Abstract
Genes at the Rp1 rust resistance locus of maize confer race-specific resistance to the common rust fungus Puccinia sorghi. Three variant genes with nonspecific effects (HRp1 -Kr1N, -D*21 and -MD*19) were found to be generated by intragenic crossing over within the LRR region. The LRR region of most NBS-LRR encoding genes is quite variable and codes for one of the regions in resistance gene proteins that controls specificity. Sequence comparisons demonstrated that the Rp1-Kr1N recombinant gene was identical to the N-terminus of the rp1-kp2 gene and C-terminus of another gene from its HRp1-K grandparent. The Rp1-D*21 recombinant gene consists of the N-terminus of the rp1-dp2 gene and C-terminus of the Rp1-D gene from the parental haplotype. Similarly, a recombinant gene from the Rp1-MD*19 haplotype has the N-terminus of an rp1 gene from the HRp1-M parent and C-terminus of the rp1-D19 gene from the HRp1-D parent. The recombinant Rp1 -Kr1N, -D*21 and -MD*19 genes activated defense responses in the absence of their AVR proteins triggering HR (hypersensitive response) in the absence of the pathogen. The results indicate that the frequent intragenic recombination events that occur in the Rp1 gene cluster not only recombine the genes into novel haplotypes, but also create genes with nonspecific effects. Some of these may contribute to nonspecific quantitative resistance but others have severe consequences for the fitness of the plant.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The majority of disease resistance genes that have been characterized have been found to code for NBS-LRR proteins that confer race-specific resistance (McHale et al. 2006; Bozkurt et al. 2007). However, shifts in pathogen populations have proven that race-specific resistance is typically not durable. Quantitative disease resistance is considered more durable than simply inherited resistance but is controlled by multiple genes making it more difficult to manipulate genetically and it is generally not clear if the individual genes have nonspecific effects (Hooker 1967; Young 1996; Rosewarne et al. 2008). A few resistance genes have been identified that appear to have race-nonspecific effects, but they are not NBS-LRR genes (Brueggeman et al. 2002; Krattinger et al. 2009; Fu et al. 2009). Race-nonspecific resistance is considered to be the most durable type of resistance. It can sometimes be controlled by a small number of loci and sometimes confers resistance to multiple pathogens (Krattinger et al. 2009).
Typical R proteins function to perceive pathogens by a direct or indirect interaction with specific AVR effectors. Once this recognition occurs, the R protein activates defense responses, typically including a rapid HR response (Greenberg et al. 1994). Aberrant NBS-LRR proteins have been generated that activate defense responses independently of their AVR proteins and therefore trigger HR in the absence of the pathogen. Such autoactive genes have been created by mutations at specific sites (Bendahmane et al. 2002; Shirano et al. 2002; Howles et al. 2005) or engineering truncated proteins or hybrid proteins by swapping domains from homologs or alleles (Hwang et al. 2000; Hwang and Williamson 2003; Frost et al. 2004; Howles et al. 2005; Rairdan and Moffett 2006; Zhang and Gassman 2007). These observations have led to the perception that regions of NBS-LRR proteins have roles as negative regulators of the defense signaling processes controlled by other regions of the protein (Belkhadir et al. 2004b). If these interdomain interactions are interfered with by the presence of the AVR protein or specific alterations of the protein itself, constitutive signaling can result.
Overexpression of NBS-LRR genes (Oldroyd and Staskawicz 1998; Tao et al. 2000; Bendahmane et al. 2002; Belkhadir et al. 2004a; Zhang et al. 2004; Chern et al. 2005) can also cause spontaneous activation of defense responses indicating a low level of AVR-independent basal activity of these proteins. Expression of the flax NBS-LRR gene L6 in tobacco has also been shown to cause spontaneous defense responses (Frost et al. 2004), but this may also be due to increased expression of the protein.
