Abstract
Aspirin-exacerbated respiratory disease (AERD) is associated with severe asthma and aspirin can cause asthma to worsen, often in the form of a severe and sudden attack. The oral aspirin challenge is the gold standard to confirm the diagnosis of AERD, but it is time consuming and produces serious complications in some cases. Therefore, more efficient and practical method is needed to predict AERD patients. The aim of the present study was to identify AERD-related gene expression in peripheral blood mononuclear cells (PBMCs) and examine the diagnostic potential of these candidate gene(s) for predicting AERD. To do this, RNAs from 24 subjects with AERD and 18 subjects with aspirin-tolerant asthma (ATA) were subjected to microarray analysis of ~34,560 genes. In total, 10 genes were selected as candidate gene markers by applying p ≤ 0.001(t test) and ≥8-fold change, and to correct for multiple comparisons, the false discovery rate analyses were performed. By applying multiple logistic regression analysis, among possible 1,023 models (210–1), a model consisting of CNKSR3, SPTBN2, and IMPACT was selected as candidate set, because this set showed the best AUC (0.98) with 88 % sensitivity and 89 % specificity. For validation, mRNA levels by real-time PCR on PBMCs from two population sets in a gene-chip study and another replication sample, 20 AERD, 20 ATA, and 8 normal controls, were significantly different between groups with 100 % sensitivity and 100 % specificity in each of the two population sets. However, IMPACT gene did not differentiate between AERD and normal controls. The set of the two genes (CNKSR3 and SPTBN2) showed the best AUC (0.96) with 88 % sensitivity and 94 % specificity in a gene-chip study sample. In addition, this set showed perfect discriminative power with AUC (1.0, 100 % sensitivity and 100 % specificity) in each of the two population sets: the gene-chip samples and the replication samples. It also showed perfect discrimination for AERD from NC (AUC: 1.0) and ATA from NC (AUC: 1.0). In conclusion, we developed the two gene markers (CNKSR3 and SPTBN2) of PBMC which differentiate between AERD and ATA with a perfect discriminative power. These gene markers may be an efficient and practical method for predicting AERD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aspirin-exacerbated respiratory disease (AERD) refers to the aggressive mucosal inflammatory disease combined with precipitation of asthma and rhinitis attacks which occurs after ingestion of aspirin or most non-steroidal anti-inflammatory drugs. In 5–10 % of persons with asthma, aspirin can cause asthma to worsen, often in the form of a severe and sudden attack. The oral aspirin challenge (OAC) is the gold standard to confirm the diagnosis of AERD, but it is a time-consuming procedure and produces serious complications in some cases. So, the more effective and safe prediction method is needed.
This syndrome is characterized by aspirin hypersensitivity, asthma, and chronic rhinosinusitis with nasal polyposis (Samter and Beers 1967). Aspirin has been widely used for its antipyretic, anti-inflammatory, and antithrombotic properties. Both beneficial and detrimental effects are mediated primarily through the inhibition of COX-1 and/or COX-2 (Lee and Stevenson 2011). A two-compartment model has been proposed, in which both augmentation of cysteinyl leukotriene (CysLT) production and overexpression of the CysLT receptor on inflammatory cells occur within the respiratory tract (Sousa et al. 2002). The CysLT receptor is selectively antagonized by several currently available leukotriene modifiers, including montelukast, pranlukast, and zafirlukast (Leff 2001). However, clinical studies have demonstrated that the response to these medications is incomplete (Szefler et al. 2005), suggesting the presence of alternative pathways leading to AERD.
One possibility is that acetylsalicylic acid (ASA) has other direct effects on the immune and inflammatory mediators of critical inflammatory cells. ASA inhibits activation of the transcription factor NFκB (Kopp and Ghosh 1994), regulates IL4 expression by altering the availability of transcription factors to the regulatory elements in the IL4 promoter (Kim et al. 2010a, b, c), and inhibits IL4- and IL13-induced activation of STAT6 (Perez-G et al. 2002). Moreover, associations with AERD have been demonstrated in several immune and inflammatory genes other than those in arachidonate pathways (Kim et al. 2010a, b, c, 2011; Pasaje et al. 2011; Lee et al. 2010a, b; Oh et al. 2009). These observations support the idea that the multiple effects of ASA are derived from its ability to regulate a network of biochemical and cellular events that are more complex than was initially thought.
