Abstract
We investigated the genetic basis for the derivation of pink coloration in petals from blue flowers in cultivated gentians. Using a revertant blue-flower phenotype that arose spontaneously from a pink-flowered cultivar, we sought to elucidate the molecular mechanism of flower color restoration caused by a suppressor mutation. Detailed sequencing analysis identified three novel deficient flavonoid 3′,5′-hydroxylase (F3′5′H) alleles in pink-flowered gentians in addition to two mutations identified previously (Nakatsuka et al. in Mol Genet Genomics 275:231–241, 2006). Among the deficient alleles, one allele that contained a novel miniature inverted-repeat transposable element (GtMITE1) insertion in an intron of F3′5′H was shown to cause missplicing, resulting in abnormal F3′5′H transcripts and the pink-flower phenotype. The other two mutations were identified as a single-nucleotide insertion and gypsy-Ty3 retrotransposon (Tgt1) insertion within exon 1 and exon 2 of the F3′5′H gene, respectively. The blue-flowered revertant mutant contained a single-nucleotide spontaneous mutation immediately 3′ of the TAA target site duplication and the GtMITE1 insertion, which caused restoration of normal splicing of F3′5′H and the normal blue-flower phenotype. Transient expression assays in gentian flowers in vivo demonstrated that normal F3′5′H splicing pattern was recovered from missplicing induced by the GtMITE1 insertion by the single-nucleotide substitution. These findings extend our knowledge of genomic evolution by transposable elements and spontaneous mutations in Gentiana species of economic and medical importance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Adaptation to environmental conditions is frequently accompanied with genotypic changes in the course of evolution. Evolutionary adaptation is highly important for most land plants, because they cannot move to avoid stress. In recent integrated genome-wide investigations of several plant species, comparative analysis of the genome confirms that complex genomic rearrangements cause loss or duplication of genes that affect gene functions (Flowers and Purugganan 2008; Al-Dous et al. 2011). Evolution of multigene families is also considered to occur through repetitive gene duplication via mechanisms such as unequal crossing over or gene conversion. For example, transcription factors such as MADS-box, MYB and basic Helix-Loop-Helix (bHLH) genes have diversified considerably in the past 100–600 million years (Chen and Rajewsky 2007). In another example regarding flavonoid biosynthetic genes, five dihydroflavonol 4-reductase (DFR) genes form a cluster within a 38-kb region in the Lotus japonicus genome (Shimada et al. 2005). On the other hand, a variety of mutations that alter gene functions are also revealed by the study of genomic parasites such as repetitive sequences, i.e., transposable elements, and spontaneous or induced mutations (Bennetzen 2000; Hoshino et al. 2001; Henikoff and Comai 2003). These genomic changes contribute to advantageous or disadvantageous novel traits and affect plant evolution through natural selection. However, little information on genomic evolution is available in most floricultural plants except for the well-studied petunia, morning glory and snapdragon; therefore, more studies are necessary for many floricultural species.
Transposable elements are generally categorized into two classes according to their transposition intermediates (Feschotte et al. 2002). Class I elements comprise long terminal-repeat (LTR) retrotransposons, short interspersed nuclear elements (SINE), and long interspersed nuclear elements (LINE), transposing via RNA intermediates by a copy-and-paste mechanism. In contrast, class II elements include superfamilies, such as hAT, CACTA, and Mutator-like elements, and are characterized by terminal inverted repeats and transpose via DNA intermediates. As a particular group of class II elements, miniature inverted-repeat transposable elements (MITEs) have been reported in many eukaryotes. MITEs were first discovered in association with genes of several grass species (Bureau and Wessler 1992, 1994a). MITEs are also abundant genomic components in a wide range of higher plant species and in several animal genomes, including Caenorhabditis elegans, insects, human, and zebrafish (reviewed by Feschotte et al. 2002). MITEs were also previously isolated and characterized from rice (Oki et al. 2008), sorghum (Bureau and Wessler 1994b), barley (Lyons et al. 2008), Arabidopsis (Le et al. 2000), alfalfa (Charrier et al. 1999), morning glory (Johzuka-Hisatomi et al. 1999; Hoshino et al. 2001), and potato (Momose et al. 2010). MITEs are characterized by their small size (between 100 and 300 bp), high copy number, high AT content, a lack of coding capacity, the presence of short terminal repeats, and their capacity to form secondary structures (Feschotte et al. 2002; Casacuberta and Santiago 2003).
In many cases, isolation and characterization of transposable elements has been conducted using anthocyanin pigmentation mutations in maize, snapdragon, petunia, and morning glory, because visible selection is feasible (Gerats et al. 1990; Dooner et al. 1991; Luo et al. 1991; Clegg and Durbin 2000). Gentians are one of the most important ornamental flowers in Japan. Cultivated gentians are mainly bred from two Japanese-endemic Gentiana species comprising Gentiana triflora and G. scabra. Both species are perennial and show high levels of genomic heterozygosity, owing to their outcrossing breeding system. Japanese gentians are diploid (2n = 26) and are reported to have a large genome (2C = 9.11–11.75 pg) (Mishiba et al. 2009).
The flowers of Japanese gentians naturally exhibit a vivid blue color (Fig. 1a), and accumulate complex polyacylated anthocyanins, such as gentiodelphin and albireodelphin (Goto et al. 1982; Hosokawa et al. 1997). Pink-flowered gentians also contain gentiocyanin as a major anthocyanin (Hosokawa et al. 1995). The biosynthetic pathway for these anthocyanins has been characterized in gentian flowers and almost all of the structural genes necessary for their biosynthesis have been cloned by several research groups (Nishihara et al. 2008). We cloned and characterized UDP-glucose:anthocyanin 5-O-glucosyltransferase (A5GT) and anthocyanin biosynthetic regulator genes from blue-flowered gentian (Nakatsuka et al. 2008a, b). Gentiodelphin and gentiocyanin are derived from delphinidin and cyanidin skeletons, and contribute to blue and pink-flower colors, respectively. Flavonoid 3′,5′-hydroxylase (F3′5′H) is a key enzyme that synthesizes delphinidin precursors and has been isolated from several blue- and violet-flowered plants, such as petunia, Catharanthus roseus, and gentian (Holton et al. 1993; Tanaka et al. 1996; Kaltenbach et al. 1999). Deficiency of the F3′5′H gene was reported to cause pink flowers to emerge from blue-flowered petunia (Snowden and Napoli 1998). We also reported that the transposable elements inserted into the F3′5′H gene of G. scabra caused pink coloration by disrupting the accumulation of normal F3′5′H transcripts (Nakatsuka et al. 2006). In that case, two different transposable elements, GsTRIM1 and dTgs1, were identified in two independent lines, belonging to class I terminal-repeat retrotransposons in miniature (TRIM) and class II hAT transposable elements, respectively. However, our previous study indicated that other uncharacterized mutated f3′5′h genes existed in pink-flowered gentians, because Southern blot analysis suggested at least four copies of F3′5′H exist in G. triflora, although the copy number varied in the gentian cultivars during the breeding process (Nakatsuka et al. 2006). In addition, the pink-flower phenotype is recessive (Kakizaki et al. 2009), although the exact mode of inheritance remains unknown in most cases.
