Abstract
Genome-wide association studies have identified two SNPs (rs402710 and rs401681) of CLPTM1L at chromosome 5p15.33 as a new lung cancer (LC) susceptibility locus in populations of European descent. Since then, the relationship between these SNPs and LC has been reported in various ethnic groups; however, these studies have yielded inconsistent results. To investigate this inconsistency, we performed a meta-analysis of 27 studies involving a total of 60,828 cases and 109,135 controls for the two polymorphisms to evaluate its effect on genetic susceptibility for LC. An overall random-effects per-allele odds ratio of 1.14 (95 % CI 1.11–1.16, P < 10−5) and 1.15 (95 % CI 1.12–1.19, P < 10−5) was found for the rs401681 and rs402710 polymorphism, respectively. Significant results were also observed for under dominant and recessive genetic models. After stratified by ethnicity, significant associations were found among Caucasians and East Asians. In the subgroup analysis by sample size, significantly increased risks were found for these polymorphisms in all genetic models. In addition, we find both rs402710 and rs401681 conferred significantly greater risks for adenocarcinoma and squamous cell carcinoma when stratified by histological type of tumors. Furthermore, associations of these polymorphisms with LC risk were observed among current smokers and former smokers, as well as never smokers. Our findings demonstrated that rs402710 and rs401681 are risk-conferring factors for the development of lung cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer is the most common cause of cancer death worldwide, with over one million cases annually (Ferlay et al. 2007). The pathogenesis of lung cancer is known to be influenced by smoking, air pollution, exposure to hazardous substances (e.g., asbestos, chromium, nickel, inorganic arsenic, mustard gas, tar, etc.). Among all the known environmental risk factors, smoking is considered to be the most important cause of lung cancer; smoking increases 20-fold susceptibility to develop lung cancer, and 80–90 % cases of lung cancer relate to smoking (Ruano-Ravina et al. 2003). Although cigarette smoking is the major risk factor for lung cancer, genetic factors also affect lung cancer susceptibility (Matakidou et al. 2005). For example, increased lung cancer rates are observed in several genetic syndromes, including Li-Fraumeni syndrome, hereditary retinoblastoma, familial breast cancer, and Bloom syndrome (Strong et al. 1984; Li et al. 1988; Sanders et al. 1989; German 1993; Johannsson et al. 1996). Despite intense efforts in the past decades, researchers have still not identified specific biomarkers for lung cancer risk assessment and prognosis prediction.
Lung cancer is classified into two main histological groups: small cell lung cancer (SC) and non-small cell lung cancer; the latter includes adenocarcinoma (AD) and squamous cell carcinoma (SQ), along with large cell lung cancer (LC). Worldwide, adenocarcinoma is the most frequently identified histological type, and the relative proportion of lung cancer due to this histology has steadily risen. Demographic, etiologic, clinical, and molecular characteristics of the lung cancer subtypes have been reported (Gabrielson 2006). Although family history of lung cancer has been associated with histological subtypes (Ambrosone et al. 1993; Gao et al. 2009), the inherited susceptibility factors that affect specific histologies are unknown.
Recently, three genome-wide association studies (GWAS) of lung cancer and subsequent pooled GWAS analyses identified inherited susceptibility variants on chromosome 15q25 (Amos et al. 2008; Hung et al. 2008; Thorgeirsson et al. 2008), 5p15 (McKay et al. 2008; Wang et al. 2008; Rafnar et al. 2009), and 6p21.5 (Wang et al. 2008). The 5p15.33 region comprises two candidate susceptibility genes, telomerase reverse transcriptase (TERT) and cleft lip and palate transmembrane 1-like (CLPTM1L). The IARC (McKay et al. 2008) and the Institute of Cancer Research (ICR) and MD Anderson groups (Wang et al. 2008) reported that two different SNPs at 5p15.33 (rs402710 and rs401681, respectively), which are in strong linkage disequilibrium (D′ = 1.00, r 2 = 0.66), are associated with the risk of lung cancer (IARC study: P = 2 × 10−7; ICR and MD Anderson groups: P = 8 × 10−9). CLPTM1L plays a role in apoptosis and has been found to be upregulated in cisplatin-resistant cell lines (Yamamoto et al. 2001).
Replication of initial genome-wide association findings is considered a gold standard for reporting genotype–phenotype associations. Over the past few years, considerable efforts have been devoted to exploring the relationships between the two SNPs (rs402710 and rs401681) of CLPTM1L at 5p15.33 and lung among various populations. However, existing studies have yielded inconsistent results. These disparate findings may be due partly to insufficient power, false-positive results, and publication biases. The interpretation of these studies has been further complicated by the use of different populations, or histological types. To help clarify the inconsistent findings, we conducted a comprehensive meta-analysis to quantify the overall risk of rs402710 and rs401681 polymorphism of CLPTM1L on developing lung cancer.
Materials and methods
Literature search strategy
Genetic association studies published before the end of January 2014 on lung cancer and the two SNPs (rs402710 and rs401681) at 5p15.33 were identified through a search of PubMed, Web of Science, EMBASE, SCOPUS, and Cochrane databases using combinations of the following keywords: ‘CLPTM1L,’ ‘rs402710,’ ‘rs401681,’ ‘5p15,’ ‘5p15.33,’ ‘polymorphism’ or ‘variant,’ and ‘lung cancer’ or ‘lung carcinoma’ or ‘lung neoplasm’ without restriction on language. We replaced one of those search terms each time until all possible combination mode was searched to avoid any missing literature. The titles and abstracts of potential articles were screened to determine their relevance, and any clearly irrelevant studies were excluded. The full texts of the remaining articles were read to determine whether they contained information on the topic of interest. Furthermore, reference lists of primary studies and review articles were also reviewed by a manual search to identify additional relevant publications.