The Rp1 rust resistance locus of maize consists of a cluster of NBS-LRR genes that confer race-specific resistance to the common rust fungus Puccinia sorghi (Collins et al. 1999). These genes often mispair in meiosis and recombine unequally which re-assorts them into new combinations creating haplotypes with novel gene combinations and sometimes novel genes. Several different Rp1 recombinant haplotypes have been selected by their nonparental phenotypes. These variants consist of haplotypes with nonparental race specificities (Richter et al. 1995), haplotypes with reduced levels of resistance with parental race specificities (Sun et al. 2001) and haplotypes with spontaneous necrotic phenotypes with nonspecific rust resistance reactions (Hu et al. 1996; Hu et al. 1997). Some of these phenotypes are due to the novel combination of rp1 genes but many of the phenotypes are the result of the generation of new Rp1 genes by intragenic recombination (Sun et al. 2001; Smith and Hulbert 2005). The term “spontaneous necrotic phenotype” indicates the variants described in this work exhibited necrotic spotting resistance phenotypes similar to the hypersensitive response (HR), however without inoculation. Rp1 haplotypes with spontaneous necrotic phenotypes and nonspecific rust resistance reactions were the focus of this investigation. The phenotypes of three haplotypes have been described (Hu et al. 1996). The HRp1-Kr1N variant exhibited a diffuse necrosis without necrotic spotting which is not inducible by rust inoculation. Necrosis begins at the tip of the leaf once the leaf is fully expanded then progresses down towards the base of the leaf. In addition, HRp1-Kr1N maintains the parental, Rp1-Kr1, resistance specificity. The HRp1-D*21 and HRp1-MD*19 variants exhibited necrotic spotting phenotypes without inoculation (Sun et al. 2001), but rust inoculation rapidly induces the necrotic spots. HRp1-D*21 confers a nonspecific resistance with all common rust biotypes tested (P. sorghi).
This analysis will shed light on the factors at the Rp1 locus that control race-specific and nonspecific interactions and condition cell death activating plant defenses in the absence of the pathogen.
Materials and methods
Library construction and screening
A HRp1-Kr1N genomic DNA library was constructed and screened following standard procedures (Maniatis et al. 1982). This haplotype is typically lethal as a homozygote, but homozygous seedlings sometimes reached the 3–4 leaf stage when cultured under summer greenhouse conditions where high temperatures (>28 C) are maintained at day and night. Genomic DNA isolated from a HRp1-Kr1N homozygous maize line was partially digested with Sau3a (NEB, Beverly, MA) and size fractionated on a sucrose gradient. DNA fragments ranging from 9 to 23 kb in size were ligated into a Lambda DASH II replacement vector and packaged using a Lambda DASH II/BamHI Vector Kit as described by the manufacturer (Stratagene, LaJolla, CA). An rp1 3′ probe (amplified with primer pair P22 and 4890R) was used to screen the genomic library for cross-hybridizing clones. Bacteriophage growth, purification and DNA extraction was performed using standard molecular protocols (Maniatis et al. 1982).
DNA sequence alignment of all clones was performed using the GCG Seqweb version 2 program or the Baylor College of Medicine (searchlauncher.bcm.tmc.edu), Basic Local Alignment Search Tool. The sequences from the 5′ and 3′ regions of genes or full-length gene sequences were aligned and analyzed, to identify a recombinant gene, determine the recombination exchange point and verify correct construction of transformation constructs. All sequencing was performed at the Kansas State University Sequencing Facility.