Gentian materials and F3′5′H expression. a Blue-flowered cultivar ‘Maciry’. b Pink-flowered line ‘NWP’. c Pink-flowered cultivar ‘Maerchen Ashiro’. The blue-flowered stems originated from a spontaneous bud mutation. d The variegated flowers shown in c. e Northern blot analysis of F3′5′H. Total RNAs from the petals of ‘Maciry’ (lane 1), ‘NWP’ (lane 2), and ‘Maerchen Ashiro’ (lane 3) were electrophoresed on a 1% agarose gel and transferred to a nylon membrane. The blot was hybridized with a DIG-labeled F3′5′H cDNA fragment
In this study, we investigated the cause of flower color mutation in two pink-flowered hybrid gentians bred from G. triflora and G. scabra. In particular, we attempted to reveal the genetic relationship of F3′5′H alleles in both species. The results clearly identified three novel mutated f3′5′h alleles, one with a T-insertion and the others harboring novel transposable element insertions, which are responsible for the pink coloration of petals in these gentians. In addition, the revertant blue-flower phenotype originating from a pink-flowered cultivar was analyzed to elucidate the molecular mechanism of flower color restoration. We confirmed that a single-nucleotide substitution within an intron region counteracted the effect of transposable element insertion and functioned as a suppressor mutation. The involvement of MITEs in genome evolution in gentian is also discussed.
Materials and methods
Plant materials
Cultivated Japanese gentians bred from G. triflora and G. scabra were used in this study. Two pink-flowered gentian lines/cultivars, namely the breeding line ‘NWP’ and cultivar ‘Maerchen Ashiro’, were kindly provided by Nishiwaga town, Iwate, Japan, and the Hachimantai Floricultural Research and Development Center, respectively. The blue-flowered cultivars ‘Maciry’ and ‘Alta’ were obtained from the Iwate Agricultural Research Center. A blue-flowered bud mutant derived from the pink-flowered cultivar ‘Maerchen Ashiro’, which emerged in the field at the Hachimantai Floricultural Research and Development Center, was also used. ‘Maciry’ is derived from G. triflora and ‘Alta’ is derived from G. scabra. ‘NWP’ and ‘Maerchen Ashiro’ are interspecific hybrids selected from repeated crosses between G. triflora and G. scabra. A double haploid line, ‘Aki6PS’, derived from anther culture of G. triflora was used for genetic analysis of F3′5′H genes (Doi et al. 2010).
Expression analysis of F3′5′H genes in gentians
Northern blot analysis was performed to examine the expression levels of the F3′5′H gene in the petals of gentian plants. Total RNAs were isolated from petals of each gentian cultivar/line with the Plant RNA Isolation Reagent (Invitrogen, CA, USA). Five micrograms of total RNAs were separated on a 1% denatured agarose gel and transferred onto a Hybond N+ nylon membrane (GE Healthcare, Uppsala, Sweden). A probe for gentian cv. Maciry F3′5′H cDNA including a complete open reading frame (ORF) was labeled using the PCR DIG Probe Synthesis Kit (Roche Applied Science, IN, USA) using primers as described by Nakatsuka et al. (2006). Hybridization and detection were performed using the DIG Luminescent Detection Kit for Nucleic Acid (Roche) according to the manufacturer’s instructions. To examine which F3′5′H genes were expressed in gentian petals, reverse transcription (RT)-PCR analysis was performed using the consensus primers F6 and R6 for all F3′5′H genes (Supplementary Table S1). Total RNA was isolated from blue-flowered gentian petals as described previously (Nakatsuka et al. 2006). cDNAs were synthesized from total RNA after removal of genomic DNA using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara Bio, Shiga, Japan) according to the manufacturer’s instructions. Each 50 μl reaction mixture contained 1× Ex Taq buffer, 200 μM dNTPs, 0.5 μM of each primer, 5 U Ex Taq polymerase (Takara Bio) and 1 μL cDNA template. Cycle conditions were as follows: preheating at 94°C for 90 s; 30 cycles of denaturation at 95°C for 20 s, annealing at 60°C for 40 s, and extension at 72°C for 1 min; and final extension at 72°C for 10 min. The amplified fragments were subcloned into the pCR4TOPO TA cloning vector (Invitrogen) and sequenced using the BigDye Terminator version 1.1 Cycle Sequencing Kit and an ABI 3130 genetic analyzer (Applied Biosystems Japan, Tokyo, Japan).
Determination of the sequences of F3′5′H in pink-flowered gentians
RT-PCR analysis was used to identify F3′5′H variants in the pink-flowered gentians. Total RNAs were treated with DNase to remove contamination from genomic DNA using a DNA-free Kit (Ambion, TX, USA). cDNA was synthesized with the RNA PCR Kit (AMV) version 3.0 (Takara Bio) using the oligo-dT adapter primer. Each 25 μl PCR reaction contained 1× Ex buffer, 0.2 mM dNTPs, 0.4 μM of each primer, 1.25 U Ex Taq polymerase, and 1 μl cDNA template. The primers used were F1 and R1 (Supplementary Table S1). The reaction conditions were as follows: preheating at 94°C for 2 min; 35 cycles at 95°C for 30 s, 55°C for 1 min, and 72°C for 2 min; and final extension at 72°C for 10 min. The amplified fragments were subcloned into the pCR4TOPO TA cloning vector and sequenced as described above.
To determine genomic sequences corresponding to several F3′5′H variants in pink-flowered gentians, genomic PCR analysis was also performed. Genomic DNAs were isolated from leaves of each line/cultivar using Nucleon PhytoPure (GE Healthcare). PCR reactions were performed as described above, except that the cDNA templates were substituted with genomic DNAs. To detect F3′5′H with long insertions, long-range PCR reactions were performed using Takara PrimeSTAR GXL DNA polymerase (Takara Bio) according to the manufacturer’s instructions. The primer pairs used are listed in Supplementary Table S1. Subcloning and sequencing analysis were performed as described above except the cloning vector pCR-Blunt II-TOPO (Invitrogen) was used for cloning of the long PCR products. Isolation of genomic F3′5′H sequences from blue-flowered cultivars was also performed in a similar manner by PCR. Total RNAs and genomic DNAs isolated from revertant blue flowers of ‘Maerchen Ashiro’ were also used in the above analyses.
Isolation of 5′-upstream regions of F3′5′H genes in gentian
The 5′-upstream regions of gentian F3′5′H genes were identified using inverse PCR. One microgram of genomic DNA was digested by Ase I and self-ligated with the Takara Ligation Kit version 3.0 (Takara Bio). Inverse PCR was performed in 25 μl reaction mixtures containing 100 ng ligated-genome DNA, 1× LA buffer, 2.5 μM MgCl2, 400 μM dNTPs, 0.2 μM of each primer, and 1.25 U of LA Taq polymerase (Takara Bio) using primers listed in Supplementary Table S1. The reaction conditions were as follows: preheating at 94°C for 90 s; 35 cycles at 95°C for 20 s, 60°C for 40 s, and 72°C for 3 min; and final extension at 72°C for 10 min. Amplified fragments of about 3.0 kb for ‘Maciry’ and 1.8 kb for ‘Alta’ were subcloned and sequenced as described above.
Presence of GtMITE1 and Tgt1 in gentian genome
To investigate the distribution of the transposable elements GtMITE1 and Tgt1 in the gentian genome, Southern blot analyses were performed as described previously (Nakatsuka et al. 2006). Total genomic DNAs were isolated from gentian leaf samples using a modified CTAB method. After digestion with suitable restriction enzymes, the genomic DNAs were separated on a 0.8% (w/v) agarose gel, and then transferred to nylon membranes. The membranes were probed with GtMITE1 or two Tgt1 ORFs. F3′5′H cDNA that included a complete ORF was also used as a probe.
