Abstract
Strain CFT073 is a bona fide uropathogen, whereas strains 83972 and Nissle 1917 are harmless probiotic strains of urinary tract and faecal origin, respectively. Despite their different environmental origins and dispositions the three strains are very closely related and the ancestors of 83972 and Nissle 1917 must have been very similar to CFT073. Here, we report the first functional genome profiling of Nissle 1917 and the first biofilm profiling of a uropathogen. Transcriptomic profiling revealed that Nissle 1917 expressed many UPEC-associated genes and showed that the active genomic profiles of the three strains are closely related. The data demonstrate that the distance from a pathogen to a probiotic strain can be surprisingly short. We demonstrate that Nissle 1917, in spite of its intestinal niche origin, grows well in urine, and is a good biofilm former in this medium in which it also out-competes CFT073 during planktonic growth. The role in biofilm formation of three up-regulated genes, yhaK, yhcN and ybiJ, was confirmed by knockout mutants in Nissle 1917 and CFT073. Two of these mutants CFT073∆yhcN and CFT073∆ybiJ had significantly reduced motility compared with the parent strain, arguably accounting for the impaired biofilm formation. Although the three strains have very different strategies vis-à-vis the human host their functional gene profiles are surprisingly similar. It is also interesting to note that the only two Escherichia coli strains used as probiotics are in fact deconstructed pathogens.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Escherichia coli is a versatile pathogen that is able to colonise several host-related niches. It can cause a range of diseases such as urinary tract infection (UTI), sepsis, meningitis and diarrhea. At the other side of the spectrum are harmless, commensal strains and even strains with probiotic qualities. The uropathogenic E. coli (UPEC) strain CFT073 was isolated from the blood of a hospitalised patient suffering from acute pyelonephritis and has been fully sequenced (Mobley et al. 1990; Welch et al. 2002). Escherichia coli strain Nissle 1917, serotype O6:K5:H1, is an excellent coloniser of the human gut and it has been reported that it is able to colonise and establish itself in the human intestine even in the presence of a natural resident bacterial flora (Lodinova-Zadnikova et al. 1992; Schulze and Sonnenborn 1995; Lodinova-Zadnikova and Sonnenborn 1997). The strain was originally isolated during World War I from a soldier who escaped a severe outbreak of diarrhea affecting his regiment. Nissle 1917 seems to have a beneficial effect on several types of intestinal disorders and appears to be well tolerated by humans. Nissle 1917 has been marketed as a probiotic remedy against intestinal disorders in several European countries since the 1920s (Schulze and Sonnenborn 1995; Schultz 2008). During this period of time it has been ingested by an appreciable number of people and is probably the best “field-tested” E. coli strain in humans in the world. The strain is well equipped with fitness and survival factors such as iron-uptake systems, adhesins, proteases and microcins, but lacks defined virulence factors such as haemolysin and P-fimbrial adhesin (Grozdanov et al. 2004).
Urinary tract isolates can colonise different niches and either cause pyelonephritis (kidney infection), as strain CFT073, and cystitis (bladder infection), or they can exist as commensal-like strains in the urinary tract colonising the bladder asymptomatically, i.e., as asymptomatic bacteriuria (ABU) strains. The most well-characterised ABU strain is isolate 83972 (Klemm et al. 2007). Strain 83972 was originally isolated from a young girl who had carried it for at least 3 years without symptoms (Lindberg et al. 1975; Andersson et al. 1991). The strain is well adapted for growth in the human urinary tract (Roos and Klemm 2006; Roos et al. 2006b) where it establishes long-term bacteriuria (Hull et al. 2000). It has been used for prophylactic purposes in numerous studies where it demonstrated a beneficial effect in patients with recurrent UTI who are refractory to conventional therapy (Hull et al. 2000; Sundén et al. 2006).
Bacteria often live as surface-associated communities rather than as planktonic cells, referred to as biofilms. Bacterial biofilm formation is commonly associated with many health problems (Costerton et al. 1999; O’Toole et al. 2000). The intestinal tract is heavily colonised by microorganisms and bacterial biofilm formation most likely plays an important role in colonisation in this habitat; there is a growing interest in studying biofilms in the colon—in particular regarding their role in inflammatory bowel disease (Macfarlane and Dillon 2007). In the urinary tract, infections associated with biofilms include chronic cystitis, prostatitis, and catheter- and stent-associated infections (Warren 2001). Moreover, biofilm formation seems to be a trait associated with cystitis and ABU isolates rather than pyelonephritis isolates, indicating that biofilm might be important for persistent colonisation of the urinary tract (Hancock et al. 2007; Mabbett et al. 2009). Bacterial biofilms related to infections are particularly problematic since sessile bacteria can withstand host defence mechanisms and are extremely resistant to antibiotics, biocides and hydrodynamic shear forces (Costerton et al. 1995, 1999). Therefore, understanding the molecular mechanisms by which E. coli isolates form biofilms is important for developing methods to control and prevent biofilm growth.
ABU isolate 83972 and Nissle 1917 are the only probiotic E. coli strains that have been used in humans; they are both closely related to CFT073. Also, all three strains belong to the same sequence type (ST) 73 clonal group (Welch et al. 2002; Grozdanov et al. 2004; Zdziarski et al. 2007). We recently revealed that the three strains are very similar genetically with only a few hundred genes differing between the virulent strain CFT073 and the two probiotic strains (Vejborg et al. 2010). On this background, we have compared the transcriptome profiles of the three strains and studied urine growth and biofilm formation as well as competition.
Materials and methods
Bacterial strains and media
The 83972 and CFT073 strains are well characterised UTI isolates (Mobley et al. 1990; Klemm et al. 2007). Nissle 1917 used here was a streptomycin-resistant version with growth and competition properties identical to those of wild-type Nissle 1917 (Hancock et al. 2010). In the competition between 83972 and CFT073 an ampicillin-resistant version of the 83972 strain was used; 83972amp has been described previously (Hancock et al. 2007) and has growth characteristics identical to those of the wild-type counterpart. For the flow-chamber experiments yellow-fluorescent-tagged (Yfp) versions, constructed by lambda Red-mediated combination, of Nissle 1917, CFT073 and 83972 were used; Nissle 1917yfp was a kind gift from Malin Dahl, and CFT073yfp and 83972yfp have been described previously (Ferrières et al. 2007). Human urine, LB or MOPS minimal medium supplemented with 0.2% glucose and 0.02% casamino acids (Neidhardt et al. 1974), were used in all experiments. Human urine was collected from a pool of ten healthy women volunteers who had no history of UTI or antibiotic use in the prior 2 months. For each experiment, urine was collected randomly from three to five of these individuals (pH 5.5–7.5). The urine was pooled (pH 6.5–7.0), filter sterilised, stored at 4°C, and used within the following 2–3 days.
Growth conditions and stabilisation of RNA for microarray experiments
For the planktonic growth experiments, overnight cultures of Nissle 1917, CFT073 and 83972 were grown in triplicates in pooled human urine or MOPS minimal medium until reaching exponential phase and then used for inoculation of 25 ml urine or MOPS to an OD600 of 0.05. The cultures were grown in 100-ml non-baffled Erlenmeyer flasks at 37°C and 130 rpm, and 5-ml samples for isolation of RNA were extracted from three individual cultures at mid-exponential phase (corresponding to an OD600 of approximately 0.4 and 0.5 in urine and MOPS, respectively). Extracted samples were immediately mixed with two volumes of RNAprotect™ Bacteria Reagent (QIAGEN AG), incubated for 5 min at room temperature to stabilise RNA, and centrifuged. The pellets were stored at −80°C.