Transformation constructs
A genomic fragment carrying the recombinant gene from HRp1-D*21 with approximately 2 kb of sequence upstream of the predicted coding region was subcloned from the original genomic Lambda clone (Sun et al. 2001) into the pUC19 cloning vector. The plasmid construct, designated pUCRp1-D*21 was used in stable plant transformation experiments. Two additional recombinant rp1 genes were constructed in vitro by exchanging fragments of genomic clones of the rp1-dp2 and Rp1-D genes (Fig. 1) subcloned into pUC19. The Rp1-dp2-D-1 construct carries the promoter and NBS region from the rp1-dp2 gene and the LRR regions from the Rp1-D parent. A 2.4-kb fragment carrying the promoter and NBS region was amplified from the rp1-dp2 with an rp1-dp2 specific forward primer (dp2-F1, Table 1) and a reverse common primer (2620R, Table 1). The resulting PCR product was double digested with BamHI and NsiI restriction enzymes (NEB, Beverly, MA), and gel purified. The 2.5-kb LRR region of this construct was obtained from a plasmid carrying the Rp1-D gene (Ayliffe et al. 2004) with EcoRI and NsiI. The promoter/NBS and LRR fragments were cloned into a BamHI/EcoRI digested pUC19 cloning vector. Selected clones were fully sequenced.
Recombination events that gave rise to Rp1-D*21 spontaneous necrotic variant and construction of chimeric genes Rp1-dp2-D1 and Rp1-dp2-D-2. a Recombination event between Rp1-D and rp1-dp2 paralogs gave rise to Rp1-D*21, which confers a spontaneous necrotic phenotype. b Rp1-dp2-D-1 construct contains the rp1-dp2 NBS region and LRRI-LRRII regions from the Rp1-D parental paralog. Rp1-dp2-D-2 construct contains the rp1-dp2 NBS-LRRI regions and the Rp1-D LRRII region. Vertical arrows indicate recombination exchange point. Horizontal arrows signify primer sites (dp2-f1 and 2620R)
The Rp1-dp2-D-2 construct contains the rp1-dp2 promoter, NBS-encoding region and the coding region from the first half of the LRR, with an Rp1-D fragment coding for the second half of the LRR (Fig. 1). A plasmid carrying rp1-dp2 was digested with NcoI and EcoRI to release the fragment encoding the distal half of the LRR. This region was replaced by an EcoRI-NcoI fragment from a plasmid carrying the Rp1-D gene. Selected clones were fully sequenced and aligned with the original sequences to verify the construct.
A Lambda clone (Kr1N-35-2-60) carrying the putative Rp1-Kr1N gene was partially digested with Sau3A (NEB, Beverly, MA) and subcloned into BamHI digested pUC19 cloning vector. Clones were end-sequenced with M13 primers to identify clones with a complete coding region. A clone with a full coding region was completely sequenced using 13 Rp1 primers (Table 1). This clone was then digested with EcoRI to remove the Lambda vector sequences, re-circularized and designated pKr1n-pUC19. The first 138 nucleotides of the clone is homologous to the predicted intron in the 5′ UTR of the Rp1-D gene followed by a 3,855 nucleotide ORF. To drive expression of the putative Rp1-Kr1N gene in plants, a ubiquitin promoter was added. An M13 forward primer was coupled with a reverse primer (pAHC17-R-GCATCAACATGTATACCTATCCT) designed from the intron in the 5′ region of the maize ubiquitin gene to PCR amplify the ubiquitin promoter from the pAHC17 plasmid (Christensen et al. 1992). An approximately 1.4-kb fragment that included the ubiquitin promoter (~900 bp) and approximately 500 bp of the 5′ ubiquitin intron was amplified and cloned into an Invitrogen (Carlsbad, CA) TOPO TA cloning vector. A sequence-verified clone was then digested with EcoR1, and the 1.4-kb fragment ligated into the EcoR1 in the pKr1n-pUC19 plasmid.
The maize ubiquitin promoter was used for the Rp1-Kr1N transient assay in place of the endogenous promoter as the maize Rp1 endogenous promoter may not function well in other plant species. Previous work demonstrated that maize transgenic lines expressing the Rp1-D gene under the control of the ubiquitin promoter or its endogenous promoter gave high levels of Rp1-D transcripts (Ayliffe et al. 2004). However, wheat and barley transgenic lines expressing the Rp1-D gene under the control of its endogenous promoter gave low levels of Rp1-D transcripts that were mostly truncated.