Transient expression analysis in gentian petals by particle bombardment
To investigate alternative splicing caused by a single-nucleotide substitution of the GtMITE1-inserted F3′5′H allele in ‘Maerchen Ashiro’, transient expression analysis in gentian petals was performed using particle bombardment. Genomic sequences of the GtMITE1-inserted F3′5′H allele from pink and revertant blue flowers of ‘Maerchen Ashiro’ were amplified using primer sets listed in Supplementary Table S1 and subcloned into the pCR4TOPO vector. Each insertion was double-digested by Xba I and Sac I, then cloned into the same restriction sites of pBI221 (Clontech, CA, USA) instead of the GUS gene. Particle bombardment into the petals of gentian plants cultured in vitro was performed as described by Nakatsuka et al. (2005). Five micrograms of plasmid DNA were used for each bombardment. One day post-bombardment, total RNAs were isolated from each petal and genomic DNA was removed by DNase treatment as described above. cDNAs were synthesized and F3′5′H transcripts were amplified using transgene-specific primer sets (Supplementary Table S1) under the following conditions: preheating at 94°C for 90 s; 30 cycles at 95°C for 20 s, 60°C for 40 s, and 72°C for 2 min 30 s; and final extension at 72°C for 10 min. The amplified fragments were separated on a 1.6% agarose gel and visualized by staining with ethidium bromide.
Results
Expression analysis of F3′5′H in pink-flowered gentians
A blue-flowered cultivar, ‘Maciry’ (Fig. 1a), and two pink-flowered lines/cultivars, ‘NWP’ (Fig. 1b) and ‘Maerchen Ashiro’ (Fig. 1c), were used for expression analysis of F3′5′H. Both pink-flowered gentians are interspecific hybrids derived from G. triflora and G. scabra. In G. scabra, our previous research found that two independent transposable elements (termed GsTRIM1 or dTgs1) inserted in the first exon of the F3′5′H gene caused pink coloration in petals by disrupting accumulation of normal F3′5′H transcripts (Nakatsuka et al. 2006). Therefore, we first investigated F3′5′H transcription in petal tissues of the two pink-flowered gentians using the blue-flowered ‘Maciry’ for comparison. Northern blot analysis detected no or low levels of F3′5′H transcripts in the petals of both pink-flowered plants when compared with ‘Maciry’ (Fig. 1e). In contrast, other flavonoid biosynthetic genes, such as the chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), DFR, anthocyanidin synthase (ANS), and UDP-glucose:anthocyanidin 3-O-glucosyltransferase (A3GT) genes, were expressed normally in the petals of both pink-flowered gentians (data not shown). Therefore, we deduced that these two pink-flowered gentians resulted from a deficiency of F3′5′H activity caused by reduced levels of F3′5′H transcripts.
Analysis of F3′5′H genomic sequences in blue-flowered gentians and identification of novel recessive f3′5′h alleles in pink-flowered gentians
We first attempted to identify F3′5′H genes in the blue-flowered cultivars ‘Maciry’ and ‘Alta’ as representatives of G. triflora and G. scabra, respectively. Sequencing analyses of the exons, introns and their 5′-upstream regions of F3′5′H genes revealed the presence of two different F3′5′H genes in ‘Maciry’ and one F3′5′H gene in ‘Alta’. We designated these genes F3′5′H1 and F3′5′H2, of which F3′5′H1 occurred in both cultivars (accession nos. AB586142 for ‘Maciry’, AB222604 for ‘Alta’), whereas F3′5′H2 was present only in ‘Maciry’ (accession no. AB642158). The two genes showed more than 90% sequence identity except in the promoter region (Fig. 2). Only 50.1% sequence identity existed between the F3′5′H1 and F3′5′H2 promoters in ‘Maciry’ (accession nos. AB378088 and AB642159), whereas the F3′5′H1 promoter of ‘Maciry’ shared 95.3% sequence identity with that of ‘Alta’ (accession no. AB642160). Several sequence variants were detected in the sequencing analyses probably because both cultivars were bred from several parental population and had genetic heterogeneity (data not shown). Such sequence variants apparently belonged to either F3′5′H1 or F3′5′H2 depending on the SNPs analysis. Sequencing analysis of RT-PCR products in ‘Maciry’ and ‘Alta’ amplified by the primers F6 and R6 (Supplementary Table S1) for both genes indicated that F3′5′H1 was expressed abundantly in gentian petals. No transcripts of F3′5′H2 were found in 96 sequenced fragments. Thus, F3′5′H1 is considered to be a major gene for F3′5′H activity in gentian petals, whereas F3′5′H2 is a minor gene. Tobacco plants transformed with a GUS reporter gene under control of the F3′5′H1 or F3′5′H2 promoters also indicated that the F3′5′H1 promoter had about tenfold higher activity than the F3′5′H2 promoter in petals (data not shown).
Comparison of promoter, exon and intron sequences among F3′5′H genes. Nucleotide sequence identities of gentian F3′5′H genes are shown for each region. F3′5′H1 genomic sequences from G. scabra cv. Alta (accession no. AB222604) and G. triflora cv. Maciry (accession no. AB586142), and the F3′5′H2 genomic sequence from cv. Maciry (accession no. AB642158), are compared
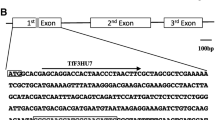
The genomic structure of the F3′5′H genes was determined by PCR using total genomic DNAs isolated from ‘NWP’ and ‘Maerchen Ashiro’ and compared with the structure of the blue-flowered cultivars ‘Maciry’ and ‘Alta’. The amplified fragments were subcloned and sequenced. One copy of F3′5′H1 of ‘Maerchen Ashiro’ contained the transposable element GsTRIM1 (accession no. AB222606; Nakatsuka et al. 2006), which was previously identified in G. scabra cv. ‘Momokorin’. It is likely that GsTRIM1 is derived from the parental G. scabra genome. In contrast, the GsTRIM1-inserted F3′5′H1 is absent from the ‘NWP’ genome (Fig. S1). Instead, f3′5′h2 harboring a single thymine nucleotide insertion at the 30-bp position from the start codon within the first exon, designated f3′5′h2 T-insertion, was present in ‘NWP’ (Fig. 3a; accession no. AB586140). As a result of this insertion, a frame shift occurred from the tenth amino acid residue of the F3′5′H protein (Fig. 3b), so this f3′5′h2 T-insertion gene is unlikely to produce functional F3′5′H enzyme. Interestingly, another copy of F3′5′H1 of ‘Maerchen Ashiro’ and ‘NWP’ contained a novel transposable element, designated GtMITE1, which was 322 bp in length and inserted in the intron of F3′5′H1 (Fig. 4a; accession no. AB586141); this mutated gene was designated f3′5′h1 GtMITE1. In addition to the GtMITE1 insertion, three nucleotide differences existed between normal F3′5′H1 and f3′5′h1 GtMITE1 . GtMITE1 has the typical features of Tourist-like MITEs (Feschotte et al. 2002), namely 3 bp (TAA) of TSD and 11 bp (GAG[T/G]ATCTCCA) of TIR (Fig. 4b). In addition, GtMITE1 is AT-rich (66.1%) and has several direct and inverted-repeat motifs. Therefore, GtMITE1 might allow the formation of a complex hairpin structure (Fig. S2). Database searches for GtMITE1 with blastn or blastx programs failed to locate similar sequences in the public databases. Southern blot analysis showed that several copies of GtMITE1 were present in gentian genomes (Fig. S3).