Inclusion and exclusion criteria
We reviewed abstracts of all citations and retrieved studies. The following criteria were used to include published studies: (1) Identification of lung cancer cases was confirmed histologically or pathologically, (2) case–control or cohort studies to evaluate the association between rs402710 or rs401681 polymorphism and lung cancer risk, and (3) genotype distribution information in cases and controls or odds ratio (OR) with its 95 % confidence interval (CI) and P value. Major reasons for exclusion of studies were (1) case-only studies or family-based studies; (2) duplicated studies; and (3) no sufficient data were reported.
Data abstraction
Data abstraction was performed independently by two reviewers, and differences were resolved by further discussion among all authors. For each study, the following characteristics were collected: the first author, publication year, ethnicity of the study population, number of cases and controls, gender, age of cases and controls, histological types, cigarette smoking status, source of control, genotyping method, and number of genotypes in cases and controls.
Quality assessment
For association studies with inconsistent results on the same polymorphisms, the methodological quality should be assessed by appropriate criteria to limit the risk of introducing bias into meta-analyses or systematic reviews. A procedure known as ‘extended-quality score’ has been developed to assess the quality of association studies. The procedure scores each paper categorizing it as having ‘high,’ ‘median,’ or ‘poor’ quality. Detailed procedure of the quality assessment was previously described (Li et al. 2006).
Statistical methods
Deviation from Hardy–Weinberg equilibrium (HWE) was examined by chi-square test with 1 degree of freedom. Crude odds ratio (ORs) with corresponding 95 % confidence intervals (CIs) was used to assess the strength of association between the 5p15.33-rs402710 and 5p15.33-rs401681 polymorphism and lung cancer risk. Additional pooled estimates were also given with corresponding results under dominant and recessive genetic models. Cochran’s chi-square-based Q statistic test was performed in order to assess possible heterogeneity between the individual studies and thus to ensure that each group of studies was suitable for meta-analysis (TanTai et al. 2014). The degree of heterogeneity was assessed using the I 2 metric (I 2 = (Q − df)/Q), which is independent of the number of studies in the meta-analysis (Chen et al. 2014). ORs were pooled according to the method of DerSimonian and Laird that takes into account the variation between studies, and 95 % CI was constructed using Woolf’s method (Woolf 1955; DerSimonian and Laird 1986). The Z test was used to determine the significance of the pooled OR. Ethnicity (Caucasian vs. East Asian), study size (≥1,000 cases or, <1,000 cases), histological types (AD, SQ, SC, and LC), and smoking behavior (former smokers or current smokers or never smokers) were pre-specified as characteristics for assessment of heterogeneity. In addition, ethnicity, histological subtype, source of controls, sample size, smoking behavior, and genotyping method were analyzed as covariates in meta-regression. Sensitivity analyses were performed to assess the stability of the results; namely, a single study in the meta-analysis was deleted each time to reflect the influence of the individual data set to the overall OR. Publication bias was assessed using Egger’s test and Begg’s funnel plots (Begg and Mazumdar 1994; Egger et al. 1997). All P values are two-sided at the P = 0.05 level. All statistical analyses were carried out with the Stata software version 10.0 (Stata Corporation, College Station, TX).
Results
Characteristics of studies
In all, we included 27 studies in this meta-analysis (Supplementary figure 1), with a total of 60,828 LC cases and 109,136 controls (McKay et al. 2008; Wang et al. 2008, 2013; Landi et al. 2009; Rafnar et al. 2009; Zienolddiny et al. 2009; Hsiung et al. 2010; Miki et al. 2010; Truong et al. 2010; Yang et al. 2010; Yoon et al. 2010; Hu et al. 2011; Jaworowska et al. 2011; Pande et al. 2011; Young et al. 2011; Bae et al. 2012; Chen et al. 2012; Ito et al. 2012; Lan et al. 2012; Shiraishi et al. 2012; Jiang et al. 2013; Ke et al. 2013; Li et al. 2013; Lu et al. 2013; Myneni et al. 2013; Zhao et al. 2013). For the rs402710 polymorphism, 16 studies were available, including a total of 31,854 cases and 41,073 controls. For the rs401681 polymorphism, 17 studies involved a total of 35,395 cases and 79,399 controls. These two polymorphisms were found to occur in frequencies consistent with HWE in the control populations of all the published studies. Fourteen studies were given high quality, and thirteen studies were given median quality. No ‘poor quality’ study was found. Characteristics of studies included in the current meta-analysis are presented in Table 1.