For the biofilm experiments, freshly grown urine cultures of Nissle 1917, CFT073 and 83972 were used for inoculation of 25 ml pooled human urine in 94 mm Petri dishes (Greiner Bio-One), each strain in triplicate. The cultures were grown statically at 37°C for 42 h and the medium was carefully poured off and replaced by fresh preheated (37°C) media twice during incubation. After 42 h the bacterial lawn on each Petri dish was quickly washed twice with 20 ml urine preheated to 37°C and immediately thereafter, 2 ml of a 1:2 mixture of PBS and RNAprotect™ Bacteria Reagent (QIAGEN AG) was poured on the lawn of attached cells and incubated for 5 min at room temperature to stabilise RNA. The stabilised mixture was then centrifuged and pellets were stored at −80°C.
RNA isolation and cDNA labelling
The procedures for RNA isolation and cDNA labeling were performed as described previously (Hancock and Klemm 2007) and according to the GeneChip® Expression Analysis Technical Manual 701023 Rev. 4 (Affymetrix, Inc., Santa Clara, CA, USA).
Microarray analysis
GeneChip E. coli Genome 2.0 Arrays (Affymetrix) were used for hybridisation of the labelled cDNA. In total, 27 samples were hybridised to 27 microarrays, i.e., nine biologically independent samples from each of the three strains were hybridised to one array each. For all three strains, i.e., Nissle 1917, CFT073 and 83972, three chips were hybridised with samples grown in three individual flasks in MOPS, three chips were hybridised with samples from cells grown in pooled human urine in three individual flasks, and three chips were hybridised with samples from biofilm cells grown in pooled human urine in three individual petri dishes. No pooling was performed and all samples were treated separately. Hybridisation, washing and staining were performed according to the GeneChip Expression Analysis Technical Manual 701023 Rev. 4 (Affymetrix), and the microarrays were scanned using the GeneChip Scanner 3000.
The 27 arrays were normalised using the invariant set normalization method (Li and Wong 2001) in dChip (http://www.dchip.org) and fold changes during biofilm growth in urine compared with planktonic growth in MOPS and in urine were calculated for each individual strain. In a third comparison, fold-changes were calculated by comparing the arrays hybridized with samples from planktonic urine with planktonic MOPS. The comparison criteria were carefully chosen to make sure that the estimation of the percentages of genes identified by chance, the empirical false discovery rate (FDR), was kept low for all comparisons in our study (<5% recommended). Permuting our samples randomly 200 times resulted in FDRs between 0.0 and 0.7% (i.e., 0–20 false positive genes) for the different comparisons. Array normalization, expression value calculation, sample comparison and FDR estimation were performed using dChip (http://www.dchip.org). Furthermore, dChip was used to perform hierarchical cluster analysis on the 27 arrays. The genes were filtered after pooling replicate arrays to generate a list for the cluster analysis using the default setting in dChip; this resulted in a list of 2,989 filtered genes consisting of genes showing variation in expression across the samples, excluding the noise from absent and non-changed genes. Subsequently hierarchical cluster analysis was performed on the 2,989 genes using the default settings. The supporting microarray data have been deposited in ArrayExpress (http://www.ebi.ac.uk/arrayexpress) with accession number E-MEXP-2609.
RT-PCR
RT-PCR to confirm DNA microarray gene expression data and was performed as described previously (Roos and Klemm 2006). The primers used in RT-PCR and PCR are listed in Supplemental Table 1.
Growth conditions for growth characteristics and competition experiments
For measuring growth characteristics, monocultures were grown in 20 ml medium inoculated with an overnight culture to an OD600 of 0.05 and grown at 37°C and 130 rpm. OD600 was measured every 30 min. Each experiment was performed in triplicates and repeated twice. For competition experiments, mixed cultures were grown in 50 ml cultures inoculated 1:1 with two freshly grown precultures of E. coli 83972, Nissle 1917 or CFT073 to an OD600 of 0.05 (corresponding to ~5 × 108 CFU of each strain). The cultures were grown at 37°C and 130 rpm, and samples were extracted after 16 h, diluted and plated on LB plates with and without antibiotic. The 1:1 starting ratio was confirmed following the same procedure. Each competition experiment was performed in triplicates and repeated three times.
Biofilm formation in microtitre plates
Cells were pre-grown in pooled human urine, LB or MOPS, and 10 μl were used to inoculate 1 ml of growth medium in 24-well flat-bottom microplates (Iwaki). The microplates were incubated statically at 37°C overnight and biofilm was monitored by crystal violet staining as described previously (Hancock et al. 2007). Each strain was assayed in three to four wells on each plate and all experiments were repeated at least five times. In each plate three to four wells were used as blanks containing sterile growth medium.
Biofilm formation in flow-cell chambers
The flow-chamber system was prepared and assembled as previously described (Christensen et al. 1999). The biofilms were cultivated at 37°C in human urine. The biofilms were inspected using a Zeiss LSM510 confocal scanning laser microscope (CSLM) (Carl Zeiss, Jena, Germany) and the pictures were edited using the IMARIS software package (Bitplane). The experiment was repeated four times.
Competition during biofilm formation in microtitre plates
The wells of a 24-well flat-bottom microtitre plate were filled with 1 ml of growth medium and inoculated with an equal number of cells of the two strains in competition to a final OD600 of 0.01. The cells were incubated statically at 37°C for 16 h and subsequently treated as previously described (Ferrières et al. 2007). The colony-forming units (CFU) of each strain in the final biofilm population were determined by plating serial dilutions of the cell suspension onto LB–agar plates with and without antibiotic. The initial 1:1 ratio was confirmed in the same way.
Construction of knock-out mutants of Nissle 1917, CFT073 and 83972
Mutants of Nissle 1917, CFT073 and 83972 were constructed using the λRed recombinase gene replacement system (Datsenko and Wanner 2000). Primers for amplification of the npt gene of pKD4 are listed in Supplemental Table 1, i.e., bssS, yhaK, yhcN and ybiJ, corresponding to the b identifiers b1016, b3106, b3238 and b0802, respectively. The correct double-crossover and recombination event was confirmed by primers listed in Supplemental Table 1.
Construction of plasmids for complementation
The yhaK, yhcN and ybiJ genes of CFT073 was amplified by PCR using primer pairs containing BamHI and SalI restriction sites (Supplemental Table 1), and cloned into pACYC184. The resulting plasmids, pYhaK (pVR6), pYhcN (pVR7) and pYbiJ (pVR8), were transformed into mutant strains of CFT073 and Nissle 1917.
Motility on urine agar plates
Each strain was stabbed into the centre of a urine agar plate (9:1 ratio of human urine and H2O) containing 0.3% (w/v) agar and incubated at 37°C in an upright position. Colony diameter was measured after 16 h of incubation. The experiment was performed in duplicates or triplicates and repeated three times in different batches of pooled human urine.
Results
Global gene expression profiling of Nissle 1917, CFT073 and 83972
Using a new comparative genomic hybridisation chip we recently found a very close genetic relationship between the uropathogen CFT073 and the two probiotics isolates Nissle 1917 and 83972 (Vejborg et al. 2010). However, such studies reveal little about the functionality of the genes. To complement the genomic findings, we wanted to compare their transcriptomes and performed microarray analysis. Comparison of the expression profiles of these three strains during planktonic growth and biofilm formation in human urine, with planktonic growth in minimal medium as control, was performed using the GeneChip® E. coli Genome 2.0 Arrays which carries probes for all genes present in CFT073.