Genomic structure and sequence of single-nucleotide-inserted F3′5′H2 gene. a The genomic structure of f3′5′h2 T-insertion in the pink-flowered line ‘NWP’. b Comparison of nucleotide and deduced amino acid sequences in the pink-flowered line ‘NWP’ and blue-flowered cultivar ‘Maciry’. The sequences of F3′5′H genes are indicated from the start codon
Molecular characterization of the GtMITE1-inserted F3′5′H1 gene. a Schematic diagram of the genomic structure and transcript of f3′5′h1 GtMITE1. Insertion of GtMITE1 within the first intron of F3′5′H1 resulted in missplicing of transcripts. Arrows indicate the position of primers used for PCR analyses in this study. b GtMITE1 sequence of f3′5′h1 GtMITE1 in pink-flowered gentians. Black uppercase, gray uppercase, and gray lowercase letters indicate GtMITE1, exon, and intron sequences of the F3′5′H1 gene, respectively. Filled arrows and underlined characters indicate terminal inverted repeats and target site duplications, respectively. Open squares indicate 3′-splice acceptor site sequences. Boldface indicates replacement of guanine by cytosine in the blue-flowered revertant of ‘Maerchen Ashiro’. c RT-PCR analysis of F3′5′H transcripts in gentians. cDNAs were synthesized from total RNAs extracted from petals and amplified by PCR using the primer pair F1 and R1 (a), as described in the “Materials and methods”. Lane 1 blue-flowered cultivar ‘Maciry’, lane 2 pink-flowered line ‘NWP’, lane 3 pink-flowered cultivar ‘Maerchen Ashiro’, lane 4 blue-flowered revertant of ‘Maerchen Ashiro’. Fragment lengths are shown on the right. d Genomic PCR and CAPS analysis of f3′5′h1 GtMITE1. Lane 1 blue-flowered cultivar ‘Maciry’, lane 2 pink-flowered line ‘NWP’, lane 3 pink-flowered cultivar ‘Maerchen Ashiro’, lane 4 blue-flowered revertant of ‘Maerchen Ashiro’. Lane 5 Acl I-digested lane 3 fragment, lane 6 Acl I-digested lane 4 fragment. Partial F3′5′H1 fragments were amplified from genomic DNAs using the primer pair F2 and R2 (a). The amplified fragments from pink-flowered and blue-flowered revertant ‘Maerchen Ashiro’ were subjected to CAPS analysis following Acl I digestion
Although ‘Maerchen Ashiro’ was expected to contain the F3′5′H2 gene in its genome, no F3′5′H2 sequences were identified under normal PCR conditions. Thus, we attempted to detect F3′5′H genes by long-range PCR using genomic DNA derived from four cultivars/lines as templates (Fig. S4). Long PCR products about 10 kb in length were amplified in each cultivar/line except ‘Alta’. Sequence analysis of ‘Maerchen Ashiro’ confirmed one recessive allele contained a novel retrotransposon inserted into the second exon of F3′5′H2 (Fig. S5; accession no. AB618204). Sequences of both ends of these long fragments indicated the same Tgt1-inserted F3′5′H (f3′5′h2 Tgt1) allele was also present in the blue-flowered cultivar ‘Maciry’ and the other pink-flowered cultivar ‘NWP’. On the other hand, it was demonstrated that G. scabra represented by ‘Alta’ did not have any F3′5′H2 gene as described above.
This transposable element, designated Tgt1 (accession no. AB618204), was 9,446 bp in length with a 2,180-bp terminal repeat and two putative ORFs (Fig. S4). ORF1 and ORF2 encoded a deduced 488 and 1,152 amino acid residues, respectively, and exhibited low similarity to several putative gypsy-Ty3 retrotransposons in plants. A sequence of 5 bp (AGCAG) of TSD was also observed in both proximity regions. Southern blot analysis using both ORF1 and ORF2 regions as a probe showed that several copies of Tgt1-related elements were present in gentian genomes.
Genetic analysis of F3′5′H alleles
Our previous and current studies identified several dominant and recessive F3′5′H alleles from blue- and pink-flowered Japanese gentians. However, the genetic relationship between F3′5′H1 and F3′5′H2 remained unknown. In the present study, we investigated the genomic structure of F3′5′H genes of the blue-flowered ‘Aki6PS’, which was a doubled haploid line produced by anther culture of G. triflora (Doi et al. 2010). ‘Aki6PS’ contained both F3′5′H1 and f3′5′h2 Tgt1 homozygotes (data not shown); therefore, F3′5′H2 was not an allele of the F3′5′H1 gene, which indicated the existence of two F3′5′H loci in G. triflora. The genotype of ‘Aki6PS’ could be represented as F3′5′H1/F3′5′H1 and f3′5′h2 Tgt1/f3′5′h2 Tgt1. The F3′5′H genotypes of the other Japanese gentian materials used in this study can be summarized as follows: for the pink-flowered gentians, ‘NWP’ f3′5′h1 GtMITE1/f3′5′h1 GtMITE1, f3′5′h2 T-insertion/f3′5′h2 Tgt1; ‘Maerchen Ashiro’ f3′5′h1 GsTRIM1/f3′5′h1 GtMITE1, f3′5′h2 Tgt1/null; and for the blue-flowered gentians, ‘Maciry’, F3′5′H1/F3′5′H1, F3′5′H2/f3′5′h2 Tgt1; ‘Alta’, F3′5′H1/F3′5′H1.
Splicing variants of f3′5′h1 GtMITE1 in pink-flowered gentians
We investigated how the novel transposable element GtMITE1 impaired the accumulation of normal F3′5′H enzyme in pink flowers. Northern blot analysis detected very weak levels of F3′5′H transcripts in the petals of ‘NWP’ and ‘Maerchen Ashiro’. We, therefore, performed a more sensitive RT-PCR analysis using total RNAs isolated from petals of each pink-flowered gentian. The primer pairs targeted the full-length coding regions (corresponding to the cDNA) in all samples (Fig. 4c). However, the fragment lengths differed between pink- and blue-flowered genotypes. The fragments amplified from both pink-flowered gentians were longer (1.2 kb) than those from the blue-flowered ‘Maciry’ (1.1 kb). Sequencing analysis confirmed that the 1.2-kb F3′5′H1 fragment was a splice variant derived from missplicing by a novel 3′-splice acceptor site within GtMITE1 (Fig. 4b). These abnormal F3′5′H1 transcripts that contained a partial sequence of GtMITE1 generated several nonsense codons and would be translated as truncated F3′5′H proteins (data not shown). These results strongly suggested that the GtMITE1 insertion into the first intron of the F3′5′H1 gene caused the significant reduction of F3′5′H expression levels. RT-PCR analysis also showed that transcripts of f3′5′h2 T-insertion and f3′5′h2 Tgt1 were undetectable in the petals of pink- and blue-flowered gentians (data not shown).
Determination of the cause of the blue-flowered revertant phenotype in ‘Maerchen Ashiro’
Among populations of the vegetatively propagated cultivar ‘Maerchen Ashiro’, we found one bud-mutated plant with blue sector petals (Fig. 1c, d). Such flower phenotypes frequently result from the transposition of transposable elements in petunia, snapdragon and morning glory (Gerats et al. 1990; Dooner et al. 1991; Clegg and Durbin 2000), so we predicted that transpositions of transposable elements are involved in this flower color restoration in gentian. Thus, mutated and normal flowers of ‘Maerchen Ashiro’ were subjected to comparative sequence analysis of the F3′5′H genes. Total genomic DNAs were isolated from petal tissue of pink and blue flowers, and the coding regions of F3′5′H genes were amplified and sequenced. The genome of ‘Maerchen Ashiro’ contains three mutated f3′5′h alleles, namely f3′5′h1 GsTRIM1, f3′5′h1 GtMITE1 and f3′5′h2 Tgt1, as described above. The genomic sequences of f3′5′h1 GsTRIM1 and f3′5′h2 Tgt1 did not change between pink and revertant blue flowers (data not shown). In contrast, a single-base substitution at a position 2.5 kb into the f3′5′h1 GtMITE1 allele was found in the revertant blue-flowered bud compared with the typical pink-flowered bud (Fig. 4b). The mutation was a point mutation, with replacement of guanine (G) by cytosine (C), immediately 3′ of the TSD (Fig. 4b). This single-base substitution accidentally produced a site for the restriction enzyme Acl I (recognition site AA|CGTT); therefore, the transversion from G to C was easily detected by cleaved amplified polymorphic sequence (CAPS) analysis. The amplified genomic DNA fragments around the GtMITE1 region from revertant blue flowers and from pink flowers were digested by the Acl I enzyme (Fig. 4d). The amplified fragment from pink-flowered ‘Maerchen Ashiro’ could not be digested by Acl I, whereas that from the blue-flowered revertant produced 450 and 150 bp restriction fragments. Because a 600-bp non-digestible fragment was still detected in the blue-flowered revertant, the reversion must have occurred in a single chromosome and hence the blue-flowered revertant carried alleles of F3′5′H1 genes with and without the substitution mutation. The single-base substitution might have occurred in particular cells that accumulate anthocyanins at early stages of bud development.
