization hypotheses.
Gain and loss of introns in the expansin superfamily
In the expansin superfamily, EXPA genes contain one [(A) or (B)] or two (A B) introns, EXPB genes contain three [(A C F) or (A B F)] or four (A C B F) introns, EXLA genes contain four (A C B F) introns, and EXLB genes contain three (A C F) or four (A C B F) introns (Sampedro et al. 2005; Carey Hepler and Cosgrove 2013). Gain and loss of introns among expansin members have often been discussed with the help of 17 phylogenetic clades that were reconstructed based on the last common ancestor of monocots and eudicots (Sampedro et al. 2005). Intron losses have been reported previously in EXPAs from Arabidopsis, poplar, and grapevine. One Arabidopsis gene (AtEXPA20) in clade EXPA-VIII was found to have lost intron B, two poplar genes (PtEXPA21 and PtEXPA22) in clade EXPA-VIII lost intron A, and one grapevine gene (VvEXPA10) in clade EXPA-IV lost intron B (Sampedro et al. 2005, 2006; DeSanto et al. 2013). Zhu et al. (2014) extensively studied the expansin superfamily in soybean, but did not assign them to phylogenetic clades; however, they suggested (Zhu et al. 2014; Fig. 2 and Additional file 1) that at least four EXPA genes (GmEXPA20, GmEXPA21, GmEXPA35, and GmEXPA43) and one EXLB gene (GmEXLB1) contained a new species-specific intron. In the present study, we found that one gene [BrEXPA1 (paralog of AtEXPA20)] in clade EXPA-VIII had lost intron B, two genes (BrEXPA2 and BrEXPA23) in clade EXPA-IX had lost intron A, and one gene (BrEXPA7) in clade EXPA-I had lost intron A. Interestingly, BrEXLA1 and BrEXLA2 in clade EXLA-I retained only the (A B) introns and lost introns (C F). In addition, one gene (BrEXPA18) in clade EXPA-IV was found to have one new intron (H) and one gene (BrEXPB6) in clade EXPB-I was identified with two new introns (H I). This is the first report of intron gain in EXPB and intron loss in EXLA genes, indicating that these losses and gains may be species specific.
Modular rearrangements in the BrEXP superfamily
Protein domains can evolve by point mutations and modular rearrangements. In most protein families, the combination of domains is always the same; i.e., neighboring domains are always arranged in the same order (Vogel et al. 2004). Expansins always have a two-domain structure (domain 1 and domain 2), so proteins that lack either of the domains were not considered as expansins (Kende et al. 2004). In this study, domains 1 and 2 were found in 54 B. rapa proteins and 52 of them strictly followed the expansin rule. One protein (BrEXPA18) contained a tandem repeat of domain 2 (Fig. 1b) and another protein (Bra016981; not consider an expansin) contained expansin-specific domains, two different macro domains, and a ribosomal protein-S26e domain (data not shown). These domain repeat and multidomain features are likely to be the results of modular rearrangements (Bjorklund et al. 2006; Weiner et al. 2008), which are driven primarily by fusion of pre-existing arrangements, single domain proteins, and domains that occur frequently at protein termini (Weiner et al. 2008). Our synteny analysis (Table 2) clearly showed that the expansin-specific domains in Bra01681 were generated from the WGT event, suggesting that Bra01681 formed after the WGT event by domain recombination.
Domain deletion in the BrEXP superfamily
In this study, four proteins (Bra033068, Bra000142, Bra025800, and Bra016473) that contained only domain 1 and one protein (Bra004891) that contained only domain 2 were identified. The synteny of the expansin-coding genes between B. rapa and A. thaliana clearly showed that WGT had generated these five proteins in B. rapa. Indeed, analysis indicated the genes that code these five proteins were orthologs and paralogs of actual expansins of Arabidopsis and B. rapa, respectively (Table 2). Thus, it is likely that during the course of evolution after WGT, these five proteins lost one of the two expansin domains. Proteins Bra033068, Bra000142, Bra025800, and Bra016473 lacked the domain (domain 2) at the C-terminus, while Bra004891 lacked the domain (domain 1) at the N-terminus. This phenomenon coincides with previous results that domain deletions frequently occur at the C-terminus (Weiner et al. 2006). Domain losses can be achieved by the introduction of start and stop codons which render the terminal domains nonfunctional, such that further shortening occurs until the whole domain is lost (Weiner et al. 2006). The Bra033068 and Bra025800 proteins were less than 100 amino acids long, while Bra000142, Bra016473, and Bra004891 had 130–170 amino acids, indicating that the domain deletion processes may have various dimensions.