Association of CLPTM1L-rs401681 polymorphism with lung cancer
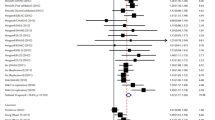
For LC risk and the rs401681 polymorphism, our meta-analysis gave an overall OR of 1.14 [95 % CI 1.11–1.16, P(Z) < 10−5; P(Q) = 0.62; Fig. 1] without statistically significant between-study heterogeneity (P > 0.05). Significantly increased LC risks were also found using dominant [OR 1.20, 95 % CI 1.12–1.30, P(Z) < 10−5; P(Q) = 0.11] and recessive model [OR 1.28, 95 % CI 1.14–1.44, P(Z) < 10−5; P(Q) = 0.008]. After adjusting for multiple testing using Bonferroni correction, all significant associations for rs401681 under different genetic models remained. In meta-regression analysis, ethnicity, sample size, genotyping method, source of controls, and genotyping method did not significantly explain such heterogeneity (P > 0.05). By contrast, histological subtype (P = 0.03) was significantly correlated with the magnitude of the genetic effect. Galbraith plot analyses of all included studies were used to assess the potential sources of heterogeneity. One study (Pande et al. 2011) was found to be contributors of heterogeneity for rs401681 polymorphism (Supplementary figure 2).
When stratifying for ethnicity, an OR of 1.14 (95 % CI 1.11–1.17, P < 10−5) and 1.13 (95 % CI 1.09–1.17, P < 10−5) resulted for the G allele, among Caucasian, and East Asian populations, respectively. Similar results were also detected using dominant and recessive genetic models (Table 2). Subsidiary analyses of sample size yielded a per-allele OR for small studies of 1.16 (95 % CI 1.11–1.22, P < 10−5), and for large studies of 1.14 (95 % CI 1.10–1.17, P < 10−5).
The association between genotypes and the risk of lung cancer was further examined by stratifying the subjects according to histological type and smoking behavior (Table 3). In subgroup analyses by histological types of LC, we found that the rs401681 polymorphism was significantly associated with lung adenocarcinoma (G allele: OR 1.49, 95 % CI 1.19–1.86, P = 0.001) and squamous cell carcinoma (G allele: OR 1.59, 95 % CI 1.23–2.06, P = 0.001), while marginal significant associations were detected for small cell carcinoma (G allele: OR 1.41, 95 % CI 1.03–1.94, P = 0.03). However, no significant associations were detected for large cell carcinoma. We investigated the relationship between the polymorphism and known environmental tobacco exposures. The effect of environmental tobacco smoke was similar for never smokers with per-allele OR of 1.20 (95 % CI 1.08–1.33, P = 0.001), compared to current smokers (OR 1.43, 95 % CI 1.05–1.95, P = 0.02) and former smokers (OR 1.23, 95 % CI 1.10–1.37, P < 10−4).
Association of CLPTM1L-rs402710 polymorphism with lung cancer
Overall, there was evidence of an association between increased risk of LC and the rs402710 polymorphism in different genetic models when all the eligible studies were pooled into the meta-analysis. Using random-effect model, the summary per-allele OR of the rs402710 G variant for LC was 1.15 [95 % CI 1.12–1.19, P(Z) < 10−5, P(Q) = 0.15; Fig. 2], with corresponding results under dominant and recessive genetic models of 1.18 [95 % CI 1.15–1.24, P(Z) < 10−5, P(Q) = 0.08] and 1.22 [95 % CI 1.18–1.27, P(Z) < 10−5, P(Q) = 0.002], respectively. In addition, for rs402710, associations were maintain statistically significant after Bonferroni correction only for multiple genetic models.
When studies were stratified for ethnicity, significant risks were found among East Asians in all genetic model [G allele: OR 1.12, 95 % CI 1.07–1.18; dominant model: OR 1.15, 95 % CI 1.07–1.23; recessive model: OR 1.18, 95 % CI 1.10–1.26]. Significant results were also found in the Caucasian populations [G allele: OR 1.17, 95 % CI 1.13–1.20; dominant model: OR 1.20, 95 % CI 1.16–1.25; recessive model: OR 1.23, 95 % CI 1.19–1.28]. By considering sample size subgroups, the OR was 1.15 (95 % CI 1.10–1.19, P < 10−5) in large studies compared to 1.17 (95 % CI 1.11–1.23, P < 10−5) in small studies. One study (Shiraishi et al. 2012) was found to be contributors of heterogeneity for rs402710 polymorphism after Galbraith plot analyses (Supplementary figure 3) (Table 4).
In the subgroup analyses by histological type, significant associations were found for lung adenocarcinoma, squamous cell carcinoma, and small cell carcinoma. However, we failed to detect any association between large cell carcinoma for the polymorphism (Table 5). In addition, associations of the polymorphism with LC risk were observed both in smokers and never smokers (Table 5). Although the formal test for heterogeneity was not significant, we conducted meta-regression as there were also grounds for considering the ethnicity, sample size, control source, histological type, and genotyping method as potential sources of heterogeneity. However, the meta-regression showed that none of these covariates significantly contributed to the heterogeneity among the individual study results except for ethnicity (P = 0.008).
Sensitivity analyses and Publication bias
A single study involved in the meta-analysis was deleted each time to reflect the influence of the individual dataset to the pooled ORs, and the corresponding pooled ORs were not qualitatively altered (Supplementary figures 4 and 5). Begg’s funnel plot and Egger’s test were performed to access the publication bias of the literatures. The shape of the funnel plots was symmetrical for these polymorphisms (Supplementary figures 6 and 7). The statistical results still did not show publication bias in these studies for rs401681 (Begg’s test, P = 0.15; Egger’s test, P = 0.14) and rs402710 (Begg’s test, P = 0.36; Egger’s test, P = 0.22).