Effect of GtMITE1 insertion on the splicing of the F3′5′H gene
To investigate whether this substitution could cause the position change of the intron 3′-splice acceptor site and function as a suppressor mutation, we performed a transient expression analysis by particle bombardment using gentian petals (Fig. 5). When the pink-flower-derived f3′5′h1 GtMITE1 genomic sequence was transiently expressed in gentian petals, one major band (1.0 kb) was detected by RT-PCR analysis using primers F4 and R4 (Fig. 5a). By contrast, transient expression of the blue-flowered revertant-derived construct produced a smaller band (0.9 kb) in addition to the 1.0-kb band (Fig. 5b). Sequence analysis confirmed that the longer band (1.0 kb) was derived from missplicing at the same splice acceptor site within GtMITE1 as determined in the F3′5′H transcripts in flowers in vivo, as shown in Fig. 4b (AG framed by rectangle). In contrast, the 0.9-kb fragments underwent normal splicing (ag framed by rectangle) and encoded a normal F3′5′H cDNA. The sequencing results are summarized in a schematic diagram (Fig. 5c). These results strongly indicated that the single-base substitution mutation caused altered splicing in f3′5′h1 GtMITE1, resulting in the revertant phenotype. However, the recovery was not perfect and 1.0 kb of abnormal F3′5′H remained in this experiment. Thus, in revertant blue flowers, both normal and abnormal F3′5′H transcripts were detected (Fig. 4c, lane 4).
Transient expression analysis of single-nucleotide substitution in the GtMITE1-inserted F3′5′H1 gene. a Schematic diagram of vectors used for transient expression analysis by particle bombardment. The genomic sequence of f3′5′h1 GtMITE1 from the pink-flowered cultivar ‘Maerchen Ashiro’ and its blue-flowered revertant were used for vector construction. The difference between the f3′5′h1 GtMITE1 sequences is a single nucleotide (G or C). Arrows indicate the positions of primers used in this study. 35Spro, Cauliflower mosaic virus 35S RNA promoter; NOSter, nopaline synthase terminator of Agrobacterium tumefaciens. b RT-PCR analysis of transcript variants by transient expression assay. Gentian petals were bombarded with the plasmids p35Sp-gF3′5′H (pink) and p35Sp-gF3′5′H (blue). After culture for 24 h, total RNAs were isolated and subjected to RT-PCR analysis using the primer pair F4 and R4 as described in the “Materials and methods”. Lane 1 bombarded with p35Sp-gF3′5′H (pink), lane 2 bombarded with p35Sp-gF3′5′H (blue), lane 3 unbombarded (negative control), lane 4 p35Sp-gF3′5′H (pink) plasmid DNA alone (positive control). The amplified fragment lengths are shown on the left side of the panel and correspond to F3′5′H transcript variants shown in c. c Schematic diagram of the structures of transcript variants. The fragment lengths are shown on the right
Discussion
Our previous report demonstrated that pink-flowered gentians resulted from interruption of the F3′5′H gene by two different transposable elements (Nakatsuka et al. 2006). The present study identified three novel mutated f3′5′h genes in the hybrid cultivar ‘Maerchen Ashiro’ and breeding line ‘NWP’, which showed insertion of a single T nucleotide and the presence of GtMITE1 or Tgt1 in the F3′5′H genes. Based on detailed sequencing analyses and pedigree analyses, it was speculated that two F3′5′H loci (F3′5′H1 and F3′5′H2) and one F3′5′H locus (F3′5′H1) are present in G. triflora and G. scabra, respectively, although an allelism test is required for final confirmation. GtMITE1 and GsTRIM1 insertions are found in F3′5′H1, whereas single-nucleotide and Tgt1 insertions are found in F3′5′H2. Expression analysis confirmed that the F3′5′H1 locus was expressed preferentially in gentian petals. This was supported by the finding that F3′5′H mRNA is derived from the F3′5′H1 locus only (Tanaka et al. 1996; Nakatsuka et al. 2006). However, all pink-flowered cultivars derived from G. triflora studied so far showed mutation in F3′5′H2 in addition to F3′5′H1, therefore, F3′5′H2 also might contribute to F3′5′H activity in gentian petals. In petunia flowers, the genetic loci Hf1 and Hf2 control F3′5′H activity. Hf1 is expressed in the limb and tube of the corolla, whereas Hf2 action is restricted to the limb (Holton et al. 1993). Given that the promoters differ greatly in sequence, F3′5′H2 might be expressed in other tissues or induced under stress conditions. Further studies are necessary to examine the role of F3′5′H2 in gentian.
The present study is the first to identify MITEs in members of the Gentianaceae. GtMITE1 belongs to the Tourist-like MITE family. GtMITE1 was inserted into an intron region of the F3′5′H1 gene and, because the TSD sequences of MITEs are TA and TAA, these elements appear to be distributed preferentially in AT-rich regions (Le et al. 2000). MITEs also exist in high copy numbers in other plant genomes, e.g., rice mPing (70 copies in ‘Nipponbare’; Jiang et al. 2003), maize Tourist (>5,000 copies; Bureau and Wessler 1992, 1994a), maize Heartbreaker (4,000 copies; Zhang et al. 2000) and alfalfa Bigfoot (1,000 to 10,000 copies; Charrier et al. 1999). However, there are also lower copy MITE families, e.g., Emigrant in Arabidopsis (Santiago et al. 2002), and MetMIT and MITRAV in Medicago truncatula (Grzebelus et al. 2009). Southern blot analysis using a GtMITE1 probe showed the presence of a relatively low copy number in the Japanese gentian genome (Fig. S3), therefore GtMITE1 also belongs to such lower-copy subfamilies. Bureau and Wessler (1994a) classified maize Tourist into four subfamilies based on their length or conserved sequence domains, and indicated that <70% nucleotide similarity is not uncommon. Therefore, GtMITE1 might also comprise other subfamily members, which could not be detected by high-stringency Southern blot analysis of the Japanese gentian genome. PCR amplification using GtMITE1 TIR as a primer revealed the presence of a number of sequences homologous to GtMITE1 in the gentian genome (data not shown). Thus, GtMITE1 family members might have caused a variety of unidentified mutations previously and contributed to genomic evolution in gentian. Further analysis of the GtMITE1 family would also provide clues to identify the autonomous element(s).
We also identified an inactive f3′5′h2 Tgt1 allele that contained an insertion of the gypsy-Ty3 LTR-retrotransposon Tgt1 in the second exon of F3′5′H2. LTR-retrotransposons are classified into two principal classes, gypsy-Ty3 and copia-Ty1, in higher plants (Levin 2002). Tgt1 showed the typical feature of the gypsy-Ty3 class, namely two polyproteins encoding their proteins in the order Gag, protease, reverse transcriptase, and integrase (Fig. S4). Other gypsy-Ty3 retrotransposons identified include del1 from Lilium henryi (Smyth et al. 1989), RIRE3 from rice (Kumekawa et al. 1999), and Tekay, Cinful, Hunk and Grande from maize (Sanmiguel and Bennetzen 1998). More than 10,000 estimated copy numbers of del1, cinful-1 and tekay existed per genome. These LTR-retrotransposons were thought to contribute to the increased size of each genome (Smyth et al. 1989; Sanmiguel and Bennetzen 1998). Southern blot analysis supported the presence of a relatively high copy number of Tgt1 in the gentian genome by long exposure (Fig. S5). Therefore, Tgt1 family members might comprise considerable regions of the gentian genome.