Transformation constructs for both transient and stable plant transformation experiments were co-bombarded with constructs carrying selectable and or visible markers. The pAHC27 plasmid (Christensen et al. 1992) was used in transient assays to express the uidA reporter gene, which encodes ß-glucuronidase (GUS) and is driven by the maize ubiquitin promoter. The pBARGUS or pAHC20 plasmids were utilized to express the Bar and GUS reporter genes in stable plant transformations. pBARGUS contains a BAR gene that encodes phosphinothricin acetyltransferase and the GUS (ß-glucuronidase) reporter gene under the control of the CaMV 35S promoter and Adh1 promoter respectively (Fromm et al. 1990). pAHC20 contains a BAR reporter gene under the control of the ubiquitin promoter.
Generation of transgenic maize and wheat plants
A maize callus culture was established from immature embryos of HiII maize grown in the green house (Armstrong and Green 1985). The HiII maize population was highly susceptible to all the P. sorghi isolates used in this study. Wheat explants were prepared as pre-cultured (3–5 days) immature embryos from the spring wheat variety ‘Bobwhite’. Transformation was performed with a particle inflow gun using tungsten as the micro carrier. Transgenic embryogenic maize callus was cultured under selection of 10 mg/l glufosinate for 4–5 months. Growing tissue was then selected, tested for GUS activity and regenerated to adult plants according to Songstad et al. (1996). Wheat callus was cultured for 2 weeks under selection of 5 mg/l glufosinate before shoot and root formation was initiated (modified from Altpeter et al. 1996). T0 plants were transferred to soil and screened for transgene integration. Selected T0 wheat plants were self-fertilized to generate T1 progeny, while T0 maize plants were crossed to HiII and H95 maize lines.
Transient assays and stable plant transformation
Maize and wheat lines stably expressing recombinant rp1 genes were made using biolistic protocols as described previously (Ayliffe et al. 2004). The maize line HiII and the wheat line Bobwhite were utilized as the recipient lines. To verify that transgenes were being transcribed, RT-PCR was performed with total RNA isolated from transgenic maize callus clones or fully expanded second leaf segments of plants carrying the transgene. Total RNA was isolated with Trizol Reagent (GIBCOBRL, Rockville, MD) as described by the manufacturer. RT-PCR was performed using a ProStar First-Strand RT-PCR Kit (Stratagene, LaJolla, CA). First strand synthesis was performed with an oligo dT primer. A Kr1N-1415F/Kr1N-2556R primer pair was used to specifically amplify Ubi-Kr1N cDNA sequences and a DF/DR3 primer pair was used to amplify Rp1-dp2-D-1 and Rp1-dp2-D-2 cDNAs (Table 1). These primers were designed from regions that were polymorphic among Rp1 sequences and the sequences were specific to each transgene.
Transient transformation procedures were also carried out using biolistic transformation methods. Particle bombardment was performed with a particle inflow gun (60 psi of helium pressure) with an inside chamber vacuum of −28 psi and a target distance of 15 cm (Finer and McMullen 1991). Tungsten metal particles (20 μg of 99.9% 1 μm M10 equv., Atlantic Equipment Engineers, Bergenfield, NJ) were used as a DNA carrier. Three micrograms (1 μg/μl) of purified selectable marker plasmid and 3 μg of plasmid carrying the Rp1 gene (1 μg/μl) were precipitated onto a 25 μl suspension of tungsten particles by adding 25 μl of 2.5 M CaCl2 and 10 μl of 100 mM spermidine to the DNA sample. Following a 5-min incubation on ice, 50 μl of supernatant was removed. The particles coated with purified plasmid remained and 2 μl of the DNA/tungsten particle sample was used for each bombardment. Three treatments were used for transient assay transformations, one experimental and two controls. The experimental treatment consisted of the co-bombardment of pAHC27 and Ubi-Kr1N plasmids. One control treatment was the co-bombardment of pAHC27 and the Kr1N-pUC19 plasmids and a second control was the pAHC27 and Ubi-Rp1-D plasmids bombarded together. The pUbi-Rp1-D (Ayliffe et al. 2004) construct contains the Rp1-D gene coding region and 3′ UTR downstream of the maize ubiquitin promoter and was utilized as a control plasmid for transient assay experiments.