To search for disease-related gene expression profiles, whole-genome gene expression microarrays have been used. Several types of tissues have been used for human microarray studies of asthma, such as bronchial epithelium, nasal mucosa, and airway inflammatory cells from both patients and healthy subjects (Lilly et al. 2005; Gelb et al. 2010; Chamberland et al. 2009; Baines et al. 2009; Guajardo et al. 2005). Although the analysis of gene expression in the airway can provide more exact information, obtaining airway tissues is not easy, especially in cases of severe asthma (Kavuru et al. 1999). Thus, peripheral blood mononuclear cells (PBMCs) have been used as an alternative (Hanselb et al. 2005; Aoki et al. 2009; Subrata et al. 2009).
Over the past decade, diagnoses of multifactorial complex diseases and their subphenotypes have been attempted using gene expression profile analysis. Although the diagnostic value of the expression of each gene alone is regarded as minimal, multigene analysis can increase predictability and potentially be used as a biomarker.
In the present study, we identified candidate genes affecting AERD using a gene expression chip. In addition, we evaluated the diagnostic value of mRNA levels of these candidate genes in the differentiation of AERD from aspirin-tolerant asthma (ATA), through additional analysis of the area under the receiver operating characteristic curve (AUC–ROC), sensitivity, and specificity (Kraft et al. 2009).
Methods
Study subjects
Subjects were recruited from Soonchunhyang University Hospital, South Korea. All subjects were Korean. Asthma was diagnosed by physicians in accordance with the definition of Global Initiative for Asthma (GINA) guidelines (Bateman et al. 2008). The details of the laboratory tests for airway reversibility, skin prick test, and oral aspirin challenge are described in an online supplement.
Oral aspirin challenge reactions were categorized into two groups as follows: (1) ≥15 % decrease in FEV1 and/or naso-ocular reactions (AERD) or (2) <15 % decrease in FEV1 without naso-ocular or cutaneous reactions (ATA). Peripheral venous blood was collected before aspirin challenge. PBMCs were separated using Ficoll–Hypaque solution. The clinical profiles of asthma patients and healthy control subjects are summarized in Table 1.
The institutional review board (SCHBC_IRB_06_05) for human studies of Soonchunhyang University Hospital approved the protocol, and written informed consent was obtained from all subjects.
cDNA synthesis and microarray hybridization
A human oligonucleotide microarray (Genomictree, Korea) containing 34,560 oligonucleotide probes was used for gene expression analysis. Synthesis of target cDNA probes and hybridization were performed according to a previously described protocol (Yang et al. 2003) and method E1 in the online repository.
Real-time PCR for candidate genes
For three genes, CNKSR family member 3 (CNKSR3), spectrin, beta, non-erythrocytic 2 (SPTBN2), and impact homolog (IMPACT), real-time PCR was performed as described in the online repository (see method E1).
Using the 2−ΔΔCt method, the data are presented as the fold change in gene expression normalized to an endogenous reference gene (β-actin, GAPDH) and relative to a control (CNKSR3-AIA, SPTBN2-AIA, and IMPACT-AIA). The fold differences in CNKSR3, SPTBN2, and IMPACT mRNA expression in each patient were also calculated by comparing their 2−ΔΔCt.
Statistical methods
For microarray analysis, background-corrected values for each probe on oligonucleotide array were extracted using GenePix Pro 4.0 software (Molecular Devices). Detection of p values and normalization were performed for the extracted values. Statistical significance of the microarray data was calculated by the t test using R (ver. 2.8.1; http://www.r-project.org/). In addition, other statistical analysis was performed using the false discovery rate (FDR) to correct for multiple comparisons in multiple hypotheses testing (Korbinian Strimmer 2008) using R. The detailed process is described in the statistical methods section of the supplementary material (see method E1 in the online repository).
For the diagnostic values of gene expression in the discrimination of AERD from ATA, we selected candidate genes that satisfied the criteria of p < 0.001 and a fold-change value of ≥8-fold between the two groups. After filtering with the p value and fold-change value, a power set was made with the 10 candidate genes and multiple logistic regression (MLR) analysis was performed with models in the power set. ROC curves for all models were obtained, and AUCs were calculated (Bradley 1997; Fawcett 2006). For the analysis of mRNA levels of real-time PCR, we also applied the same t test method.