The f3′5′h1 GtMITE1 mutated allele in pink-flowered gentians showed reduced expression levels and produced mRNA variants of F3′5′H with several nonsense codons (Figs. 1e, 4c). mRNAs that encode truncated protein isoforms are known to be degraded by the NMD pathway (Mcglincy and Smith 2008). NMD is one of several RNA surveillance pathways that ensure the fidelity of gene expression by degrading mRNAs that lack the proper arrangement of translational signals. The primary function of NMD is thought to be the removal of errors in gene expression that might otherwise lead to the accumulation of potentially toxic truncated proteins. In mammals, a stop codon is recognized as premature if it is located >20–24 nucleotides upstream of an exon–exon junction (Isken and Maquat 2007). The nonsense codon positions in f3′5′h1 GtMITE1 did not conform to this mammalian rule, therefore, NMD might occur independently of splicing in gentian as well as yeast and fruit fly (Behm-Ansmant et al. 2007). NMD might also contribute to the degradation of F3′5′H transcripts derived from the f3′5′h2 T-insertion or f3′5′h2 Tgt1 alleles in a similar fashion.
Transposition of rice MITE mPing was induced by anther culture (Kikuchi et al. 2003) and in particular strains (Nakazaki et al. 2003), and is regulated by the autonomous MITEs Ping and Pong (Jiang et al. 2003; Yang et al. 2007). A transpositionally active MITE belonging to the Stowaway family was also identified in rice (Yang et al. 2009). More recently, Momose et al. (2010) found the first active Stowaway MITEs in the dicotyledonous potato by studying somaclonal variation of tuber skin color. Currently, we have no evidence for transposition of GtMITE1. Therefore, the determination of transposition conditions and identification of autonomous MITEs require further study.
Although we did not reveal excision or transposition of GtMITE1, we analyzed one blue-flowered revertant from the pink-flowered cultivar ‘Maerchen Ashiro’ and characterized this bud mutant in detail. Interestingly, this blue-flowered revertant had a single-nucleotide substitution (from TAAG to TAAC) immediately 3′ of the TSD of GtMITE1 in f3′5′h1 GtMITE1 (Fig. 4b). This change partially restored missplicing caused by GtMITE1 insertion to normal splicing, resulting in normal F3′5′H transcripts (Fig. 4c). The correlation between the single-nucleotide substitution and F3′5′H splicing variants was demonstrated by transient expression analysis in gentian petals (Fig. 5). Spontaneous DNA damage, such as nucleotide substitution, insertion, and deletion, occurs as an inevitable consequence of the chemical nature of DNA and its aqueous environment, or as a result of metabolic errors (Britt 1999). The T-insertion in the f3′5′h2 T-insertion allele might also be derived from such mutation, although the spontaneous mutation rate is not known in gentian.
Plant intron 5′- and 3′-splice site sequences (GT and AG, respectively) are highly conserved. Introns are removed in a two-step cleavage–ligation reaction, where the first step involves cleavage at the 5′ splice site with formation of an intron lariat at an adenosine nucleotide (the branchpoint), usually 18–40 nucleotides upstream of the 3′ splice site (Brown 1996; Brown et al. 1996). From over 1,000 Arabidopsis introns, consensus sequences for branchpoints have been shown to be [C/T]100T100[A/G]64A100[C/T]70 (Brown 1996; Brown et al. 1996). The sequences around the 3′ TSD of GtMITE1 exhibited CTAA G at 19 nucleotides upstream of the 3′ splice site (Fig. 4b). The CTAA C nucleotide sequence arising from the nucleotide substitution corresponds to the consensus branchpoint sequences of Arabidopsis. Thus, the appearance of a novel branchpoint by this random nucleotide substitution might induce normal pre-mRNA splicing and recovery of translation in f3′5′h1 GtMITE1. In higher plants, many intragenic and extragenic mutations that affected splicing have been reported. In the case of the rice waxy mutation, a single-base mutation at the 5′ splice site of the first intron induced low-level expression of the Wx b allele (Isshiki et al. 1998). Brassica rapa FLOWERING LOCUS C (FLC) showed three alternative splicing patterns owing to a mutation at the splicing site (Yuan et al. 2009). A splice site mutation is also known in maize in which a single-base mutation in the 5′-terminal of the intron activated two cryptic splice sites and alters the splicing pattern from extant splice sites (Lal et al. 1999). Arabidopsis apetala3-1, which affected floral morphology, contained a single-base substitution located 2 bp from the 3′ end of exon 5, resulting in temperature-dependent splicing defects (Sablowski and Meyerowitz 1998). This ap3-1 mutation was suppressed by the intragenic suppressor mutation ap3-11, which created a novel branchpoint (Yi and Jack 1998). Natural mutations associated with suppression of normal gene expression frequently occur and contribute to the evolution of new functions in some cases. However, more significantly, in this study the mutation caused by a transposable element was recovered accidentally by a single-nucleotide substitution mutation that modified the splice donor or acceptor site. The point mutation that affected flower color is a unique case that recovered an original branchpoint, which was formerly suppressed by GtMITE1 insertion.
Conclusion
In summary, we identified three novel mutant f3′5′h alleles in pink-flowered Japanese gentians. Tgt1 and GtMITE1 are novel transposable elements identified in Gentiana species. Tgt1 has a sequence that belongs to gypsy Ty3-retrotransposable elements and might be useful for future elucidation of genomic evolution and transition mechanisms in Gentiana species. Mutation by GtMITE1 induced a change in petal color and characterization of a revertant also revealed a change in splicing position owing to a spontaneous single-base suppressor mutation. Although Gentiana contains many valuable species utilized as ornamentals or for medical purposes, little information on genomic evolution in the genus is available. Further characterization of these transposable elements and spontaneous mutations will increase our knowledge of the genomic constitution and evolution in gentians.