Figure 1 displays genes that were significantly up- and down-regulated in the three strains and reveal the similarities in expression between the strains. In all comparisons, Nissle 1917 and CFT073 shared a larger number of up- or down regulated genes than Nissle 1917 and 83972 or 83972 and CFT073. It should be noted that due to the chip design there are genes found in Nissle 1917 and/or 83972 that are not present as probes on the microarray wherefore the number of significantly changed genes for Nissle 1917 and 83972 could potentially be higher. However, despite that all genes in CFT073 are represented on the microarray the number of genes significantly changed in this strain was not larger than for the other two strains.
Venn diagrams displaying the number of significantly up- and down-regulated genes in Nissle 1917, CFT073 and 83972 between different growth conditions, i.e., exponential growth in MOPS (‘MOPS’), exponential growth in human urine (‘Urine’) and biofilm formation in human urine (‘Biofilm’). a and b show up- and down-regulation during growth in urine planktonic and biofilm, respectively, compared with MOPS (baseline), and c shows up- and down-regulation in urine biofilm compared with urine planktonic growth (baseline)
Hierarchical clustering analysis of the 27 microarrays used for hybridisation of the three strains was performed (Fig. 2). In planktonic MOPS and biofilm urine, Nissle 1917 and CFT073 clustered together before clustering with 83972. On the other hand, in planktonic urine, Nissle 1917 and 83972 clustered together before clustering with CFT073, although the distance was not large between the three planktonic urine samples. It is interesting to note that the two UTI isolates do not cluster together before clustering with Nissle 1917 in any of the growth conditions indicating a closer relationship of the functional genomes between either one of the UTI isolates and Nissle 1917 than between the two UTI isolates.
Correlation matrix of the expression data from the 27 microarrays obtained by hierarchical cluster analysis (treated as nine samples pooling replicate arrays). The cluster analysis was performed on 2,989 genes filtered by dChip (to exclude the noise from absent and non-changed genes) using the default settings. ‘MOPS’, cells grown planktonically in minimal lab medium; ‘Urine’, cells grown planktonically in pooled human urine; ‘Biofilm’, cells grown in biofilms on petri dishes in pooled human urine
Iron acquisition and uptake systems
Several iron acquisition systems associated with fitness/virulence of UPEC strains have been identified in Nissle 1917, i.e. salmochelin, aerobactin, haemin and yersiniabactin (Grozdanov et al. 2004). All these systems were up-regulated during planktonic and biofilm growth in urine of Nissle 1917 (Fig. 3). The iroBCDEN, sitABCD, iucABCD and iutA genes were significantly up-regulated both in urine planktonic growth and biofilm in all three strains up to 56-fold. The chuASTWXYU genes were up-regulated in urine planktonic growth in all strains. The yersiniabactin system encoded on the genes on high pathogenicity island (HPI) was highly up-regulated during urine growth of Nissle 1917 in planktonic and biofilm growth (2.2–66-fold). The fec system, which is a citrate-dependent iron uptake system found in K-12 but missing in CFT073 and other UPEC strains, was up-regulated in Nissle 1917 (2.7–11-fold); these genes showed no change in strain 83972 during the conditions investigated here, but have previously been reported up-regulated in vivo in the urinary tract (Roos and Klemm 2006).
Signal intensity of iron acquisition and uptake genes of Nissle 1917, CFT073 and 83972 during planktonic growth in human urine. The signal intensity is calculated from triplicate microarrays of each strain using dChip. For Nissle 1917 all genes except fhuB were significantly up-regulated in urine compared with minimal medium. For CFT073 all genes except the fec genes and c2426-c2434 were significantly induced. For 83972 all genes except fhuB, fecBCDE, feoB and the majority of the yersiniabactin genes were significantly induced
Genes up-regulated in urine biofilms
In total, 277 genes were up-regulated during biofilm formation in human urine in all three strains compared with planktonic growth in human urine (Fig. 1c). Of these 277 biofilm-specific genes, 150 encoded hypothetical proteins. Twenty of the two hundred and seventy-seven genes were previously identified as belonging to the thirty most up-regulated genes during biofilm formation in urine of two UTI isolates (Hancock and Klemm 2007) (Table 1).
Up-regulation of stress genes is a common trend in biofilm transcriptome studies (Wood 2009) and apart from the cold and heat shock genes (cspAG and ibpAB) several other stress-related genes were up-regulated in Nissle 1917, CFT073 and 83972 during biofilm formation compared with planktonic growth in urine, i.e., bhsA, oxyS, hmp, ffs, ytfE, grxA, trxC, yhaK and marR were significantly induced (Table 1).
UPEC-associated adhesins are down-regulated in urine biofilm
Three fimbrial species are strongly associated with UTI E. coli isolates, viz. type 1 fimbriae, P fimbriae and F1C fimbriae, of these type 1 fimbriae in particular are associated with biofilm formation as well. In 83972 all three are non-functional due to a large deletion in the fim cluster and several point mutations in the pap and foc cluster (Klemm et al. 2006; Roos et al. 2006a). The fim genes encoding type 1 fimbriae have been reported up-regulated in CFT073 grown in vivo in mice compared with static LB cultures (Snyder et al. 2004). Interestingly, none of the fim genes were up-regulated in CFT073, Nissle 1917 or 83972 during planktonic growth in urine compared with planktonic growth in minimal lab medium. Instead, in CFT073 the fimAIC genes were down-regulated 3.7–5.8-fold and the regulatory fimB gene was down-regulated 9.9-fold. Similarly, the fimAIC and fimB genes were down-regulated in urine biofilms of CFT073 2.1–6.5-fold and 6.5-fold, respectively, compared with planktonic growth in MOPS, in accordance with this, the regulatory fimE gene, promoting OFF orientation of the fim switch, was induced 2.1-fold in urine biofilms. Also, Nissle 1917 showed significant up-regulation 3.0-fold of the fimE gene during biofilm formation in urine (all other fim genes showed no change and no significant expression levels in all three conditions in Nissle 1917).
Expression of F1C fimbriae has been reported to play a role in biofilm formation of Nissle 1917 (Lasaro et al. 2009). However, in urine the genes encoding F1C fimbriae were significantly down-regulated in Nissle 1917 biofilms compared with planktonic growth (3.1–26-fold). In CFT073, the focA gene was down-regulated 16-fold in biofilm compared with planktonic growth and the regulatory sfaB gene was repressed 70-fold. However, all genes showed the highest expression during planktonic growth in urine in both Nissle 1917 and CFT073; compared with planktonic growth in MOPS, the foc/sfa genes were up-regulated 1.8–6.5-fold in Nissle 1917 and CFT073. Taken together, the results indicate that expression of F1C fimbriae does not play an important role during late stages of biofilm growth of Nissle 1917 or CFT073 in human urine.
The pap gene cluster is virtually absent in Nissle 1917, only a fragment of papA and the intact papI gene are present (Grozdanov et al. 2004). In CFT073, however, the whole pap gene cluster was up-regulated 2.5–7.6-fold in planktonic urine compared with planktonic MOPS, but showed no up-regulation in biofilms, instead it was down-regulated 2.5–47-fold, suggesting that P fimbriae have no role during biofilm growth of CFT073.