Leaf segments from fully expanded seedlings were placed on H20/agar plates (8 g/500 mL) for transient transformation assays. Leaf sections were bombarded twice with DNA/tungsten particle samples. Plates containing bombarded leaf segments were placed in a growth chamber at 23°C overnight. Following incubation, a GUS histochemical assay was done to determine the viability of transformed cells (Jefferson 1987). Leaf segments were submerged in GUS assay stain and agitated gently overnight at 37°C. Leaf segments were destained for 4–6 h by gently shaking leaves in 100% EtOH for 1 h at 37°C followed by several incubations of 75% EtOH, changing the 75% EtOH solution every hour until the leaf segments were destained. GUS stained cells were then visualized under a dissecting microscope and counted to obtain numerical data for statistical analysis.
Statistical analyses
Statistical analysis of numerical data obtained from transient assay experiments was performed using SAS statistical program version 8.01 (SAS®, SAS Institute Inc., SAS Campus Cary NC, USA). SAS was used to test whether cells bombarded with the Ubi-Kr1N plasmid had fewer cells expressing the reporter gene than control bombardments in transient transformation experiments. The P (probability) value was set to 0.0001. All other parameters were set to default.
Results
Characterization of the Rp1-D*21 recombinant gene
The HRp1-D*21 variant conferring a spontaneous necrotic phenotype was originally identified in an Rp1-D/Rp1-D x rp1/rp1 testcross population (Pryor 1987). Analysis of the variant haplotype indicated it was derived from an unequal crossing over event in which the most distal gene in the haplotype, Rp1-D, recombined with the gene that was second from the proximal end of the array, rp1-dp2 (Sun et al. 2001). The resulting haplotype therefore carries the rp1-dp1 gene and a recombinant gene, which consists of the 5′ portion of the rp1-dp2 gene (NBS and first 280 amino acids of the LRRI) and the 3′ portion of the Rp1-D gene (remaining 480 amino acids coding for part of the LRRI-LRRII region).
To determine if the recombinant gene in HRp1-D*21 conferred the nonspecific resistance phenotype, transgenic maize lines were made expressing the gene. PCR analysis confirmed genomic integration of the putative Rp1-D*21 gene in six T0 plants, two of which produced T1 families by crossing to the HiII maize line. Both plants appeared normal before transplanting into soil but developed chlorotic spots soon within a week of transplanting to the greenhouse (Fig. 4d). T1 progeny of both lines segregated for the Rp1-D*21 spontaneous necrotic phenotype described for plants carrying a native copy of the gene (Pryor 1993; Hu et al. 1996). The necrotic spotting phenotype segregated 24:42 and 39:45 (necrotic spots: normal). As with lines carrying the native copy of the gene, necrotic spots did not appear until leaf tissue was fully expanded and they typically appeared first near the tip of seedling leaves. Rp1-D*21 transgenics responded rapidly to inoculation with rust isolates with a hypersensitive response even in leaves that were not fully expanded. This hypersensitive reaction was observed with P. sorghi as well as P. triticina confirming the recombinant gene confers a nonspecific response to multiple rust species.