References
Al-Dous EK, George B, Al-Mahmoud ME, Al-Jaber MY, Wang H, Salameh YM, Al-Azwani EK, Chaluvadi S, Pontaroli AC, Debarry J, Arondel V, Ohlrogge J, Saie IJ, Suliman-Elmeer KM, Bennetzen JL, Kruegger RR, Malek JA (2011) De novo genome sequencing and comparative genomics of date palm (Phoenix dactylifera). Nat Biotechnol 29:521–527
Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E (2007) A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J 26:1591–1601
Bennetzen JL (2000) Transposable element contributions to plant gene and genome evolution. Plant Mol Biol 42:251–269
Britt AB (1999) Molecular genetics of DNA repair in higher plants. Trends Plant Sci 4:20–25
Brown JW (1996) Arabidopsis intron mutations and pre-mRNA splicing. Plant J 10:771–780
Brown JW, Smith P, Simpson CG (1996) Arabidopsis consensus intron sequences. Plant Mol Biol 32:531–535
Bureau TE, Wessler SR (1992) Tourist: a large family of small inverted repeat elements frequently associated with maize genes. Plant Cell 4:1283–1294
Bureau TE, Wessler SR (1994a) Mobile inverted-repeat elements of the tourist family are associated with the genes of many cereal grasses. Proc Natl Acad Sci USA 91:1411–1415
Bureau TE, Wessler SR (1994b) Stowaway: a new family of inverted repeat elements associated with the genes of both monocotyledonous and dicotyledonous plants. Plant Cell 6:907–916
Casacuberta JM, Santiago N (2003) Plant LTR-retrotransposons and MITEs: control of transposition and impact on the evolution of plant genes and genomes. Gene 311:1–11
Charrier B, Foucher F, Kondorosi E, d’Aubenton-Carafa Y, Thermes C, Kondorosi A, Ratet P (1999) Bigfoot. A new family of MITE elements characterized from the Medicago genus. Plant J 18:431–441
Chen K, Rajewsky N (2007) The evolution of gene regulation by transcription factors and microRNAs. Nat Rev 8:93–103
Clegg MT, Durbin ML (2000) Flower color variation: a model for the experimental study of evolution. Proc Natl Acad Sci USA 97:7016–7023
Doi H, Takahashi R, Hikage T, Takahata Y (2010) Embryogenesis and doubled haploid production from anther culture in gentian (Gentiana triflora). Plant Cell Tissue Organ Cult 102:27–33
Dooner HK, Robbins TP, Jorgensen RA (1991) Genetic and developmental control of anthocyanin biosynthesis. Annu Rev Genet 25:173–199
Feschotte C, Zhang X, Wessler SR (2002) Miniature inverted-repeat transposable elements and their relationship to established DNA transposons. In: Craig NL, Craigie R, Gellert M, Lambowitz AM (eds) Mobile DNA II. ASM Press, Washington, DC, pp 1147–1158
Flowers JM, Purugganan MD (2008) The evolution of plant genomes: scaling up from a population perspective. Curr Opin Genet Dev 18:565–570
Gerats AG, Huits H, Vrijlandt E, Marana C, Souer E, Beld M (1990) Molecular characterization of a nonautonomous transposable element (dTph1) of petunia. Plant Cell 2:1121–1128
Goto T, Kondo T, Tamura H, Imagawa H, Iino H, Takeda K (1982) Structure of gentiodelphin, an acylated anthocyanin isolated from Gentiana makinori, that is stable in dilute aqueous solution. Tetrahedron Lett 23:3695–3698
Grzebelus D, Gladysz M, Macko-Podgorni A, Gambin T, Golis B, Rakoczy R, Gambin A (2009) Population dynamics of miniature inverted-repeat transposable elements (MITEs) in Medicago truncatula. Gene 448:214–220
Henikoff S, Comai L (2003) Single-nucleotide mutations for plant functional genomics. Ann Rev Plant Biol 54:375–401
Holton TA, Brugliera F, Lester DR, Tanaka Y, Hyland CD, Menting JG, Lu CY, Farcy E, Stevenson TW, Cornish EC (1993) Cloning and expression of cytochrome P450 genes controlling flower colour. Nature 366:276–279
Hoshino A, Johzuka-Hisatomi Y, Iida S (2001) Gene duplication and mobile genetic elements in the morning glories. Gene 265:1–10
Hosokawa K, Fukushi E, Kawabata J, Fujii C, Ito T, Yamamura S (1995) Three acylated cyaniding glucosides in pink flowers of Gentiana. Phytochemistry 40:941–944
Hosokawa K, Fukushi E, Kawabata J, Fujii C, Ito T, Yamamura S (1997) Seven acylated anthocyanins in blue flowers of Gentiana. Phytochemistry 45:167–171
Isken O, Maquat LE (2007) Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev 21:1833–1856
Isshiki M, Morino K, Nakajima M, Okagaki RJ, Wessler SR, Izawa T, Shimamoto K (1998) A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5′ splice site of the first intron. Plant J 15:133–138
Jiang N, Bao Z, Zhang X, Hirochika H, Eddy SR, McCouch SR, Wessler SR (2003) An active DNA transposon family in rice. Nature 421:163–167
Johzuka-Hisatomi Y, Hoshino A, Mori T, Habu Y, Iida S (1999) Characterization of the chalcone synthase genes expressed in flowers of the common and Japanese morning glories. Genes Genet Syst 74:141–147
Kakizaki Y, Nakatsuka T, Kawamura H, Abe Y, Yamamura S, Nishihara M (2009) Development of codominant DNA marker distinguishing pink from blue flowers in Gentiana scabra. Breed Res 11:9–14 (in Japanese)
Kaltenbach M, Schroder G, Schmelzer E, Lutz V, Schroder J (1999) Flavonoid hydroxylase from Catharanthus roseus: cDNA, heterologous expression, enzyme properties and cell-type specific expression in plants. Plant J 19:183–193
Kikuchi K, Terauchi K, Wada M, Hirano HY (2003) The plant MITE mPing is mobilized in anther culture. Nature 421:167–170
Kumekawa N, Ohtsubo H, Horiuchi T, Ohtsubo E (1999) Identification and characterization of novel retrotransposons of the gypsy type in rice. Mol Gen Genomics 260:593–602
Lal S, Choi JH, Shaw JR, Hannah LC (1999) A splice site mutant of maize activates cryptic splice sites, elicits intron inclusion and exon exclusion, and permits branch point elucidation. Plant Physiol 121:411–418
Le QH, Wright S, Yu Z, Bureau T (2000) Transposon diversity in Arabidopsis thaliana. Proc Natl Acad Sci USA 97:7376–7381
Levin HL (2002) Newly identified retrotransposons of the Ty3/gypsy class in fungi, plants and vertebrates. In: Craig NL, Craigie R, Gellert M, Lambowitz AM (eds) Mobile DNA II. ASM Press, Washington, D. C
Luo D, Coen ES, Doyle S, Carpenter R (1991) Pigmentation mutants produced by transposon mutagenesis in Antirrhinum majus. Plant J 1:59–69
Lyons M, Cardle L, Rostoks N, Waugh R, Flavell AJ (2008) Isolation, analysis and marker utility of novel miniature inverted repeat transposable elements from the barley genome. Mol Gen Genomics 280:275–285
McGlincy NJ, Smith CW (2008) Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci 33:385–393
Mishiba K, Yamane K, Nakatsuka T, Nakano Y, Yamamura S, Abe J, Kawamura H, Takahata Y, Nishihara M (2009) Genetic relationships in the genus Gentiana based on chloroplast DNA sequence data and nuclear DNA content. Breeding Sci 59:119–127
Momose M, Abe Y, Ozeki Y (2010) Miniature inverted-repeat transposable elements of Stowaway are active in potato. Genetics
Nakatsuka T, Nishihara M, Mishiba K, Yamamura S (2005) Two different mutations are involved in the formation of white-flowerd gentian plants. Plant Sci 169:949–958
Nakatsuka T, Nishihara M, Mishiba K, Hirano H, Yamamura S (2006) Two different transposable elements inserted in flavonoid 3′, 5′-hydroxylase gene contribute to pink flower coloration in Gentiana scabra. Mol Gen Genomics 275:231–241
Nakatsuka T, Haruta KS, Pitaksutheepong C, Abe Y, Kakizaki Y, Yamamoto K, Shimada N, Yamamura S, Nishihara M (2008a) Identification and characterization of R2R3-MYB and bHLH transcription factors regulating anthocyanin biosynthesis in gentian flowers. Plant Cell Physiol 49:1818–1829
Nakatsuka T, Sato K, Takahashi H, Yamamura S, Nishihara M (2008b) Cloning and characterization of the UDP-glucose:anthocyanin 5-O-glucosyltransferase gene from blue-flowered gentian. J Exp Bot 59:1241–1252