Gene ontology and pathway analyses
Gene ontology analysis, which is used to determine the biological process, molecular function, and molecular class of a gene or the tendencies of selected genes, was performed using the David Database (http://david.abcc.ncifcrf.gov/) (Huang et al. 2009). To investigate whether differentially expressed genes belonged to specific pathways, the KEGG pathway database was used (Kanehisa and Goto 1999).
Results
Comparison of gene expression between AERD and ATA
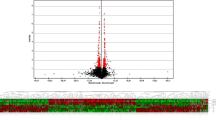
To identify genes that may be related to AERD, we applied high-throughput gene expression microarray consisting of 34,560 featured genes to RNA samples obtained from ATA (n = 18) and AERD (n = 24) subjects. The overall research approach is depicted in Figure E1 in the online repository. To evaluate the overall difference in gene expression levels in PBMCs between AERD and ATA, we calculated the gene expression level using a volcano plot. We selected genes with a p value of <0.01 and a ≥2-fold change. Volcano plots of significance against fold-change values for each gene revealed that the expression levels were quite different between AERD and ATA (Fig. 1a), and we identified 248 genes that showed a significant increase in gene expression and 70 genes that showed a significant decrease in AERD versus ATA. We also performed the FDR analysis. All q values of the selected 318 genes were <0.05 (see Table E1 in the online repository). For the selected 10 genes, q values of 9 genes were <0.007 and that of one gene was <0.01 (HMGCS2). In our experiment, the order of genes sorted by p value was the same as that of genes sorted by q value (value of the Pearson’s product-moment correlation: 0.97, p value < 2.2e−16). The heat map of these differentially expressed genes revealed distinctive RNA expression profiles of PBMCs (Fig. 1a). The upregulated and downregulated genes were perfectly classified by the hierarchical clustering method.
Gene ontology and pathway analyses
We determined the tendency of the 318 genes listed in Table E2 in the online repository, using gene ontology (Table 2) and pathway (Table 3) analyses. We searched biological processes using the David Database and searched pathways using the KEGG pathway Database. In 318 genes, PLA2G10 and GGT6 were found related to arachidonic acid pathway which was known to be related to AERD. PLA2G10 was upregulated (fold change: 2.4, p value: 0.0004) and GGT6 was downregulated (fold change: 0.28, p value: 0.009) (Figure E2 in the online repository). For top 10 genes, only 3 genes had pathway information (TCF7L1, CNKSR3 and HMGCS2) but for other 7 genes, we could not found information for arachidonic acid pathway, asthma or aspirin.
Selection of the best model for predicting AERD using MLR analysis and ROC curve analysis of candidate genes
Since 318 genes are too numerous for the selection of gene markers, we applied the more stringent criteria of p ≤ 0.001 and a fold change of ≥8 to differentiate gene expression between AERD and ATA. Consequently, we identified 10 genes (Fig. 1b; Table 4). Gene expression of CNKSR3, HMGCS2, SPTBN2, and PPP1R15A was upregulated in the PBMCs of the AERD groups, whereas that of DCST2, IMPACT, MLSTD1, WFDC9, SPEF2, CCDC15, and 7CF7L1 was downregulated in the ATA groups.
Using the 10 genes as candidate gene markers, we performed a MLR analysis for all elements of the power set of candidate genes and measured the 1,023 AUCs of ROC curves to select AERD-specific diagnostic marker gene(s) to discriminate from ATA. First, we made 1,023 (210−1) models for all genes by MLR and measured all p values of the coefficients of genes for each model. We separated the 1,023 models into 10 groups (Groups 1–10). Group n indicates the models that were made of the number of genes for MLR analysis. Among the 1,023 models, only 81 models satisfied our criteria: all p values of coefficients for genes <0.05 of each model. All data for AUCs and p values are presented in the supplementary table (see Table E2 in the online repository). As the number of analyzed genes increased, the p value of coefficients decreased (Figure E3 in the online repository). Only the models made of <3 genes showed the average log of p values of <0.05 (Figure E3 in the online repository).
AUCs of ROCs were calculated for Groups 1, 2, and 3. The top five AUC values of each group, consisting of 1, 2, or 3 genes, respectively, are presented in Fig. 2a. Combinations of three genes showed better AUC values than one or two genes. One of the best AUC values was observed for the combination of the three genes, PPP2R15A, SPEF2, and TCF7L1 (AUC: 0.9907), and their ROC curves (Fig. 2).