Interestingly, while the three UPEC-associated fimbrial gene clusters were down-regulated during biofilm growth in urine, a number of other fimbrial or putative fimbrial genes showed up-regulation in urine biofilms compared with urine planktonic growth. The yadC gene encoding a putative fimbrial-like protein was up-regulated 7.0–11-fold in Nissle 1917, CFT073 and 83972 biofilms; the adjacent gene yadK showed 2.2–3.4-fold up-regulation in the three strains. The aufDEG genes belonging to the aufA-G fimbrial gene cluster showed up-regulation 1.8–2.5-fold in biofilms compared with planktonic growth in MOPS in both CFT073 and Nissle 1917.
UPEC-associated genes expressed in Nissle 1917
Grozdanov et al. have previously reported high similarity between the pathogenicity islands of Nissle 1917 and those of CFT073 (Grozdanov et al. 2004) and recently, comparative genomic hybridisation (CGH) analysis showed that a wide range of virulence or fitness factors were present in Nissle 1917 and 83972 (Vejborg et al. 2010). Here, the expression data revealed expression of several virulence-associated factors. The urovirulence-associated iha gene was up-regulated in Nissle 1917 both during growth in planktonic urine and urine biofilm. The curli genes csgDEF were up-regulated 1.7–2.3-fold in Nissle 1917 grown in planktonic urine. The microcin genes mchBCDF were up-regulated in human urine 1.8–5.0-fold in Nissle 1917. The capsule-encoding gene kpsM was down-regulated in both planktonic urine and urine biofilms of Nissle 1917, while the kpsT gene showed no change; the kspMT genes showed down-regulation in CFT073 planktonic urine and urine biofilm.
Chen et al. have identified 29 genes that are under positive selection only in UPEC strains (Chen et al. 2006). We analysed the expression levels of these 29 UPEC-specific genes in Nissle 1917. Of the 29 genes, 28 turned out to be significantly changed (up- or down-regulated) demonstrating that these genes are expressed by Nissle 1917 during at least one of the growth conditions. The exception was the yojI gene, encoding a hypothetical ABC transporter, which showed no significant change and low expression during the three conditions tested in Nissle 1917. However, this gene was filtered present in Nissle 1917 by our CGH analysis (data not shown). In strain 83972, 25 of these 29 genes have previously been filtered present based on expression data analysis (Hancock et al. 2008b); here, we found that 22 of the genes were significantly changed, i.e., expressed during at least one growth condition. Also, the CGH analysis revealed that the remaining four genes, which were previously filtered ‘absent’ based on expression data analysis, are most likely present in 83972 (data not shown). These results support the previous studies where Nissle 1917 has shown to be closely related to UPEC strains—in particular to the CFT073 strain; moreover, the results reveal that the majority of UPEC-related genes present in Nissle 1917 and 83972 are also expressed by the strains grown under the conditions investigated here.
Differences between a pathogen and two probiotic strains: putative virulence factors
Genes that are up-regulated or highly expressed during growth in urine of the pathogenic strain CFT073 but that are either down-regulated or show no change and low expression levels in the two probiotic strains Nissle 1917 and 83972 could be regarded as potential virulence factors. There was only one gene that was up-regulated in CFT073 and down-regulated in Nissle 1917 and 83972 during planktonic growth in urine compared with minimal medium, i.e., c0651 encoding a putative methyltransferase. Analysis of genes that were significantly higher expressed in CFT073 grown in urine compared with the two probiotic isolates Nissle 1917 and 83972 resulted in a total number of 282 genes, whereof the 50 highest expressed are displayed in Table 2. These genes included the idnRTODK genes encoding proteins involved transport and catabolism of l-idonate which were up-regulated up to 41-fold. The majority of the 282 genes are located on pathogenicity islands of CFT073. The pap genes encoding P fimbriae belonged to this group of genes specifically expressed in the pathogenic strain, as did the ireA gene encoding a siderophore receptor associated with virulence of UPEC isolates which was induced 8.4-fold. The aec-35 gene encoding a putative transcriptional regulator which has been associated with virulence in avian pathogenic E. coli (Chouikha et al. 2006) was one of the highest expressed of the CFT073-specific genes (Table 2). In addition, the aec-35 to aec-37 gene cluster, associated with carbohydrate assimilation, was significantly induced in CFT073 grown in urine, while no expression was seen in the two probiotic strains. The 282 genes also included the haemolysin genes, hlyACD, and three colibactin genes, clbALM. Of the 50 genes listed in Table 2, 22 are genes encoding hypothetical proteins located on genomic islands and phage regions. Five pairs of adjacent genes are found among these: c3205-6, c3694-5, c4509-10, c4545-6 and c5163-4. Potentially, these genes found significantly higher expressed in the uropathogen CFT073 during growth in human urine compared with the two probiotic strains Nissle 1917 and 83972 could be important for the virulence in the human urinary tract.
Comparing biofilm formation in urine with planktonic growth in urine resulted in only three genes that were up-regulated in CFT073 but down-regulated in Nissle 1917 and 83972, i.e., araB, recA and yqjC (encoding a ribulokinase, recombinase A and a hypothetical protein, respectively), and 186 genes that were up in CFT073 biofilms but showed no change in the two probiotic strains. The 25 most up-regulated of these 186 genes are displayed in Supplemental Table 2. Only one gene associated with virulence was identified among these genes, i.e., the hlyE gene encoding a pore-forming toxin, haemolysin E, first identified in E. coli K-12 (Oscarsson et al. 1996) which was induced 2.2-fold in CFT073 biofilm. Interestingly, many more virulence factors were identified highly expressed exclusively in CFT073 grown in urine compared with expression in urine biofilms, suggesting that human urine induces expression of a considerably larger number of virulence factors than the biofilm growth mode.
Highly expressed genes during growth in urine: similarities and differences
Analysis of the 50 highest expressed genes in each of the strains CFT073, Nissle 1917 and 83972 revealed some interesting differences between the three strains. Of the 50 highest expressed genes in CFT073 grown planktonically in human urine (Supplemental Table 3), the majority were highly expressed in Nissle 1917 and 83972 as well; 24 and 22 of the genes belonged to the 50 highest expressed in Nissle 1917 and 83972, respectively. These included genes involved in iron acquisition (sitA, iucC, iucD and iroB) and central metabolism (icdA, mdh, sucB and sucC), as well as ompA and sodA. However, 12 genes that were highly expressed in CFT073 showed significantly lower expression in the other two strains. These were the fucI, fucU, tnaA, glcB, idnD and xylABFGH genes and the two papA variants. All the fucAPIK genes involved in transport and degradation of l-fucose were significantly higher expressed in CFT073 compared with Nissle 1917 and 83972.
Analysis of the 50 highest expressed genes in Nissle 1917 grown planktonically in human urine identified 12 genes as significantly lower expressed in CFT073 and 83927; these were the hisABCDFG, flgBCE, fliC and rbsBD genes involved in histidine biosynthesis, flagellar biosynthesis and ribose transport and degradation. In fact, all flagellar flg genes (flgA-N) showed significantly higher expression in urine of Nissle 1917 than in CFT073 and 83972 (6.9–182-fold). Similarly, the fliC-T, flhAB and motAB genes all involved in flagellar biosynthesis were higher expressed in urine growth of Nissle 1917 (2.6–119-fold) compared with CFT073 and 83972.
Analysis of the 50 highest expressed genes in strain 83972 grown planktonically in human urine only identified two genes that were significantly lower expressed in CFT073 and Nissle 1917, i.e., ygbJ and ygbM. Interestingly, the adjacent genes were all significantly higher expressed in 83972 than the other two strains; the ygbJKLMN genes, encoding hypothetical proteins (b2736-40), were all 16–52-fold higher expressed in 83972.