Nakazaki T, Okumoto Y, Horibata A, Yamahira S, Teraishi M, Nishida H, Inoue H, Tanisaka T (2003) Mobilization of a transposon in the rice genome. Nature 421:170–172
Nishihara M, Nakatsuka T, Mizutani-Fukuchi M, Tanaka Y, Yamamura S (2008) Gentians: From gene cloning to molecular breeding. In: Jaime A, da Silva T (eds) Floricultural and ornamental biotechnology V. Global Science Books, UK, pp 57–67
Oki N, Yano K, Okumoto Y, Tsukiyama T, Teraishi M, Tanisaka T (2008) A genome-wide view of miniature inverted-repeat transposable elements (MITEs) in rice, Oryza sativa ssp. japonica. Genes Genet Syst 83:321–329
Sablowski RWM, Meyerowitz EM (1998) Temperature-sensitive splicing in the floral homeotic mutant apetala3–1. Plant Cell 10:1453–1463
Sanmiguel P, Bennetzen JL (1998) Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Annal Bot 82:37–44
Santiago N, Herraiz C, Goni JR, Messeguer X, Casacuberta JM (2002) Genome-wide analysis of the Emigrant family of MITEs of Arabidopsis thaliana. Mol Biol Evol 19:2285–2293
Sato K, Hamada M, Asai K, Mituyama T (2009) CENTROIDFOLD: a web server for RNA secondary structure prediction. Nucleic Acids Res 37:W277–W280
Shimada N, Sasaki R, Sato S, Kaneko T, Tabata S, Aoki T, Ayabe S (2005) A comprehensive analysis of six dihydroflavonol 4-reductases encoded by a gene cluster of the Lotus japonicus genome. J Exp Bot 56:2573–2585
Smyth DR, Kalitsis P, Joseph JL, Sentry JW (1989) Plant retrotransposon from Lilium henryi is related to Ty3 of yeast and the gypsy group of Drosophila. Proc Natl Acad Sci USA 86:5015–5019
Snowden KC, Napoli CA (1998) Psl: a novel Spm-like transposable element from Petunia hybrida. Plant J 14:43–54
Tanaka Y, Yonekura K, Fukuchi-Mizutani M, Fukui Y, Fujiwara H, Ashikari T, Kusumi T (1996) Molecular and biochemical characterization of three anthocyanin synthetic enzymes from Gentiana triflora. Plant Cell Physiol 37:711–716
Yang G, Zhang F, Hancock CN, Wessler SR (2007) Transposition of the rice miniature inverted repeat transposable element mPing in Arabidopsis thaliana. Proc Natl Acad Sci USA 104:10962–10967
Yang G, Nagel DH, Feschotte C, Hancock CN, Wessler SR (2009) Tuned for transposition: molecular determinants underlying the hyperactivity of a Stowaway MITE. Science 325:1391–1394
Yi Y, Jack T (1998) An intragenic suppressor of the Arabidopsis floral organ identity mutant apetala3-1 functions by suppressing defects in splicing. Plant Cell 10:1465–1477
Yuan YX, Wu J, Sun RF, Zhang XW, Xu DH, Bonnema G, Wang XW (2009) A naturally occurring splicing site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time. J Exp Bot 60:1299–1308
Zhang Q, Arbuckle J, Wessler SR (2000) Recent, extensive, and preferential insertion of members of the miniature inverted-repeat transposable element family Heartbreaker into genic regions of maize. Proc Natl Acad Sci USA 97:1160–1165
Acknowledgments
The authors thank Nishiwaga town and the Iwate Agricultural Research Center for providing gentian cultivars. We also thank Ms Y. Kakizaki, R. Horikiri and C. Yoshida, Iwate Biotechnology Research Center, for technical assistance. This research was supported by the Japan Society for the Promotion of Science KAKENHI (18580010) and in part by a ‘Research project for utilizing advanced technologies in agriculture, forestry and fisheries’ (No. 2040) from the Ministry of Agriculture, Forestry and Fisheries of Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M.-A. Grandbastien.
Nucleotide sequence data reported are available in the DDBJ/EMBL/GenBank databases under accession numbers AB586140, AB586141, AB586142, AB618204, AB642158, AB642159 and AB642160.
Electronic supplementary material
Below is the link to the electronic supplementary material.
438_2011_652_MOESM2_ESM.ppt
Fig. S1 Confirmation of the presence of GsTRIM1 in gentian breeding lines and cultivars. (a) Genomic DNAs isolated from four gentian cultivars/lines were subjected to PCR analysis to identify the presence or absence of GsTRIM1 within F3′5′H1 genes, as described by Nakatsuka et al. (2006). Arrows indicate the positions of primers used in this analysis. (b) Fragments 800 bp and 350 bp in length represent the presence and absence of GsTRIM1 in exon 1 of the F3′5′H1 genes, respectively. Lane 1, blue-flowered cultivar ‘Maciry’; lane 2, pink-flowered line ‘NWP’; lane 3 pink-flowered cultivar ‘Maerchen Ashiro’; lane 4, pink-flowered cultivar ‘Momokorin’. (PPT 448 kb)
438_2011_652_MOESM3_ESM.ppt
Fig. S2 Predicted secondary structure of GtMITE1 . The secondary structure of GtMITE1 predicted by the CentroidFold web server (http://www.ncrna.org/centroidfold/) (Sato et al. 2009) from an RNA sequence. Each predicted base-pair is colored with the heat color gradation from blue to red corresponding the base-pairing probability from 0 to 1, where the base-pairing probability is the probability that a pair of bases forms a base-pair via hydrogen bonds in their secondary structures, and can be interpreted as confidence measure of predicted base-pairs. (PPT 659 kb)
438_2011_652_MOESM4_ESM.ppt
Fig. S3 Genomic Southern blot analysis of gentian breeding lines and cultivars. Total DNAs isolated from three gentian cultivars/lines were digested with the restriction enzymes EcoR I or Xba I, and transferred to a nylon membrane and hybridized with GtMITE1 (a) and F3′5′H probes (b). Lane 1, blue-flowered cultivar ‘Maciry’; lane 2, pink-flowered line ‘NWP’; lane 3, pink-flowered cultivar ‘Maerchen Ashiro’. The probes used are shown below the panels. (PPT 2040 kb)
438_2011_652_MOESM5_ESM.ppt
Fig. S4 Amplification of genomic F3′5′H genes by long-range PCR. (a) F3′5′H genes were amplified by standard (lanes 1 to 4) and long-range (lanes 5 to 12) PCR in four different cultivars/lines. Lanes 1, 5 and 9, blue-flowered cultivar ‘Maciry’; lanes 2, 6 and 10, pink-flowered line ‘NWP’; lanes 3, 7 and 11, pink-flowered cultivar ‘Maerchen Ashiro’; lanes 4, 8 and 12, blue-flowered cultivar ‘Alta’. The primers used are shown above the panel. Arrows indicate the amplified long PCR products. (b) Schematic diagram of F3′5′H genomic structure. The positions of the primers used are indicated. (PPT 489 kb)
438_2011_652_MOESM6_ESM.ppt
Fig. S5 Schematic diagram of the genomic structure and Southern blot analysis of f3′5′h Tgt1. (a) Insertion of Tgt1 within the second exon of F3′5′H in ‘Maerchen Ashiro’. Tgt1 has the features of long terminal-repeat retrotransposons, containing 5 bp of the target sequence duplication (TSD) and 2,180 bp of long terminal repeats (LTR; gray arrows). Two deduced open reading frames (ORFs) are indicated by black arrows. Primer binding sites (PBS) and polypurine tract (PPT) are also shown. (b) Southern blot analysis of Tgt1. Lane 1, pink-flowered line ‘NWP’; lane 2, pink-flowered cultivar ‘Maerchen Ashiro’. Total DNAs were digested with restriction enzymes EcoR I and transferred to a nylon membrane and hybridized with a DIG-labeled probe containing two ORFs. The arrow indicates the expected size derived from Tgt1. The duration of exposure to X-ray film is shown below the panels. (PPT 357 kb)
Rights and permissions
About this article
Cite this article
Nishihara, M., Hikage, T., Yamada, E. et al. A single-base substitution suppresses flower color mutation caused by a novel miniature inverted-repeat transposable element in gentian. Mol Genet Genomics 286, 371–382 (2011). https://doi.org/10.1007/s00438-011-0652-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-011-0652-x