Validation of gene expression with quantitative measurement of mRNA using real-time PCR
We validated the best model consisting of the three genes CNKSR3, SPTBN2, and IMPACT using real-time PCR to measure their mRNA levels in PBMCs obtained from AERD (n = 22) and ATA (n = 18), using gene-chip analysis. Quantification of CNKSR3, SPTBN2, and IMPACT mRNA levels was performed by measuring the amount of the CNKSR3, SPTBN2, and IMPACT real-time PCR product after correcting for the amount of β-actin and GAPDH. The expression levels of CNKSR3 and SPTBN2 mRNA in PBMCs were significantly higher in AERD than in ATA subjects, but that of IMPACT was significantly higher in ATA subjects (Fig. 3).
Using other replication samples that were not included in the gene-chip samples, we also performed real-time PCR to validate the result of a previous experiment. PBMCs were obtained from 20 subjects with AERD, 20 with ATA, and 8 normal controls (NCs). Their clinical data are presented in Table 1. The expression levels of CNKSR3 and SPTBN2 mRNA in PBMCs were also significantly higher in subjects with AERD than in those with ATA, while those of IMPACT were significantly higher in the latter than in the former (Fig. 4). As IMPACT gene did not clearly differentiate between AERD and NCs, we excluded IMPACT gene from the set of gene markers.
Diagnostic accuracy of the combination of CNKSR3 and SPTBN2 gene levels for predicting AERD
To evaluate the diagnostic accuracy of the combination of CNKSR3 and SPTBN2, we calculated sensitivity and specificity using a contingency table of 24 AERD and 18 ATA gene-chips data, which resulted in values of 0.88 and 0.94, respectively, with an AUC of 0.96.
For real-time PCR data, the MLR analysis was performed for the two population sets: the gene-chip samples and the replication samples. In addition, to evaluate diagnostic accuracy of a set consisting of the two gene markers between AERD and NC, between ATA and NC, the MLR analysis was also performed. The sensitivity and selectivity are summarized in Tables 5 and 6. This set showed perfect discriminative power with AUC (1.0, 100 % sensitivity and 100 % specificity) in each of the two population sets: the gene-chip samples and the replication samples. It also showed perfect discrimination for AERD from NC (AUC: 1.0) and ATA from NC (AUC: 1.0) in the replication samples.
Discussion
Acetylsalicylic acid is the medication used most commonly for pain control and for the prophylaxis of coronary artery disease and other vascular diseases (Gollapudi et al. 2004). However, AERD remains widely underdiagnosed in the asthmatic population due to insufficient awareness of the relationship between ASA ingestion and asthma exacerbation. In a multicenter study in 10 European countries of 500 patients, 15 % of the patients were unaware of their intolerance to aspirin and learned about it only after having provocation tests performed (Szczeklik et al. 2000). In addition, familial history of intolerance to aspirin was reported by only 1 % in the pan-European study (Szczeklik et al. 2000) and only 3 % in the US study (Berges-Gimeno et al. 2002).
A fairly limited number of epidemiological studies have estimated the prevalence of AERD. Estimation of the prevalence of aspirin hypersensitivity in adult asthmatic patients was reported to depend on clinical history alone or on challenge with ASA. Therefore, the identification of aspirin hypersensitivity, especially in patients who are unaware that they are hypersensitive, is essential to avoid serious complications. The diagnosis can be established with certainty, only with provocation tests using increasing doses of ASA. There are four commonly used provocation tests, with different routes of ASA administration: oral, bronchial (inhaled), intravenous, and nasal (Szczeklik and Nizankowska 2000). Although OAC is the gold standard to confirm the diagnosis, the OAC is a time-consuming procedure and produces serious complications in some cases. Therefore, the development of non-invasive methods is necessary to facilitate diagnosis while preventing the unexpected complications of aspirin use in susceptible patients.
In this study, we identified genes related to AERD using microarray analysis of PBMCs. We found that a combination of two genes, namely CNKSR3 and SPTBN2, showed the best power for predicting AERD. To the best of our knowledge, this is the first reported attempt to select disease markers for AERD using gene expression in PBMCs. In the microarray analysis of PBMCs, when a p value threshold of <0.01 and a fold change of ≥2 were applied, a total of 318 genes were selected. We also performed the FDR analysis. All 318 genes had a q value <0.05. This means that 318 genes are almost significant and only about 16 genes might be the false positives.