Discussion
The association between polymorphisms of 5p15.33 and lung cancer risk had been originally reported by McKay et al. (2008). Recently, many lung cancer GWAS and replication studies have been conducted in European populations (Wang et al. 2008; Rafnar et al. 2009; Truong et al. 2010) and to a lesser extent in East Asians (Miki et al. 2010; Yoon et al. 2010; Hu et al. 2011). However, there are significant differences in allele frequencies and the prevalence of lung cancer among different populations. It is, therefore, important to quantitatively assess the effects of the GWAS-identified markers in different ethnic populations and explore potential heterogeneity of published data. This is the most comprehensive meta-analysis that examined the two SNPs (rs402710 and rs401681) at 5p15.33 and the relationship with susceptibility for lung cancer. Its strength was based on the accumulation of published data giving greater information to detect significant differences. In total, the meta-analysis involved 27 studies that provided 60,828 LC cases and 109,135 controls.
Our results demonstrated that the rs402710 and rs401681 polymorphism is a risk factor for developing lung cancer. In the stratified analysis by ethnicity, significant associations were found in East Asians and Caucasians for these polymorphisms in all genetic models, suggesting a similar role of these variant among different ethnicity with different genetic backgrounds. However, we observed that association between these polymorphisms and risk of lung cancer in Caucasians was stronger than that in East Asians. There are several possible reasons for such differences. Firstly, the frequencies of the risk-association alleles in these polymorphisms vary between different races. For example, the G allele distribution of the rs401681 varies between Caucasians and East Asians, with a prevalence of 56 and 70 %, respectively (McKay et al. 2008; Wang et al. 2008; Hu et al. 2011; Jaworowska et al. 2011). Secondly, study design or small sample size or some environmental factors may affect the results. Most of these studies did not consider most of the important environmental factors. It is possible that variation at this locus has modest effects on lung cancer, but environmental factors may predominate in the progress of lung cancer and mask the effects of this variation. Specific environmental factors such as lifestyle and smoking have been already well studied in recent decades (Matakidou et al. 2005). In addition, different populations usually have different linkage disequilibrium patterns. A polymorphism may be in close linkage with another nearby causal variant in one ethnic population but not in another. These polymorphisms may be in close linkage with different nearby causal variants in different populations.
Stratification of tumors by histological type indicated that the association of the 2 SNPs on 5p15.33 with lung cancer risk appeared to be similar for various histological types of lung cancer. In the stratified analysis by smoking behavior, significant associations were found among never smokers, ever smokers, and former smokers for the two polymorphisms. These results were in accordance with those reported by previous genome-wide association studies in a white population. McKay et al. (2008) reported odds ratios of 1.18 for rs402710, whereas Wang et al. (2008) reported an odds ratio of 1.14 for rs401681 (in strong linkage disequilibrium with rs402710). Neither of these studies reported heterogeneity by histology, smoking status, age at diagnosis, or sex. The magnitudes of these associations are consistent with our findings.
The two SNPs rs401681 and rs402710 are next to each other in the CLPTM1L gene and are strongly linked. CLPTM1L, named for its similarity to a gene implicated in susceptibility to cleft lip palate, was identified through screening for cisplatin (CDDP) resistance-related genes and was found to be upregulated in CDDP-resistant ovarian tumor cell lines and to induce apoptosis in CDDP-sensitive cells (Yamamoto et al. 2001). The CLPTM1L gene is well conserved and expressed in various tissues, including lung tissue.
Survival of cells undergoing DNA damage from genomic instability or genotoxic stress leads to the accumulation of genetic lesions characterizing human tumors. Abrogation of cell-cycle checkpoint and apoptotic safeguards against replication of damaged DNA is a hallmark of cancer. The result of protection against DNA damage-induced apoptosis includes both vulnerability to unchecked somatic mutation and decreased sensitivity to radiotherapy and chemotherapy. The previously referenced study by Zienolddinny et al. (2009) suggests that CLPTM1L polymorphisms may affect DNA damage accumulation. Thus, it is plausible that protection from apoptosis by CLPTM1L contributes to accumulation of DNA damage, thereby conferring susceptibility to tumorigenesis. An alternative hypothesis is that CLPTM1L plays a role in recognition or repair of DNA damage, affecting the accumulation of such damage. Another is that CLPTM1L directly influences the amount of DNA damage that is incurred by genotoxic agents. Recently, James et al. (2012) suggests that CLPTM1L expression does not directly affect levels of acute DNA damage incurred by cisplatin or nitrosamine 4-(methyl-nitrosa-mino)-1-(3-pyridyl)-1-butanone (NNK). Moreover, the current study demonstrates that CLPTM1L has an apoptotic role downstream of DNA damage and through regulation of Bcl-xL expression, which is likely to affect accumulation of DNA damage. The observed difference in accumulation of Bcl-xL upon modulation of CLPTM1L expression, along with reconstitution of an apoptosis-resistant phenotype with expression of exogenous Bcl-xL, provides evidence that CLPTM1L acts upstream of Bcl-xL to confer resistance to genotoxic stress-induced apoptosis.
Limitations also inevitably existed in this meta-analysis. First, our meta-analysis is based on unadjusted estimates, whereas a more precise analysis could be performed if individual data were available, which would allow for an adjustment estimate. To be made, however, this approach requires the authors of all of the published studies to share their data. Second, the subgroup meta-analyses considering interactions between the 2 SNPs and histological type, as well as smoking behavior, were performed on the basis of a fraction of all the possible data to be pooled, so selection bias may have occurred and our results may be overinflated. Nevertheless, the total number of subjects included in this part of the analysis comprises the largest sample size so far. Finally, only published studies were included in this meta-analysis. Therefore, publication bias may have occurred, even though the use of a statistical test did not show it.