These results indicate that the three strains employ different carbohydrate and amino acid metabolic systems even though grown in the same medium, i.e., pooled human urine. For instance, CFT073 showed induced levels of genes for import and degradation of d-xylose and l-fucose (during planktonic growth in urine, but not during biofilm formation in urine), while Nissle 1917 showed high expression of genes involved in ribose transport and degradation (in urine both planktonically and biofilm).
Competition in planktonic growth mode
Growth characteristics were determined for Nissle 1917, CFT073 and 83972 in three different media, i.e., MOPS minimal medium, pooled human urine and LB rich medium. All strains grew well in all media, also Nissle 1917, a faecal isolate, performed well in urine (Fig. 4a). Pairwise competition of the strains was performed in MOPS, urine and LB. Strains were competed against each other with an initial ratio 1:1 in shake flasks at 37°C for 16 h. It turned out that even though the better growing strain won the competition in most cases (Figs. 4a, 5a), this was not always the case. Even though CFT073 showed significantly better growth characteristics in urine, i.e., reaching a higher final cell density and displaying a higher growth rate, than Nissle 1917, it was significantly out-competed by Nissle 1917 in urine where it constituted only 11% of the population at the end of the experiment. Another exception was seen in the competition between 83972 and CFT073. In LB, 83972 displayed better growth characteristics than that of CFT073 but was still out-competed constituting only 26% of the mixed population at the end of the competition. Interestingly, both probiotic strains out-competed CFT073 in urine. Our results show that even though growth rate is an important characteristic for competition in the planktonic growth mode, there are most certainly other factors that play a great role in competition between two strains—factors that seem to be strain specific as well as media dependent.
Growth characteristics and biofilm formation of strains Nissle 1917, 83972 and CFT073 in minimal lab medium (MOPS), pooled human urine and rich lab medium (LB). a Growth curves shown as means of triplicates for each strain. Error bars indicate standard deviation σ(n−1). b Biofilm formation in microtitre plates shown as means of at least triplicates. Error bars indicate standard deviation σ(n−1). Strains CFT073 and Nissle 1917 formed significantly less biofilm compared with strain 83972 in all three media (paired t test, P < 0.001). c Biofilm flow-chamber graphs of 83972, CFT073 and Nissle 1917 after 24 h in human urine. The scale bars represent 10 μm
Competition in a planktonic growth mode and in b biofilm growth mode. In all competitions two of the strains Nissle 1917, CFT073 and 83972 were mixed with a starting ratio 1:1. After 16 h of competition, cells were extracted and CFU determined. Results are means of triplicates from three independent experiments and error bars indicate standard deviation σ(n−1). Asterisks indicate results where one strain significantly outcompeted the other (paired t test, P < 0.01)
Competition during biofilm growth
Comparing the biofilm-forming capability of the three strains in three different media revealed no significant differences in biofilm formation of a specific strain. In all three media, i.e., MOPS, human urine and LB, the ABU isolate 83972 was the best biofilm former, while Nissle 1917 formed significantly more biofilm than the UPEC isolate CFT073 in all media tested (Fig. 4b). Interestingly, although Nissle 1917 and 83972 were better biofilm formers in all three growth media, they were not able to out-compete CFT073 during biofilm formation in all cases (Fig. 5b). On the contrary, CFT073 out-competed Nissle 1917 in all three media constituting 71, 64 and 81% of the population after 16 h of co-culture in MOPS, urine and LB, respectively. As observed previously (Ferrières et al. 2007), 83972 managed to out-compete CFT073 during biofilm formation in urine; however, no significant difference was observed in MOPS, and in LB, CFT073 greatly out-competed 83972. Despite the fact that 83972 formed significantly more biofilm than Nissle 1917 in all three media, out-competition was only observed in urine where 83972 constituted 77% of the population at the end of the competition experiment. Taken together, the results reveal that the individual biofilm formation of a strain does not correlate with the ability to compete; also, the media greatly influences the outcome of a competition experiment.
YhaK, YhcN and YbiJ significantly influence biofilm formation
Four genes, which were highly up-regulated during biofilm formation in all three strains (Table 1), were selected for knock-out mutagenesis. Four, three and three positive knock-out mutants of Nissle 1917, CFT073 and 83972, respectively, were obtained and subsequently analysed for biofilm formation in microtitre plates in urine and minimal medium (Fig. 6). None of the mutants showed impaired planktonic growth in urine or minimal medium compared with wild-type strain. Three mutants of Nissle 1917, i.e., Nissle 1917∆yhaK, Nissle 1917∆yhcN and Nissle 1917∆ybiJ, reduced biofilm formation in human urine by 15, 29 and 35%, respectively, compared with wild-type. Two mutants of CFT073, i.e. CFT073∆yhcN and CFT073∆ybiJ, reduced biofilm formation in human urine by 25 and 34%, respectively. In minimal lab medium, the majority of the mutants also significantly reduced biofilm formation albeit to a lesser extent (Fig. 6b); however, in the case of Nissle 1917∆yhaK, no significant reduction in biofilm formation was observed in minimal medium. It turned out that the two mutants CFT073∆yhcN and CFT073∆ybiJ displayed significantly reduced motility in urine–agar plates (Fig. 7). Despite that all genes selected for knock-out mutagenesis were induced in all three strains during biofilm growth, none of the three mutants of 83972, i.e. 83972∆yhaK, 83972∆yhcN and 83972∆ybiJ, showed any significant change in biofilm formation compared with wild-type (Fig. 6).
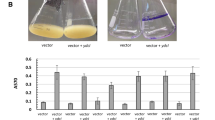
Biofilm formation in polystyrene microtitre plates in a human urine and b minimal medium of knock-out mutants of Nissle 1917, CFT073 and 83972. The cells were incubated for 16 h at 37°C and A595 was measured after staining with crystal violet and washing with PBS. Values are shown relative to wild-type and are means of at least three independent experiments. Error bars indicate standard deviation (σn−1). The mutants that formed significantly less biofilm compared with corresponding wild-type strain are indicated with asterisks (paired t test, *P < 0.05 and **P < 0.01)
Motility on urine agar plates of CFT073, CFT073∆yhcN and CFT073∆ybiJ. Both mutants showed impaired motility compared with parent and CFT073∆ybiJ showed signs of aggregation (enlarged panels). CFT073∆yhcN showed reduced motility by 34% (paired t test, P < 0.01) and CFT073∆ybiJ was non-motile. CFT073∆yhcN and CFT073∆ybiJ complemented with plasmids expressing yhcN and ybiJ showed motility similar to that of the wild-type, i.e. 1.03 ± 0.20 and 1.12 ± 0.24 (mean ± SE) of that of CFT073(pACYC184), respectively (paired t test, P > 0.90)
The biofilm-related gene bssS has been shown to regulate biofilm through quorum sensing (Domka et al. 2006). Here, Nissle 1917∆bssS showed a 10% significant reduction in biofilm formation in minimal medium compared with wild-type, while no significant difference was observed for CFT073∆bssS; furthermore, neither mutant showed any significant difference in biofilm formation in human urine despite up-regulation of the gene in this medium (2.7- and 3.7-fold for the two strains) (Fig. 6).
To fulfill molecular Koch’s postulates, all mutants that showed significantly reduced biofilm formation compared with wild-type were complemented with a plasmid expressing the corresponding deleted gene. None of the five plasmid-complemented mutants showed any significant reduction in biofilm formation or motility (Fig. 7) compared with wild-type (pACYC184) (paired t test, P > 0.05).