Since 318 genes are too numerous for the development of gene markers, we narrowed down the number of candidate gene markers by applying more stringent criteria (p < 0.001 and ≥8-fold change). Finally, 10 candidate genes were selected and all q values of the FDR analysis were <0.07 except a gene (HMGCS2). This means that the selected 10 candidate genes are true positive significantly. Since the complex dependencies between the 10 selected genes were unknown (Kruskal and Tanur 1978), we made all 1,023 possible models using the 10 genes and measured the p values to examine the validity of the models. Interestingly, in only 80 of the 1,023 models were all of the p values of the coefficients <0.05. So, we compared AUC values for only genes or combinations of genes with p values of <0.05. After performing MLR analysis, the best model (PPP1R15A, SPEF2, and TCF7L1) and the second best model (CNKSR3, IMPACT, and SPTBN2) showed almost perfect discrimination.
We measured the mRNA levels of the selected PPP1R15A, SPEF2, and TCF7L1 genes using real-time PCR with gene-chip samples, but the results of two genes (SPEF2 and TCF7L1) were not compatible with the results of gene-chip data (data not shown). Thus, we chose the second combinational model of three genes (CNKSR3, IMPACT, and SPTBN2), for which the AUC values of the two models were 0.99 and 0.98, respectively, in the gene-chip data. Considering that an AUC value of 1 indicates perfect classification, these values indicate a very high predictability of the markers.
When CNKSR3, IMPACT, and SPTBN2 gene were used as candidate genes, the AUC of ROC curve was ‘1.0’. This means that three genes have enough discriminative power AERD from ATA. We also performed real-time PCR with another replication sample set (including NC samples). In Fig. 4, IMPACT gene did not differentiate between AERD and NC well (p value: 0.03). So, the IMPACT gene was excluded from candidate gene set. When we performed real-time PCR for functional validation of the two selected genes, significant p values for each gene were obtained. In addition, the combination of the mRNA levels of the two genes showed perfect accuracy for diagnosing AERD from ATA (Table 5). The two genes also showed perfect discrimination for diagnosing AERD from NC, and diagnosing ATA from NC (Table 6). Since the real-time PCR method is cheaper and faster than a gene-chip, the measurement of the mRNA levels of these two genes may be a good genetic biomarker.
The approach of selecting gene markers using ROC curves has been applied to the selection of biomarkers for other diseases, such as gastric cancer, knee osteoarthritis, and lung cancer (Chong et al. 2010; Peat et al. 2010; Rom et al. 2010). However, to the best of our knowledge, this approach has not been previously applied in the study of AERD for genetic markers.
In previous studies, the most upregulated and/or downregulated genes have been selected and used as candidates, followed by logistic regression analysis and measurement of the AUC–ROC curve (Zhao et al. 2010; Kalady et al. 2010). However, such models might not show the best discriminative performance as disease markers; models that consisted of >2 genes showed a more discriminative performance, as shown in Fig. 2. In contrast, we selected several candidate genes that satisfied our criteria. We applied MLR analysis for all possible 2n−1 models and selected the model that has an AUC that was comparable to that of the top model among all models. In our experiment, a model consisting of three genes showed the highest performance.
To date, the functions of these genes used as diagnostic markers for AERD in the present study have not been revealed in AERD, which is a limitation of our study. CNKSR3 is known to be a direct mineralocorticoid receptor target gene that plays a key role in the regulation of the epithelial sodium channel (Ziera et al. 2009). SPTBN2 regulates the glutamate-signaling pathway by stabilizing the glutamate transporter EAAT4 at the surface of the plasma membrane. Mutations in this gene cause a form of spinocerebellar ataxia, SCA5, characterized by neurodegeneration, progressive locomotor incoordination, dysarthria, and uncoordinated eye movements (Perkins et al. 2010). SPTBN2 also plays a critical role in hepatocyte proliferation and transitional phenotypes in TGF-β signaling (Thenappan et al. 2010). Although we could found two genes for predicting AERD from ATA, we could not found the functional relationship of the two genes to asthma and aspirin. However, the new candidate genes highlight the broad scope of the pathogenesis behind AERD.
In previous AERD-genotyping studies, although the aspirin hypersensitivity in two ethically distant population of Slavonics (Dekker et al. 1997) and Korean (Choi et al. 2004) had highly increased frequency of HLA-DPB1 locus allele*0301, in our gene-chip data, the expression value of HLA-DPB1 did not show significant difference (fold change: 1.03, p value: 0.699).