In conclusion, our results suggest that the rs402710 and rs401681 polymorphism at 5p13.33 may contribute to increased risk of lung cancer. However, additional large studies are needed to validate our findings. Moreover, gene–gene and gene–environment interactions should also be considered in future studies.
References
Ambrosone CB, Rao U, Michalek AM, Cummings KM, Mettlin CJ (1993) Lung cancer histologic types and family history of cancer. Analysis of histologic subtypes of 872 patients with primary lung cancer. Cancer 72:1192–1198
Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, Sullivan K, Matakidou A, Wang Y, Mills G, Doheny K, Tsai YY, Chen WV, Shete S, Spitz MR, Houlston RS (2008) Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet 40:616–622
Bae EY, Lee SY, Kang BK, Lee EJ, Choi YY, Kang HG, Choi JE, Jeon HS, Lee WK, Kam S, Shin KM, Jin G, Yoo SS, Lee J, Cha SI, Kim CH, Jung TH, Park JY (2012) Replication of results of genome-wide association studies on lung cancer susceptibility loci in a Korean population. Respirology 17:699–706
Begg CB, Mazumdar M (1994) Operating characteristics of a rank correlation test for publication bias. Biometrics 50:1088–1101
Chen XF, Cai S, Chen QG, Ni ZH, Tang JH, Xu DW, Wang XB (2012) Multiple variants of TERT and CLPTM1L constitute risk factors for lung adenocarcinoma. Genet Mol Res 11:370–378
Chen KJ, Fan F, Wang Y, Wei GT, Hu L, Xu F (2014) GSTT1 null genotype contributes to hepatocellular carcinoma risk: a meta-analysis. Tumour Biol 35:213–218
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7:177–188
Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634
Ferlay J, Autier P, Boniol M, Heanue M, Colombet M, Boyle P (2007) Estimates of the cancer incidence and mortality in Europe in 2006. Ann Oncol 18:581–592
Gabrielson E (2006) Worldwide trends in lung cancer pathology. Respirology 11:533–538
Gao Y, Goldstein AM, Consonni D, Pesatori AC, Wacholder S, Tucker MA, Caporaso NE, Goldin L, Landi MT (2009) Family history of cancer and nonmalignant lung diseases as risk factors for lung cancer. Int J Cancer 125:146–152
German J (1993) Bloom syndrome: a Mendelian prototype of somatic mutational disease. Medicine 72:393–406
Hsiung CA, Lan Q, Hong YC, Chen CJ, Hosgood HD, Chang IS, Chatterjee N, Brennan P, Wu C, Zheng W, Chang GC, Wu T, Park JY, Hsiao CF, Kim YH, Shen H, Seow A, Yeager M, Tsai YH, Kim YT, Chow WH, Guo H, Wang WC, Sung SW, Hu Z, Chen KY, Kim JH, Chen Y, Huang L, Lee KM, Lo YL, Gao YT, Kim JH, Liu L, Huang MS, Jung TH, Jin G, Caporaso N, Yu D, Kim CH, Su WC, Shu XO, Xu P, Kim IS, Chen YM, Ma H, Shen M, Cha SI, Tan W, Chang CH, Sung JS, Zhang M, Yang TY, Park KH, Yuenger J, Wang CL, Ryu JS, Xiang Y, Deng Q, Hutchinson A, Kim JS, Cai Q, Landi MT, Yu CJ, Park JY, Tucker M, Hung JY, Lin CC, Perng RP, Boffetta P, Chen CY, Chen KC, Yang SY, Hu CY, Chang CK, Fraumeni JF Jr, Chanock S, Yang PC, Rothman N, Lin D (2010) The 5p15.33 locus is associated with risk of lung adenocarcinoma in never-smoking females in Asia. PLoS Genet 6:e1001051
Hu Z, Wu C, Shi Y, Guo H, Zhao X, Yin Z, Yang L, Dai J, Hu L, Tan W, Li Z, Deng Q, Wang J, Wu W, Jin G, Jiang Y, Yu D, Zhou G, Chen H, Guan P, Chen Y, Shu Y, Xu L, Liu X, Liu L, Xu P, Han B, Bai C, Zhao Y, Zhang H, Yan Y, Ma H, Chen J, Chu M, Lu F, Zhang Z, Chen F, Wang X, Jin L, Lu J, Zhou B, Lu D, Wu T, Lin D, Shen H (2011) A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat Genet 43:792–796
Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, Chen C, Goodman G, Field JK, Liloglou T, Xinarianos G, Cassidy A, McLaughlin J, Liu G, Narod S, Krokan HE, Skorpen F, Elvestad MB, Hveem K, Vatten L, Linseisen J, Clavel-Chapelon F, Vineis P, Bueno-de-Mesquita HB, Lund E, Martinez C, Bingham S, Rasmuson T, Hainaut P, Riboli E, Ahrens W, Benhamou S, Lagiou P, Trichopoulos D, Holcátová I, Merletti F, Kjaerheim K, Agudo A, Macfarlane G, Talamini R, Simonato L, Lowry R, Conway DI, Znaor A, Healy C, Zelenika D, Boland A, Delepine M, Foglio M, Lechner D, Matsuda F, Blanche H, Gut I, Heath S, Lathrop M, Brennan P (2008) A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 452:633–637