Conclusion
In this study, we identified 39 BrEXPA, 9 BrEXPB, 2 BrEXLA, and 3 BrEXLB genes in the B. rapa genome. In silico analyses revealed that (1) more than 50 % of the duplicated expansin genes were lost in B. rapa after the WGT event, (2) BrEXPs underwent strong purifying selection, (3) segmental duplication and fragment recombination accelerated the evolution of BrEXPs, and (4) BrEXPs experienced intron gain and loss, domain deletion, domain fusion, and domain repeat. This study has extended our knowledge about the impact of WGT on the BrEXP superfamily and revealed many new species-specific structural characteristics of BrEXPs. Furthermore, it has provided basic resources for studying the functions of BrEXPs. We believe that the functional characteristics of BrEXPA20 (paralog of BrEXPA7 and ortholog of AtEXPA1), BrEXPA18 (paralog of BrEXPA20 and ortholog of AtEXPA4), and BrEXPB6 (tandem repeat of BrEXPB5 and ortholog of AtEXPB4) will help in understanding the importance and/or consequences of intron gain/loss and domain repeat to the functions of the expansins.
References
Barker MS, Baute GJ, Liu SL (2012) Duplications and turnover in plant genomes. In: Wandel et al (ed) Plant genome diversity, 1st edn. Springer, Vienna, pp 155–169
Bjorklund AK, Ekman D, Elofsson A (2006) Expansion of protein domain repeats. PLoS Comp Biol 2(8):e114
Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 8:433–438
Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4(1):10
Carey Hepler NK, Cosgrove DJ (2013) Selaginella moellendorffii has a reduced and highly conserved expansin superfamily with genes more closely related to angiosperms than to bryophytes. BMC Plan Biol 13:4
Cheng F, Manda´kova´ T, Wu J, Xie Q, Lysak MA, Wang X (2013) Deciphering the diploid ancestral genome of the mesohexaploid Brassica rapa. Plant Cell 25:1541–1554
Cosgrove DJ (2000) Expansive growth of plant cell walls. Plant Physiol Biochem 38:109–124
DeSanto S, Vannozzi A, Tornielli GB, Fasoli M, Venturini L, Pezzotti M, Zenoni S (2013) Genome-wide analysis of the expansin gene superfamily reveals grapevine-specific structural and functional characteristics. PLoS One 8(4):e62206
DeSmet R, Adams KL, Vandepoele K, Montagu MCEV, Maere S, DePeer YV (2013) Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants. Proc Natl Acad Sci USA 110:2898–2903
Kende H, Bradford K, Brummell D, Cho HT et al (2004) Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol Biol 55:311–314
Lee TH, Tang H, Wang X, Paterson AH (2012) PGDD: a database of gene and genome duplication in plants. Nucl Acids Res. doi:10.1093/nar/gks1104
Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P (2010) RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463
Masterson J (1994) Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264:421–424
McQueen-Mason S, Cosgrove DJ (1994) Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc Natl Acad Sci USA 91:6574–6578
McQueen-Mason S, Durachko DM, Cosgrove DJ (1992) Two endogenous proteins that induce cell wall extension in plants. Plant Cell 4:1425–1433
Sampedro J, Cosgrove DJ (2005) The expansin superfamily. Genome Biol 6:242
Sampedro J, Lee Y, Carey RE, DePamphilis C, Cosgrove DJ (2005) Use of genomic history to improve phylogeny and understanding of births and deaths in a gene family. Plant J 44:409–419
Sampedro J, Carey RE, Cosgrove DJ (2006) Genome histories clarify evolution of the expansin superfamily: new insights from the poplar genome and pine ESTs. J Plant Res 119:11–21
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mole Biol Evol 30:2725–2729
Veitia RA (2005) Gene dosage balance: deletions, duplications and dominance. Trends Genet 21:33–35
Vogel C, Berzuini C, Bashton M, Gough J, Teichmann SA (2004) Supra-domains: evolutionary units larger than single protein domains. J Mol Biol 336:809–823
Wang X, Wang H, Wang J, Sun R et al (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–1039
Weiner J, Beaussart F, Bornberg-Bauer E (2006) Domain deletions and substitutions in the modular protein evolution. FEBS J 273:2037–2047
Weiner J, Moore Ad, Bornberg-Bauer E (2008) Just how versatile are domains? Evol Biol 8:285–299
Yu Z, Kang B, He X, Lv S, Bai Y et al (2011) Root hair-specific expansins modulate root hair elongation in rice. Plant J 66:725–734
Zhu Y, Wu N, Song W, Yin G, Qin Y, Yan Y, Hu Y (2014) Soybean (Glycine max) expansin gene superfamily origins: segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol 14:93–112
Acknowledgments
This work was supported by the Rural Development Administration (Korea) through the Rural Program for Agricultural Science and Technology Development (Project No. PJ010025) and the Bio-green 21 Program (Project No. PJ009068).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Krishnamurthy, P., Hong, J.K., Kim, J.A. et al. Genome-wide analysis of the expansin gene superfamily reveals Brassica rapa-specific evolutionary dynamics upon whole genome triplication. Mol Genet Genomics 290, 521–530 (2015). https://doi.org/10.1007/s00438-014-0935-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-014-0935-0