In summary, a ~34,560 gene, genome-wide mRNA expression study of PBMCs from AERD and ATA subjects demonstrated differential levels of expression of 348 genes using criteria of p ≤ 0.01 and a fold change of ≥2. Applying more stringent criteria of p ≤ 0.001 and a fold change of ≥8, 10 genes were selected as candidate gene markers for AERD and ATA. Using AUC–ROC curves, we identified a genetic marker model consisting of CNKSR3, IMPACT and SPTBN2 genes. This diagnostic value was validated with quantitative measurement of mRNA using real-time PCR in a replication population. The IMPACT gene was excluded because of weak discriminative power and the CNKSR and SPTBN2 were selected finally. This combination of gene markers showed perfect classification.
In conclusion, we identified the two gene markers (CNKSR3 and SPTBN2) which differentiate between AERD and ATA with a perfect discriminative power. These gene markers also differentiate well between AERD and NC (AUC: 1), and between ATA and NC (AUC: 1). Because of obtaining airway tissues is not easy, especially in cases of severe asthma, these gene markers from the PBMC may be diagnostically useful for predicting AERD.
References
Aoki T, Matsumoto Y, Hirata K, Ochiai K, Okada M, Ichikawa K, Shibasaki M, Arinami T, Sumazaki R, Noguchi E (2009) Expression profiling of genes related to asthma exacerbations. Clin Exp Allergy 39(2):213–221
Baines KJ, Wood LG, Gibson PG (2009) The nutrigenomics of asthma: molecular mechanisms of airway neutrophilia following dietary antioxidant withdrawal. OMICS 13(5):355–365
Bateman ED, Hurd SS, Barnes PJ, Bousquet J, Drazen JM, FitzGerald M, Gibson P, Ohta K, O’Byrne P, Pedersen SE, Pizzichini E, Sullivan SD, Wenzel SE, Zar HJ (2008) Global strategy for asthma management and prevention: GINA executive summary. Eur Respir J 31:143–178
Berges-Gimeno MP, Simon RA, Stevenson DD (2002) The natural history and clinical characteristics of aspirin-exacerbated respiratory disease. Ann Allergy Asthma Immunol 89:474
Bradley AP (1997) The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recogn 30(7):1145–1159
Chamberland A, Madore AM, Tremblay K, Laviolette M, Laprise C (2009) A comparison of two sets of microarray experiments to define allergic asthma expression pattern. Exp Lung Res 35(5):399–410
Choi JH, Lee KW, Oh HB, Lee KJ, Suh YJ, Park CS, Park HS (2004) HLA association in aspirin-intolerant asthma: DPB1*0301 as a strong marker in a Korean population. J Allergy Clin Immunol 113(3):562–564
Chong PK, Lee H, Zhou J, Liu SC, Loh MC, Wang TT, Chan SP, Smoot DT, Ashktorab H, So JB, Lim KH, Yeoh KG, Lim YP (2010) ITIH3 is a potential biomarker for early detection of gastric cancer. J Proteome Res 9(7):3671–3679
Dekker JW, Nizankowska E, Schmitz-Schumann M, Pile K, Bochenek G, Dyczek A, Cookson WO, Szczeklik A (1997) Aspirin-induced asthma and HLA-DRB1 and HLA-DPB1 genotypes. Clin Exp Allergy 27(5):574–577
Fawcett T (2006) An introduction to ROC analysis. Pattern Recogn 27(8):861–874
Gelb MH, Altemeier WA, Beyer RP, Aitken ML, Henderson WR Jr (2010) Transglutaminase 2, a novel regulator of eicosanoid production in asthma revealed by genome-wide expression profiling of distinct asthma phenotypes. PLoS ONE 55(1):e8583
Gollapudi RR, Teirstein PS, Stevenson DD, Simon RA (2004) Aspirin sensitivity: implications for patients with coronary artery disease. JAMA 292(24):3017–3023
Guajardo JR, Schleifer KW, Daines MO, Ruddy RM, Aronow BJ, Wills-Karp M, Hershey GK (2005) Altered gene expression profiles in nasal respiratory epithelium reflect stable versus acute childhood asthma. J Allergy Clin Immunol 115:243–251