Ito H, McKay JD, Hosono S, Hida T, Yatabe Y, Mitsudomi T, Brennan P, Tanaka H, Matsuo K (2012) Association between a genome-wide association study-identified locus and the risk of lung cancer in Japanese population. J Thorac Oncol 7:790–798
James MA, Wen W, Wang Y, Byers LA, Heymach JV, Coombes KR, Girard L, Minna J, You M (2012) Functional characterization of CLPTM1L as a lung cancer risk candidate gene in the 5p15.33 locus. PLoS ONE 7:e36116
Jaworowska E, Trubicka J, Lener MR, Masojć B, Złowocka-Perłowska E, McKay JD, Renard H, Oszutowska D, Wokołorczyk D, Lubiński J, Grodzki T, Serwatowski P, Nej-Wołosiak K, Tołoczko-Grabarek A, Sikorski A, Słojewski M, Jakubowska A, Cybulski C, Lubiński J, Scott RJ (2011) Smoking related cancers and loci at chromosomes 15q25, 5p15, 6p22.1 and 6p21.33 in the Polish population. PLoS One 6:e25057
Jiang M, Wu H, Qin C (2013) Genetic variant rs401681 at 5p15.33 modifies susceptibility to lung cancer but not esophageal squamous cell carcinoma. PLoS ONE 8:e84277
Jin G, Xu L, Shu Y, Tian T, Liang J, Xu Y, Wang F, Chen J, Dai J, Hu Z, Shen H (2009) Common genetic variants on 5p15.33 contribute to risk of lung adenocarcinoma in a Chinese population. Carcinogenesis 30:987–990
Johannsson O, Ostermeyer EA, Håkansson S, Friedman LS, Johansson U, Sellberg G, Brøndum-Nielsen K, Sele V, Olsson H, King MC, Borg A (1996) Founding BRCA1 mutations in hereditary breast and ovarian cancer in southern Sweden. Am J Hum Genet 58:441–450
Ke J, Zhong R, Zhang T, Liu L, Rui R, Shen N, Sun Y, Liu L, Cheng L, Miao XP (2013) Replication study in Chinese population and meta-analysis supports association of the 5p15.33 locus with lung cancer. PLoS One 8:e62485
Lan Q, Hsiung CA, Matsuo K, Hong YC, Seow A, Wang Z, Hosgood HD 3rd, Chen K, Wang JC, Chatterjee N, Hu W, Wong MP, Zheng W, Caporaso N, Park JY, Chen CJ, Kim YH, Kim YT, Landi MT, Shen H, Lawrence C, Burdett L, Yeager M, Yuenger J, Jacobs KB, Chang IS, Mitsudomi T, Kim HN, Chang GC, Bassig BA, Tucker M, Wei F, Yin Z, Wu C, An SJ, Qian B, Lee VH, Lu D, Liu J, Jeon HS, Hsiao CF, Sung JS, Kim JH, Gao YT, Tsai YH, Jung YJ, Guo H, Hu Z, Hutchinson A, Wang WC, Klein R, Chung CC, Oh IJ, Chen KY, Berndt SI, He X, Wu W, Chang J, Zhang XC, Huang MS, Zheng H, Wang J, Zhao X, Li Y, Choi JE, Su WC, Park KH, Sung SW, Shu XO, Chen YM, Liu L, Kang CH, Hu L, Chen CH, Pao W, Kim YC, Yang TY, Xu J, Guan P, Tan W, Su J, Wang CL, Li H, Sihoe AD, Zhao Z, Chen Y, Choi YY, Hung JY, Kim JS, Yoon HI, Cai Q, Lin CC, Park IK, Xu P, Dong J, Kim C, He Q, Perng RP, Kohno T, Kweon SS, Chen CY, Vermeulen R, Wu J, Lim WY, Chen KC, Chow WH, Ji BT, Chan JK, Chu M, Li YJ, Yokota J, Li J, Chen H, Xiang YB, Yu CJ, Kunitoh H, Wu G, Jin L, Lo YL, Shiraishi K, Chen YH, Lin HC, Wu T, Wu YL, Yang PC, Zhou B, Shin MH, Fraumeni JF Jr, Lin D, Chanock SJ, Rothman N (2012) Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat Genet 44:1330–1335
Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, Mirabello L, Jacobs K, Wheeler W, Yeager M, Bergen AW, Li Q, Consonni D, Pesatori AC, Wacholder S, Thun M, Diver R, Oken M, Virtamo J, Albanes D, Wang Z, Burdette L, Doheny KF, Pugh EW, Laurie C, Brennan P, Hung R, Gaborieau V, McKay JD, Lathrop M, McLaughlin J, Wang Y, Tsao MS, Spitz MR, Wang Y, Krokan H, Vatten L, Skorpen F, Arnesen E, Benhamou S, Bouchard C, Metspalu A, Vooder T, Nelis M, Välk K, Field JK, Chen C, Goodman G, Sulem P, Thorleifsson G, Rafnar T, Eisen T, Sauter W, Rosenberger A, Bickeböller H, Risch A, Chang-Claude J, Wichmann HE, Stefansson K, Houlston R, Amos CI, Fraumeni JF Jr, Savage SA, Bertazzi PA, Tucker MA, Chanock S, Caporaso NE (2009) A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet 85:679–691
Li FP, Fraumeni JF Jr, Mulvihill JJ, Blattner WA, Dreyfus MG, Tucker MA, Miller RW (1988) A cancer family syndrome in twenty-four kindreds. Cancer Res 48:5358–5362
Li DW, Collier DA, He L (2006) Meta-analysis shows strong positive association of the neuregulin 1 (NRG1) gene with schizophrenia. Hum Mol Genet 15:1995–2002