RT-PCR
The garD, rspB, yhaK, yhcN and yjfY transcripts, which were highly expressed and induced in all three strains either in urine planktonic growth (garD and rspB) or in urine biofilms (yhaK, yhcN and yjfY), were selected for RT-PCR. After 15–45 cycles of PCR, products of the transcripts were visualised on agarose gels. The strength of the gel band from each sample corresponded well to the actual signal intensities recorded from the corresponding microarray. 16S was used as a normalising internal standard and was detected with similar intensity in all samples.
Discussion
Escherichia coli, a versatile microbe capable of colonisation of the intestinal tract as well as the urinary tract, can cause life-threatening disease or exist in peaceful co-existence with the host as a commensal. Examples are known of commensal strains that show fitness advantages over the pathogenic strains, making them useful as probiotics. Here, we compare three seemingly very different E. coli strains, (1) a probiotic gut-colonising strain, Nissle 1917, (2) a probiotic bladder-colonising strain, 83972, and (3) a bona fide uropathogen capable of urinary tract colonisation and sepsis, CFT073. Both probiotic strains have been shown to out-compete pathogenic strains in their respective habitats and they are both able to express numerous fitness factors. Both Nissle 1917 and 83972 are harmless vis-à-vis the host. But both have armamentaria of fitness factors, and these are presumably the major cause of their success as probiotics. Despite the benign characteristics of Nissle 1917 and 83972, we show that they express numerous genes, which are often referred to as virulence genes. It should be pointed out that the data do not reveal whether the virulence-associated genes expressed by the two probiotic strains are in fact functional or not. Genes can be inactivated through point mutations rendering them dysfunctional. Genome sequencing of Nissle 1917 and 83972 could provide this information. However, the information reported herein complements sequencing of these strains—genes can also be silenced not only due to lesions in the actual gene and its promoter but also due to mutation of genes encoding regulatory factors. The data presented here provides insight in the active genome of these strains helping us to better distinguish between fitness factors and ‘true’ virulence factors.
All whole-transcriptome profiling studies on E. coli biofilms have up to now been performed on K-12 (Schembri et al. 2003; Beloin et al. 2004; Ren et al. 2004; Junker et al. 2006) or asymptomatic bacteriuria strains (Hancock and Klemm 2007). This is the first study analysing global gene expression in biofilms of a pathogenic strain, i.e., the UPEC isolate CFT073. Furthermore, this is the first global gene expression profiling analysis of the E. coli isolate Nissle 1917 used in the probiotic drug Mutaflor®. The gene expression data revealed that many fitness/urovirulence genes were expressed in UPEC strain CFT073 as well as in the two probiotic strains Nissle 1917 and 83972. In the case of genes involved in iron acquisition Nissle 1917 seemed particularly well equipped compared with the two UTI isolates; while many gene clusters involved in iron uptake were induced during growth in human urine in all three strains, the yersiniabactin gene cluster and the citrate-dependent iron uptake system encoded by the fec genes were induced only in Nissle 1917 (Fig. 3). Induction of all these iron systems could provide the explanation for that Nissle 1917 grows well in urine and outcompetes CFT073 during planktonic growth in this medium (Figs. 4a, 5a).
The virulence-associated iha, vat and sat genes, the mchBCD microcin genes and the colibactin genes were induced in all three strains grown in urine. As previously mentioned, the virulence-associated genes expressed by the two probiotic strains might be functional or not—this remains to be investigated in order to determine whether they are fitness factors or ‘true’ virulence factors. On the other hand, a few genes induced and/or highly expressed in CFT073, but not in the two probiotic strains, stand out as potential virulence factors, i.e., the ireA and pap genes encoding an iron siderophore receptor important for virulence (Russo et al. 2001; Hagan and Mobley 2007) and P fimbriae, respectively, also the hlyAC haemolysin genes, aec-35 and picU were induced only in CFT073.
The high pathogenicity island (HPI) is widespread and found in a range of bacterial species, in both pathogenic and non-pathogenic E. coli (Johnson et al. 2001). Interestingly, although the HPI gene cluster of strain CFT073 contains in-frame stop codons and the strain does not produce detectible yersiniabactin (Brzuszkiewicz et al. 2006), it was recently discovered that the HPI of CFT073 significantly contributed to the colonisation of the mouse kidney; an HPI mutant was significantly outcompeted by wild-type CFT073 (exhibiting a 1-log-unit decrease) in the kidneys following transurethral cochallenge in a mouse model (Lloyd et al. 2009). In line with this, in the present study we found the ferric yersiniabactin receptor gene fyuA up-regulated 25-fold in CFT073 grown in human urine, indicating that FyuA might play a role for the fitness of the strain in urine despite the mutations in the yersiniabactin synthesis genes. Recently, knockout mutants of fyuA have shown significant reduction of biofilm formation in human urine (Hancock et al. 2008a); nevertheless, the exact role of the receptor in the absence of yersiniabactin remains to be elucidated. Transcriptomics data from in vivo samples of 83972 showed an up-regulation of fyuA up to 25-fold indicating a role for this iron uptake receptor in the human urinary tract (Roos and Klemm 2006).
Gene expression analysis identified three new genes playing a significant role in biofilm formation. Nissle 1917 carrying a deletion in the yhaK gene, encoding a bicupin which may be involved in sensing oxidative stress sensing (Gurmu et al. 2009), formed significantly less biofilm in human urine. Single knock-out mutants of YhcN and YbiJ in Nissle 1917 and CFT073 displayed significantly reduced biofilm formation in both minimal medium and human urine. yhcN and ybiJ belong to the same family (viz. the YhcN family) of genes encoding small low-molecular-weight proteins as the recently identified bhsA and bsmA genes which also have been shown to be involved in E. coli biofilm formation (Zhang et al. 2007; Weber et al. 2009). The bhsA and bsmA genes both influence biofilm through stress response; in addition, bhsA affects the surface hydrophobicity and bsmA flagellar motility (Zhang et al. 2007; Weber et al. 2009). Here, CFT073∆yhcN and CFT073∆ybiJ showed reduced motility compared with parent strain in urine–agar plates. In agreement with this, motility has been shown play a significant role in biofilm formation by influencing the architecture of E. coli biofilms where reduced motility resulted in significantly reduced biofilm mass and smoother colonies (Wood et al. 2006). Furthermore, decreased biofilm formation of a non-motile mutant has been observed in numerous studies (Kirov et al. 2004; Lemon et al. 2007; Merritt et al. 2007; Kim et al. 2008) and is in line with the observation that motility is required for the initial attachment in biofilm formation (O’Toole and Kolter 1998). Thus, expression of flagella might be crucial in the attachment stage and subsequently down-regulated to promote a sessile life style, which is in agreement with our finding here—also, no flagellar genes were expressed in our 42-h-old biofilms.