Hanselb NN, Hilmera SC, Georasb SN, Copece LM, Guob J, Irizarryc RA, Diettebd GB (2005) Oligonucleotide microarray analysis of peripheral-blood lymphocytes in severe asthma. J Lab Clin Med 145:263–274
Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protoc 4(1):44–57
Kalady MF, Dejulius K, Church JM, Lavery IC, Fazio VW, Ishwaran H (2010) Gene signature is associated with early stage rectal cancer recurrence. J Am Coll Surg 211(2):187–195
Kanehisa M, Goto S (1999) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 27(1):29–34
Kavuru MS, Dweik RA, Thomassen MJ (1999) Role of bronchoscopy in asthma research. Clin Chest Med 20:153–189
Kim BS, Park SM, Uhm TG, Kang JH, Park JS, Jang AS, Uh ST, Kim MK, Choi IS, Cho SH, Hong CS, Lee YW, Lee JY, Choi BW, Park HS, Park BL, Shin HD, Chung IY, Park CS (2010a) Effect of single nucleotide polymorphisms within the interleukin-4 promoter on aspirin intolerance in asthmatics and interleukin-4 promoter activity. Pharmacogenet Genomics 20(12):748–758
Kim JH, Park BL, Cheong HS, Bae JS, Park JS, Jang AS, Uh ST, Choi JS, Kim YH, Kim MK, Choi IS, Cho SH, Choi BW, Park CS, Shin HD (2010b) Genome-wide and follow-up studies identify CEP68 gene variants associated with risk of aspirin-intolerant asthma. PLoS ONE 5(11):e13818
Kim JY, Kim JH, Park BL, Cheong HS, Park JS, Jang AS, Uh ST, Choi JS, Kim YH, Kim MK, Choi IS, Cho SH, Choi BW, Park CS, Shin HD (2010c) Putative association of SMAPIL polymorphisms with risk of aspirin intolerance in asthmatics. J Asthma 47(9):959–965
Kim JH, Cha JY, Cheong HS, Park JS, Jang AS, Uh ST, Kim MK, Choi IS, Cho SH, Park BL, Bae JS, Park CS, Shin HD. KIF3A (2011) A cilia structural gene on chromosome 5q31, and its polymorphisms show an association with aspirin hypersensitivity in asthma. J Clin Immunol 31(1):112–121
Kopp E, Ghosh S (1994) Inhibition of NF-kappa B by sodium salicylate and aspirin. Science 265:956–959
Korbinian Strimmer (2008) fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics 24(12):1461–1462
Kraft P, Wacholder S, Cornelis MC, Hu FB, Hayes RB, Thomas G, Hoover R, Hunter DJ, Chanock S (2009) Beyond odds ratios—communicating disease risk based on genetic profiles. Nat Rev Genet 10:264–269
Kruskal WH, Tanur JM (eds) (1978) Linear hypotheses, international encyclopedia of statistics. Colm Macmillan Publishers, London
Lee RU, Stevenson DD (2011) Aspirin-exacerbated respiratory disease: evaluation and management. Allergy Asthma Immunol Res 3(1):3–10
Lee JS, Kim JH, Bae JS, Kim JY, Park TJ, Pasaje CF, Park BL, Cheong HS, Park JS, Uh ST, Kim MK, Choi IS, Cho SH, Choi BW, Park CS, Shin HD (2010a) Association analysis of UBE3C polymorphisms in Korean aspirin-intolerant asthmatic patients. Ann Allergy Asthma Immunol 105(4):307–312
Lee JS, Kim JH, Bae JS, Kim JY, Park TJ, Pasaje CF, Park BL, Cheong HS, Uh ST, Park JS, Jang AS, Kim MK, Choi IS, Park CS, Shin HD (2010b) Association of CACNG6 polymorphisms with aspirin-intolerance asthmatics in a Korean population. BMC Med Genet 11:138
Leff AR (2001) Regulation of leukotrienes in the management of asthma: biology and clinical therapy. Annu Rev Med 52:1–14
Lilly CM, Tateno H, Oguma T, Israel E, Sonna LA (2005) Effects of allergen challenge on airway epithelial cell gene expression. Am J Respir Crit Care Med 171:579–586
Oh SH, Park SM, Park JS, Jang AS, Lee YM, Uh ST, Kim YH, Choi IS, Kim MK, Park BL, Shin HD, Park CS (2009) Association analysis of peroxisome proliferator-activated receptors gamma gene polymorphisms with aspirin hypersensitivity in asthmatics. Allergy Asthma Immunol Res 1(1):30–35