Li C, Yin Z, Wu W, Li X, Ren Y, Zhou B (2013) Genetic Variations in TERT-CLPTM1L Genes and Risk of Lung Cancer in Chinese Women Nonsmokers. PLoS ONE 8:e64988
Lu X, Ke J, Luo X, Zhu Y, Zou L, Li H, Zhu B, Xiong Z, Chen W, Deng L, Lou J, Wang X, Zhang Y, Wang Z, Miao X, Cheng L (2013) The SNP rs402710 in 5p15.33 is associated with lung cancer risk: a replication study in Chinese population and a meta-analysis. PLoS ONE 8:e76252
Matakidou A, Eisen T, Houlston RS (2005) Systematic review of the relationship between family history and lung cancer risk. Br J Cancer 93:825–833
McKay JD, Hung RJ, Gaborieau V, Boffetta P, Chabrier A, Byrnes G, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, McLaughlin J, Shepherd F, Montpetit A, Narod S, Krokan HE, Skorpen F, Elvestad MB, Vatten L, Njølstad I, Axelsson T, Chen C, Goodman G, Barnett M, Loomis MM, Lubiñski J, Matyjasik J, Lener M, Oszutowska D, Field J, Liloglou T, Xinarianos G, Cassidy A, EPIC Study, Vineis P, Clavel-Chapelon F, Palli D, Tumino R, Krogh V, Panico S, González CA, Ramón Quirós J, Martínez C, Navarro C, Ardanaz E, Larrañaga N, Kham KT, Key T, Bueno-de-Mesquita HB, Peeters PH, Trichopoulou A, Linseisen J, Boeing H, Hallmans G, Overvad K, Tjønneland A, Kumle M, Riboli E, Zelenika D, Boland A, Delepine M, Foglio M, Lechner D, Matsuda F, Blanche H, Gut I, Heath S, Lathrop M, Brennan P (2008) Lung cancer susceptibility locus at 5p15.33. Nat Genet 40:1404–1406
Miki D, Kubo M, Takahashi A, Yoon KA, Kim J, Lee GK, Zo JI, Lee JS, Hosono N, Morizono T, Tsunoda T, Kamatani N, Chayama K, Takahashi T, Inazawa J, Nakamura Y, Daigo Y (2010) Variation in TP63 is associated with lung adenocarcinoma susceptibility in Japanese and Korean populations. Nat Genet 42:893–896
Myneni AA, Chang SC, Niu R, Liu L, Ochs-Balcom HM, Li Y, Zhang C, Zhao B, Shi J, Han X, Li J, Su J, Cai L, Yu S, Zhang ZF, Mu L (2013) Genetic polymorphisms of TERT and CLPTM1L and risk of lung cancer—a case–control study in a Chinese population. Lung Cancer 80:131–137
Pande M, Spitz MR, Wu X, Gorlov IP, Chen WV, Amos CI (2011) Novel genetic variants in the chromosome 5p15.33 region associate with lung cancer risk. Carcinogenesis 32:1493–1499
Rafnar T, Sulem P, Stacey SN, Geller F, Gudmundsson J, Sigurdsson A, Jakobsdottir M, Helgadottir H, Thorlacius S, Aben KK, Blöndal T, Thorgeirsson TE, Thorleifsson G, Kristjansson K, Thorisdottir K, Ragnarsson R, Sigurgeirsson B, Skuladottir H, Gudbjartsson T, Isaksson HJ, Einarsson GV, Benediktsdottir KR, Agnarsson BA, Olafsson K, Salvarsdottir A, Bjarnason H, Asgeirsdottir M, Kristinsson KT, Matthiasdottir S, Sveinsdottir SG, Polidoro S, Höiom V, Botella-Estrada R, Hemminki K, Rudnai P, Bishop DT, Campagna M, Kellen E, Zeegers MP, de Verdier P, Ferrer A, Isla D, Vidal MJ, Andres R, Saez B, Juberias P, Banzo J, Navarrete S, Tres A, Kan D, Lindblom A, Gurzau E, Koppova K, de Vegt F, Schalken JA, van der Heijden HF, Smit HJ, Termeer RA, Oosterwijk E, van Hooij O, Nagore E, Porru S, Steineck G, Hansson J, Buntinx F, Catalona WJ, Matullo G, Vineis P, Kiltie AE, Mayordomo JI, Kumar R, Kiemeney LA, Frigge ML, Jonsson T, Saemundsson H, Barkardottir RB, Jonsson E, Jonsson S, Olafsson JH, Gulcher JR, Masson G, Gudbjartsson DF, Kong A, Thorsteinsdottir U, Stefansson K (2009) Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet 41:221–227
Ruano-Ravina A, Figueiras A, Barros-Dios JM (2003) Lung cancer and related risk factors: an update of the literature. Public Health 117:149–156
Sanders BM, Jay M, Draper GJ, Roberts EM (1989) Non-ocular cancer in relatives of retinoblastoma patients. Br J Cancer 60:358–365
Shiraishi K, Kunitoh H, Daigo Y, Takahashi A, Goto K, Sakamoto H, Ohnami S, Shimada Y, Ashikawa K, Saito A, Watanabe S, Tsuta K, Kamatani N, Yoshida T, Nakamura Y, Yokota J, Kubo M, Kohno T (2012) A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nat Genet 44:900–903
Strong LC, Herson J, Haas C, Elder K, Chakraborty R, Weiss KM, Majumder P (1984) Cancer mortality in relatives of retinoblastoma patients. J Natl Cancer Inst 73:303–311