Expression levels of the three genes highly expressed in all three strains, yhaK, yhcN and ybiJ, affecting biofilm formation did not correspond with the biofilm-forming ability of the strains. Two of the genes showed highest expression levels in 83972 which also formed most biofilm; on the other hand, all three genes showed the lowest expression levels in Nissle 1917 which made significantly more biofilm than CFT073. Despite the high expression levels in 83972, the three knock-out mutants of 83972 showed no reduction in biofilm formation compared with wild-type, whereas three and two of the genes affected biofilm in Nissle 1917 and CFT073, respectively. These results suggest that the mechanisms underlying the biofilm formation are different in the three different strains, as supported by the significantly different capabilities to form biofilm (Fig. 4b, c). Additional mechanisms involved in biofilm formation of 83972 most likely cover for the lack of any of these three genes—if they at all are involved in biofilm formation of this strain (although indicated by the up-regulation). Also, 83972 is non-motile in urine (Hancock and Klemm 2007), wherefore any effect on motility through lost expression of yhcN and ybiJ will have no impact. Altogether our data suggest that, despite the close genetic relationship between the three E. coli strains, as well as the large similarity in gene expression profiles and the identical conditions for biofilm formation, the biofilm forming ability of the strains is influenced by different mechanisms. To date, many genes have been identified to be involved in biofilm formation of E. coli—however, the majority of these have been investigated in a single strain background and under a few or a single environmental condition. Here, we clearly show that biofilm formation is a trait dependent not only on surface conditions, hydrodynamics, temperature and growth media, but is also highly dependent on seemingly minor strain differences.
The YhcN family consists of nine paralogous small proteins that are strongly predicted to have signal peptides facilitating their transport across the inner membrane and the probable ancestor of the YhcN family is thought to have been involved in self-identification or colony organisation by cell–cell contacts or intercellular signalling (Rudd et al. 1998). With our results, four of these nine proteins have been shown to influence biofilm formation. Seven of the members share a motif in their N-terminal domains that one of the members, YjfY, lacks (Rudd et al. 1998). Interestingly, four members sharing this motif all affect biofilm formation (BhsA, BsmA, YhcN and YbiJ) while knockout mutants of YjfY (i.e., CFT073ΔyjfY and Nissle 1917ΔyjfY) did not show any affect on biofilm formation (data not shown).
This is the first report on Nissle 1917’s ability to grow and compete in human urine. Although Nissle 1917 has been reported to confer probiotic effects in the intestinal tract, no report on the effects of Nissle 1917 entering the human urinary tract has been published. A recently published study using a rat transurethral infection model indicated that Nissle 1917 only has a limited potential for colonising the bladder and kidney (similar to that of an avirulent laboratory E. coli K-12 strain) with no lethal effects on the rats (Sonnenborn and Schulze 2009). However, the strain was able to ascend to the kidneys, which is never observed with 83972. Studies on the effects of Nissle 1917 in the human intestinal tract lack information regarding incidents of urinary tract infection in the subjects digesting Nissle 1917. However, considering the transcriptomic closeness to CFT073 and expression of several uropathogenic-associated genes it cannot be ruled out that Nissle 1917 might cause symptoms in the human urinary tract and resemble CFT073 rather than the asymptomatic strain 83972.
Although the three strains have very different strategies vis-à-vis the human host their functional gene profiles are surprisingly similar. From our data it appears that only a few hundred active genes seem to separate the highly pathogenic strain CFT073 from the two probiotic strains. It is also clear that both Nissle 1917 and 83972 are descendants of uropathogens that resembled CFT073 closely but have mellowed out and become commensals through genome erosion. The fitness genes seem to have stayed intact. This is also reflected in the fact that although Nissle 1917 is a gut isolate it has not lost its ability to grow well in urine and can even outcompete CFT073. It is also interesting, and sobering, to note that the only two E. coli strains used as probiotics are in fact deconstructed pathogens.
References
Andersson P, Engberg I, Lidin-Janson G, Lincoln K, Hull R, Hull S, Svanborg C (1991) Persistence of Escherichia coli bacteriuria is not determined by bacterial adherence. Infect Immun 59:2915–2921
Beloin C, Valle J, Latour-Lambert P, Faure P, Kzreminski M, Balestrino D, Haagensen JA, Molin S, Prensier G, Arbeille B, Ghigo JM (2004) Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol Microbiol 51:659–674
Brzuszkiewicz E, Brüggemann H, Liesegang H, Emmerth M, Ölschläger T, Nagy G, Albermann K, Wagner C, Buchrieser C, Emody L, Gottschalk G, Hacker J, Dobrindt U (2006) How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc Natl Acad Sci USA 103:12879–12884
Chen SL, Hung CS, Xu J, Reigstad CS, Magrini V, Sabo A, Blasiar D, Bieri T, Meyer RR, Ozersky P, Armstrong JR, Fulton RS, Latreille JP, Spieth J, Hooton TM, Mardis ER, Hultgren SJ, Gordon JI (2006) Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc Natl Acad Sci USA 103:5977–5982
Chouikha I, Germon P, Brée A, Gilot P, Moulin-Schouleur M, Schouler C (2006) A selC-associated genomic island of the extraintestinal avian pathogenic Escherichia coli strain BEN2908 is involved in carbohydrate uptake and virulence. J Bacteriol 188:977–987
Christensen BB, Sternberg C, Andersen JB, Palmer RJ Jr, Nielsen AT, Givskov M, Molin S (1999) Molecular tools for study of biofilm physiology. Methods Enzymol 310:20–42
Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM (1995) Microbial biofilms. Annu Rev Microbiol 49:711–745
Costerton JW, Stewart PS, Greenberg EP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645
Domka J, Lee J, Wood TK (2006) YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl Environ Microbiol 72:2449–2459
Ferrières L, Hancock V, Klemm P (2007) Biofilm exclusion of uropathogenic bacteria by selected asymptomatic bacteriuria Escherichia coli strains. Microbiology 153:1711–1719
Grozdanov L, Raasch C, Schulze J, Sonnenborn U, Gottschalk G, Hacker J, Dobrindt U (2004) Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J Bacteriol 186:5432–5441
Gurmu D, Lu J, Johnson KA, Nordlund P, Holmgren A, Erlandsen H (2009) The crystal structure of the protein YhaK from Escherichia coli reveals a new subclass of redox sensitive enterobacterial bicupins. Proteins 74:18–31
Hagan EC, Mobley HL (2007) Uropathogenic Escherichia coli outer membrane antigens expressed during urinary tract infection. Infect Immun 75:3941–3949
Hancock V, Klemm P (2007) Global gene expression profiling of asymptomatic bacteriuria Escherichia coli during biofilm growth in human urine. Infect Immun 75:966–976
Hancock V, Ferrières L, Klemm P (2007) Biofilm formation by asymptomatic and virulent urinary tract infectious Escherichia coli strains. FEMS Microbiol Lett 267:30–37
Hancock V, Ferrières L, Klemm P (2008a) The ferric yersiniabactin uptake receptor FyuA is required for efficient biofilm formation by urinary tract infectious Escherichia coli in human urine. Microbiology 154:167–175
Hancock V, Seshasayee AS, Ussery DW, Luscombe NM, Klemm P (2008b) Transcriptomics and adaptive genomics of the asymptomatic bacteriuria Escherichia coli strain 83972. Mol Gen Genet 279:523–534