Pasaje CF, Kim JH, Park BL, Cheong HS, Kim MK, Choi IS, Cho SH, Hong CS, Lee YW, Lee JY, Koh IS, Park TJ, Lee JS, Kim Y, Bae JS, Park CS, Shin HD (2011) A possible association of EMID2 polymorphisms with aspirin hypersensitivity in asthma. Immunogenetics 63(1):13–21
Peat G, Thomas E, Duncan R, Wood L (2010) Is “false-positive” clinical diagnosis of knee osteoarthritis just the early diagnosis of pre-radiographic disease? Arthritis Care Res (Hoboken) 62(10):1502–1506
Perez-G M, Melo M, Keegan AD, Zamorano J (2002) Aspirin and salicylates inhibit the IL-4 and IL-13 induced activation of STAT6. J Immunol 168:1428–1434
Perkins EM, Clarkson YL, Sabatier N, Longhurst DM, Millward CP, Jack J, Toraiwa J, Watanabe M, Rothstein JD, Lyndon AR, Wyllie DJ, Dutia MB, Jackson M (2010) Loss of beta-III spectrin leads to Purkinje cell dysfunction recapitulating the behavior and neuropathology of spinocerebellar ataxia type 5 in humans. J Neurosci 30(14):4857–4867
Rom WN, Goldberg JD, Addrizzo-Harris D, Watson HN, Khilkin M, Greenberg AK, Naidich DP, Crawford B, Eylers E, Liu D, Tan EM (2010) Identification of an autoantibody panel to separate lung cancer from smokers and nonsmokers. BMC Cancer 10(1):234
Samter M, Beers RF (1967) Concerning the nature of intolerance to aspirin. J Allergy 40:281–293
Sousa AR, Parikh A, Scadding G, Corrigan CJ, Lee TH (2002) Leukotriene receptor expression on nasal mucosal inflammatory cells in aspirin-sensitive rhinosinusitis. N Engl J Med 347:1493–1499
Subrata LS, Bizzintino J, Mamessier E, Bosco A, McKenna KL, Wikström ME, Goldblatt J, Sly PD, Hales BJ, Thomas WR, Laing IA, LeSouëf PN, Holt PG (2009) Interactions between innate antiviral and atopic immunoinflammatory pathways precipitate and sustain asthma exacerbations in children. J Immunol 15183(4):2793–2800
Szczeklik A, Nizankowska E (2000) Clinical features and diagnosis of aspirin induced asthma. Thorax 55(Suppl 2):S42–S44
Szczeklik A, Nizankowska E, Duplaga M (2000) Natural history of aspirin-induced asthma. AIANE investigators. European network on aspirin-induced asthma Eur Respir J 16(3):432–436
Szefler SJ, Phillips BR, Martinez FD, Chinchilli VM, Lemanske RF Jr, Strunk RC, Zeiger RS, Larsen G, Spahn JD, Bacharier LB, Bloomberg GR, Guilbert TW, Heldt G, Morgan WJ, Moss MH, Sorkness CA, Taussig LM (2005) Characterization of within-subject responses to fluticasone and montelukast in childhood asthma. J Allergy Clin Immunol 115:233–242
Thenappan A, Li Y, Kitisin K, Rashid A, Shetty K, Johnson L, Mishra L (2010) Role of transforming growth factor beta signaling and expansion of progenitor cells in regenerating liver. Hepatology 51(4):1373–1382
Yang SH, Kim JS, Oh TJ, Kim MS, Lee SW, Woo SK, Cho HS, Choi YH, Kim YH, Rha SY, Chung HC, An SW (2003) Genome-scale analysis of resveratrol-induced gene expression profile in human ovarian cancer cells using a cDNA microarray. Int J Oncol 22:741–750
Zhao H, Shen J, Medico L, Wang D, Ambrosone CB, Liu S (2010) A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS ONE 5(10):e13735
Ziera T, Irlbacher H, Fromm A, Latouche C, Krug SM, Fromm M, Jaisser F, Borden SA (2009) Cnksr3 is a direct mineralocorticoid receptor target gene and plays a key role in the regulation of the epithelial sodium channel. FASEB J 23(11):3936–3946
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
S. Shin and J. S. Park equally contributed as the first author.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shin, S., Park, J.S., Kim, YJ. et al. Differential gene expression profile in PBMCs from subjects with AERD and ATA: a gene marker for AERD. Mol Genet Genomics 287, 361–371 (2012). https://doi.org/10.1007/s00438-012-0685-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-012-0685-9