TanTai J, Shen Y, Zhao H (2014) Quantitative assessment of the influence of common variations on 6p21 and lung cancer risk. Tumour Biol 35:689–694
Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, Stacey SN, Bergthorsson JT, Thorlacius S, Gudmundsson J, Jonsson T, Jakobsdottir M, Saemundsdottir J, Olafsdottir O, Gudmundsson LJ, Bjornsdottir G, Kristjansson K, Skuladottir H, Isaksson HJ, Gudbjartsson T, Jones GT, Mueller T, Gottsäter A, Flex A, Aben KK, de Vegt F, Mulders PF, Isla D, Vidal MJ, Asin L, Saez B, Murillo L, Blondal T, Kolbeinsson H, Stefansson JG, Hansdottir I, Runarsdottir V, Pola R, Lindblad B, van Rij AM, Dieplinger B, Haltmayer M, Mayordomo JI, Kiemeney LA, Matthiasson SE, Oskarsson H, Tyrfingsson T, Gudbjartsson DF, Gulcher JR, Jonsson S, Thorsteinsdottir U, Kong A, Stefansson K (2008) A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 452:638–642
Truong T, Hung RJ, Amos CI, Wu X, Bickeböller H, Rosenberger A, Sauter W, Illig T, Wichmann HE, Risch A, Dienemann H, Kaaks R, Yang P, Jiang R, Wiencke JK, Wrensch M, Hansen H, Kelsey KT, Matsuo K, Tajima K, Schwartz AG, Wenzlaff A, Seow A, Ying C, Staratschek-Jox A, Nürnberg P, Stoelben E, Wolf J, Lazarus P, Muscat JE, Gallagher CJ, Zienolddiny S, Haugen A, van der Heijden HF, Kiemeney LA, Isla D, Mayordomo JI, Rafnar T, Stefansson K, Zhang ZF, Chang SC, Kim JH, Hong YC, Duell EJ, Andrew AS, Lejbkowicz F, Rennert G, Müller H, Brenner H, Le Marchand L, Benhamou S, Bouchardy C, Teare MD, Xue X, McLaughlin J, Liu G, McKay JD, Brennan P, Spitz MR (2010) Replication of lung cancer susceptibility loci at chromosomes 15q25, 5p15, and 6p21: a pooled analysis from the International Lung Cancer Consortium. J Natl Cancer Inst 102:959–971
Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan J, Matakidou A, Qureshi M, Dong Q, Gu X, Chen WV, Spitz MR, Eisen T, Amos CI, Houlston RS (2008) Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet 40:1407–1409
Wang H, Zhao Y, Ma J, Zhang G, Mu Y, Qi G, Fang Z, Wang L, Fan Q, Ma Z (2013) The genetic variant rs401681C/T is associated with the risk of non-small cell lung cancer in a Chinese mainland population. Genet Mol Res 12:67–73
Woolf B (1955) On estimating the relation between blood group and disease. Ann Hum Genet 19:251–253
Yamamoto K, Okamoto A, Isonishi S, Ochiai K, Ohtake Y (2001) A novel gene, CRR9, which was up-regulated in CDDP-resistant ovarian tumor cell line, was associated with apoptosis. Biochem Biophys Res Commun 280:1148–1154
Yang P, Li Y, Jiang R, Cunningham JM, Li Y, Zhang F, de Andrade M (2010) A rigorous and comprehensive validation: common genetic variations and lung cancer. Cancer Epidemiol Biomarkers Prev 19:240–244
Yoon KA, Park JH, Han J, Park S, Lee GK, Han JY, Zo JI, Kim J, Lee JE, Takahashi A, Kubo M, Nakamura Y, Lee JS (2010) A genome-wide association study reveals susceptibility variants for non-small cell lung cancer in the Korean population. Hum Mol Genet 19:4948–4954
Young RP, Hopkins RJ, Whittington CF, Hay BA, Epton MJ, Gamble GD (2011) Individual and cumulative effects of GWAS susceptibility loci in lung cancer: associations after sub-phenotyping for COPD. PLoS ONE 6:e16476
Zhao Z, Li C, Yang L, Zhang X, Zhao X, Song X, Li X, Wang J, Qian J, Yang Y, Jin L, Chen H, Lu D (2013) Significant association of 5p15.33 (TERT-CLPTM1L genes) with lung cancer in Chinese Han population. Exp Lung Res 39:91–98
Zienolddiny S, Skaug V, Landvik NE, Ryberg D, Phillips DH, Houlston R, Haugen A (2009) The TERT-CLPTM1L lung cancer susceptibility variant associates with higher DNA adduct formation in the lung. Carcinogenesis 30:1368–1371
Acknowledgments
This work was supported by National Nature Science Foundation of China (81001036) and Shanghai Municipal Commission of Health and Family Planning (2010Y066).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, Dp., Yang, Cl., Zhou, X. et al. Association between CLPTM1L polymorphisms (rs402710 and rs401681) and lung cancer susceptibility: evidence from 27 case–control studies. Mol Genet Genomics 289, 1001–1012 (2014). https://doi.org/10.1007/s00438-014-0868-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-014-0868-7