Identification and characterization of the Rp1-Kr1N recombinant gene
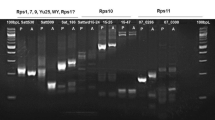
The Rp1-Kr1N variant, conferring a spontaneous necrotic phenotype, was identified in the progeny of a HRp1-Kr1 homozygote (Hu et al. 1996). The HRp1-Kr1 variant was previously recovered from an Rp1-K/rp1 × rp1/rp1 population (Richter et al. 1995) as a variant with a nonparental resistance specificity. Since the HRp1-Kr1N haplotype arose spontaneously from an HRp1-Kr1 homozygote, DNAs of the two haplotypes were compared by gel blot analysis to determine if any novel restriction fragments had been generated. Southern blot analysis of homozygous DNAs isolated from HRp1-KrN identified a novel 2.1-kb fragment when digested with HindIII and probed with an LRR probe (Rp1 3′ LRR) corresponding to the 3′ region of the Rp1-D gene (Fig. 2). The novel fragment was not found in the parental haplotype and was therefore considered to be potentially associated with the rp1 gene conferring the spontaneous necrotic phenotype. HRp1-Kr1N and HRp1-Kr1 homozygous genomic DNAs were therefore run on an agarose gel and DNA from the region corresponding to the novel fragment was gel purified and amplified with three conserved rp1 primer pairs (3535F/3990R, P19/4890R, P22/4890R; Table 1) that span the region corresponding to the rp1 LRR probe that hybridizes to the novel fragment. The three primer pairs amplified fragments of expected size from HRp1-Kr1N and HRp1-Kr1. Direct sequencing of the PCR products was then attempted, reasoning that the products from the HRp1-Kr1N line may be composed mainly of a single product (corresponding to the unique restriction fragment), while the product from the HRp1-Kr1 DNA (without the fragment) would be derived from a mixture of contaminating genes. The P19-4890R primer pair generated a PCR fragment from HRp1-Kr1N that sequenced well indicating it was derived mainly from a single gene. In contrast, no useful sequence could be generated from the HRp1-Kr1 DNA with any of the three primer pairs. Sequences obtained from the HRp1-Kr1N amplification product were aligned with sequences from 37 other genes from HRp1-K to design primers specific to the novel fragment. HRp1-Kr1N appears nearly identical to its parental HRp1-Kr1 haplotype, which is essentially indistinguishable from its parental HRp1-K haplotype (Fig. 2). Consequently, HRp1-Kr1 and HRp1-K should carry mainly the same genes. The Rp1 genes in HRp1-K have been partially characterized, but the genes from HRp1-Kr1 have not. For these reasons, HRp1-Kr1N sequences were compared to genes from its HRp1-K grandparent.
The primers Kr1N-1415F and Kr1N-2556R were designed to amplify sequences corresponding to the novel Rp1-Kr1N fragment, but none of the known sequences from HRp1-K. Genomic DNAs isolated from HRp1-K, HRp1-Kr1, HRp1-Kr1N and Hrp1-H95 (maize inbred line) were PCR amplified with the gene-specific primers to ensure primer specificity. Primers specific for the novel fragment did not amplify in the parental (HRp1-Kr1) and grandparent haplotypes (HRp1-K, Hrp1-h95). However, the primer pair amplified a fragment of expected size in the recombinant haplotype (HRp1-Kr1N).
To identify a full-length gene from HRp1-Kr1N that carries a 2.1-kb HindIII fragment, a genomic Lambda library was constructed from DNA of seedlings homozygous for HRp1-Kr1N. Forty-seven Lambda clones were identified as carrying rp1 sequences after probing with an rp1 probe. Thirty-three of the clones hybridized to both 5′ (P6-1520R) and 3′ (P22-4890R) Rp1-D gene fragment probes indicating they potentially carried full-length genes. PCR positive clones were digested with HindIII to identify clones with a 2.1-kb HindIII fragment. Four of the 27 clones carried the fragment and generated fragments of the correct size when used as templates for PCR with the gene-specific primers. The four clones were partially digested with Sau3a and subcloned into pUC19 (BamHI) to make plasmid templates for sequence analysis. Sequence analysis verified that the four Rp1-Kr1N clones were the same gene. One clone carrying the full predicted Rp1-Kr1N coding region (Kr1N-35-2-60) was fully sequenced and compared to genes from HRp1-K. Sequence comparisons demonstrated that the recombinant gene was identical to the rp1-kp2 gene on the 5′ end and another gene from HRp1-K on the 3′ end (Fig. 3).