Hancock V, Dahl M, Klemm P (2010) Probiotic Escherichia coli strain Nissle 1917 out-competes intestinal pathogens during biofilm formation. J Med Microbiol 59:392–399
Hull R, Rudy D, Donovan W, Svanborg C, Wieser I, Stewart C, Darouiche R (2000) Urinary tract infection prophylaxis using Escherichia coli 83972 in spinal cord injured patients. J Urol 163:872–877
Johnson JR, Delavari P, Kuskowski M, Stell AL (2001) Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J Infect Dis 183:78–88
Junker LM, Peters JE, Hay AG (2006) Global analysis of candidate genes important for fitness in a competitive biofilm using DNA-array-based transposon mapping. Microbiology 152:2233–2245
Kim T-J, Young BM, Young GM (2008) Effect of flagellar mutations on Yersinia enterocolitica biofilm formation. Appl Environ Microbiol 74:5466–5474
Kirov SM, Castrisios M, Shaw JG (2004) Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infect Immun 72:1939–1945
Klemm P, Roos V, Ulett GC, Svanborg C, Schembri MA (2006) Molecular characterization of the Escherichia coli asymptomatic bacteriuria strain 83972: the taming of a pathogen. Infect Immun 74:781–785
Klemm P, Hancock V, Schembri MA (2007) Mellowing out: adaptation to commensalism by Escherichia coli asymptomatic bacteriuria strain 83972. Infect Immun 75:3688–3695
Lasaro MA, Salinger N, Zhang J, Wang Y, Zhong Z, Goulian M, Zhu J (2009) F1C fimbriae play an important role in biofilm formation and intestinal colonization by the Escherichia coli commensal strain Nissle 1917. Appl Environ Microbiol 75:246–251
Lemon KP, Higgins DE, Kolter R (2007) Flagellar motility is critical for Listeria monocytogenes biofilm formation. J Bacteriol 189:4418–4424
Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol 2:1
Lindberg U, Hanson LA, Jodal U, Lidin-Janson G, Lincoln K, Olling S (1975) Asymptomatic bacteriuria in schoolgirls. II. Differences in Escherichia coli causing asymptomatic bacteriuria. Acta Paediatr Scand 64:432–436
Lloyd AL, Henderson TA, Vigil PD, Mobley HLT (2009) Genomic islands of uropathogenic Escherichia coli contribute to virulence. J Bacteriol 191:3469–3481
Lodinova-Zadnikova R, Sonnenborn U (1997) Effect of preventive administration of a nonpathogenic Escherichia coli strain on the colonization of the intestine with microbial pathogens in newborn infants. Biol Neonate 71:224–232
Lodinova-Zadnikova R, Bartakova Z, Tlaskalova H (1992) The effect of oral colonization by non-pathogenic E. coli on the immune response in neonates and possibilities of its use in the prevention of nosocomial infections in children at risk. Cesk Epidemiol Mikrobiol Imunol 42:126–132
Mabbett AN, Ulett GC, Watts RE, Tree JJ, Totsika M, Ong CL, Wood JM, Monaghan W, Looke DF, Nimmo GR, Svanborg C, Schembri MA (2009) Virulence properties of asymptomatic bacteriuria Escherichia coli. Int J Med Microbiol 299:53–63
Macfarlane S, Dillon JF (2007) Microbial biofilms in the human gastrointestinal tract. J Appl Microbiol 102:1187–1196
Merritt PM, Danhorn T, Fuqua C (2007) Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. J Bacteriol 189:8005–8014
Mobley HLT, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, Warren JW (1990) Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect Immun 58:1281–1289
Neidhardt FC, Bloch PL, Smith DF (1974) Culture Medium for Enterobacteria. J Bacteriol 119:736–747
O’Toole GA, Kolter R (1998) Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304
O’Toole G, Kaplan HB, Kolter R (2000) Biofilm formation as microbial development. Annu Rev Microbiol 54:49–79
Oscarsson J, Mizunoe Y, Uhlin BE, Haydon DJ (1996) Induction of haemolytic activity in Escherichia coli by the slyA gene product. Mol Microbiol 20:191–199
Ren D, Bedzyk LA, Thomas SM, Ye RW, Wood TK (2004) Gene expression in Escherichia coli biofilms. Appl Microbiol Biotechnol 64:515–524
Roos V, Klemm P (2006) Global gene expression profiling of the asymptomatic bacteriuria Escherichia coli strain 83972 in the human urinary tract. Infect Immun 74:3565–3575
Roos V, Schembri MA, Ulett GC, Klemm P (2006a) Asymptomatic bacteriuria Escherichia coli strain 83972 carries mutations in the foc locus and is unable to express F1C fimbriae. Microbiology 152:1799–1806
Roos V, Ulett GC, Schembri MA, Klemm P (2006b) The asymptomatic bacteriuria Escherichia coli strain 83972 out-competes UPEC strains in human urine. Infect Immun 74:615–624
Rudd KE, Ian H-S, Valerie CW, Amos B (1998) Low molecular weight proteins: a challenge for post-genomic research. Electrophoresis 19:536–544
Russo TA, Carlino UB, Johnson JR (2001) Identification of a new iron-regulated virulence gene, ireA, in an extraintestinal pathogenic isolate of Escherichia coli. Infect Immun 69:6209–6216
Schembri MA, Kjaergaard K, Klemm P (2003) Global gene expression in Escherichia coli biofilms. Mol Microbiol 48:253–267
Schultz M (2008) Clinical use of E. coli Nissle 1917 in inflammatory bowel disease. Inflamm Bowel Dis 14:1012–1018
Schulze J, Sonnenborn U (1995) Oral administration of a certain strain of live Escherichia coli for intestinal disorders? Infection 23:184–188
Snyder JA, Haugen BJ, Buckles EL, Lockatell CV, Johnson DE, Donnenberg MS, Welch RA, Mobley HL (2004) Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect Immun 72:6373–6381
Sonnenborn U, Schulze J (2009) The non-pathogenic Escherichia coli strain Nissle 1917—features of a versatile probiotic. Microb Ecol Health Dis 21:122–158
Sundén F, Håkansson L, Ljunggren E, Wullt B (2006) Bacterial interference–is deliberate colonization with Escherichia coli 83972 an alternative treatment for patients with recurrent urinary tract infection? Int J Antimicrob Agents 28S:S26–S29
Vejborg RM, Friis C, Hancock V, Schembri MA, Klemm P (2010) A virulent parent with probiotic progeny: comparative genomics of Escherichia coli strains CFT073, Nissle 1917 and ABU 83972. Mol Genet Genomics 283:469–484
Warren JW (2001) Catheter-associated urinary tract infections. Int J Antimicrob Agents 17:299–303
Weber MM, French CL, Barnes MB, Siegele DA, McLean RJ (2010) A previously uncharacterized gene, yjfO (bsmA) influences Escherichia coli biofilm formation and stress response. Microbiology 156:139–147
Welch RA, Burland V, Plunkett G 3rd, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, Stroud D, Mayhew GF, Rose DJ, Zhou S, Schwartz DC, Perna NT, Mobley HL, Donnenberg MS, Blattner FR (2002) Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA 99:17020–17024
Wood TK (2009) Insights on Escherichia coli biofilm formation and inhibition from whole-transcriptome profiling. Environ Microbiol 11:1–15
Wood TK, Gonzalez Barrios AF, Herzberg M, Lee J (2006) Motility influences biofilm architecture in Escherichia coli. Appl Microbiol Biotechnol 72:361–367
Zdziarski J, Svanborg C, Wullt B, Hacker J, Dobrindt U (2007) Molecular basis of commensalism in the urinary tract: low virulence or virulence attenuation? Infect Immun 76:695–703
Zhang XS, Garcia-Contreras R, Wood TK (2007) YcfR (BhsA) influences Escherichia coli biofilm formation through stress response and surface hydrophobicity. J Bacteriol 189:3051–3062
Acknowledgments
We thank Birthe Jul Bondo for expert technical assistance. This work was supported by grants from Lundbeckfonden (R17-A1603) and FSS (271-06-0555).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D. Andersson.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hancock, V., Vejborg, R.M. & Klemm, P. Functional genomics of probiotic Escherichia coli Nissle 1917 and 83972, and UPEC strain CFT073: comparison of transcriptomes, growth and biofilm formation. Mol Genet Genomics 284, 437–454 (2010). https://doi.org/10.1007/s00438-010-0578-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-010-0578-8