Recombination events that created Rp1-Kr1N spontaneous necrotic variant. A recombination event between the rp1-kp2 paralog and a gene from the Rp1-K haplotype generated Rp1-Kr1N recombinant gene, which confers a spontaneous necrotic phenotype. The recombinant contains the rp1-kp2 NBS-LRRI and 577 bp of LRRII region. The remainder of LRRII is from a gene in the Rp1-K haplotype. Arrows indicate recombination exchange point. SPN indicates spontaneous necrotic phenotype
Phenotype verification of the Rp1-Kr1N gene
The Rp1-Kr1N spontaneous necrotic phenotype is expressed in the absence of a pathogen (Hu et al. 1996). To determine if the gene would cause cell death when delivered into a single cell, we conducted co-bombardment experiments with a construct expressing a GUS (pAHC27) reporter gene. Three treatments were used for six separate maize transient assay transformation experiments. Each experiment consisted of three separate bombardments each with equal amounts of two separate plasmids. The experimental treatment consisted of the co-bombardment of pAHC27 and Ubi-Kr1N plasmids and tested the function of the recombinant Rp1-Kr1N gene by determining the ability of this gene to cause cell death when delivered into a single plant cell. Two control treatments were utilized. One control treatment consisted of the co-bombardment of pAHC27and Kr1N plasmids. In this treatment, the Kr1N plasmid does not have a promoter so the bombardment should therefore demonstrate how plant cells would appear if expression of the Rp1-Kr1N gene had no effect on cell viability. A second control treatment consisted of the co-bombardment pAHC27 and Ubi-Rp1-D plasmids. The Ubi-Rp1-D plasmid carries the Rp1-D gene driven by the maize ubiquitin promoter and demonstrated how plant cells would appear when expressing a functional resistance gene other than the recombinant Kr1N gene. The results indicated that co-bombardment of maize leaf sections with the recombinant gene under the control of the ubiquitin promoter and a GUS reporter gene (pAHC27: Ubi- Kr1N) effected cell viability when compared to the control treatments (Fig. 4a). Statistical analysis of numerical data from six separate transient transformation assay experiments showed there were significantly (P < 0.0001) fewer GUS expressing cells in all experiments performed in maize (B73) when compared to the two control treatments (Fig. 5). The same three treatments were also performed in transient transformation experiments in rice (cv. Fanny), big blue stem, sorghum (line BTX623), and wheat (cv. Fielder).
Rp1 recombinant gene phenotypes. a Maize Ubi-Kr1N transient assay. Co-bombardment of maize B73 seedlings with the GUS (ß-glucuronidase) reporter gene and the Rp1-Kr1N gene (Ubi-Kr1N) driven by the maize ubiquitin promoter (experimental; two left leaves) and Rp1-Kr1N with no promoter (control; two right leaves). Blue spots indicate healthy plant cells expressing the GUS reporter gene. b Rp1-D-dp2 transformation assay. Rp1-D*21 confers a nonspecific resistance and a spontaneous necrotic phenotype. It arose by an unequal crossover from an Rp1-D homozygote. Two constructs (Rp1-D-dp2-D-2; top leaf and Rp1-dp2-D-1; bottom leaf) were created using different combinations of the LRR from the two parental genes (Rp1-D and Rp1-dp2) and tested in stable transgenic plants (HiIIA background). Constructs were co-bombarded with pAHC20 plasmid as a selectable marker. A line expressing the Rp1-dp2-D-2 construct showed the Rp1-D*21 spontaneous necrotic phenotype without inoculation. cHRp1-Kr1N confers a spontaneous (uninoculated) necrosis. HRp1-Kr1N arose from an HRp1-Kr1 homozygote. HRp1-Kr1N homozygotes are lethal, while heterozygotes (Rp1-Kr1N/rp1; top leaf) develop a diffuse necrosis on the tip of the leaves that expands to the base of the leaf as they become fully expanded. rp1 homozygotes (rp1/rp1; bottom leaf) do not develop this phenotype. d T1 transgenic maize line expressing the Rp1-D*21 transgene showed a spontaneous necrotic phenotype on fully expanded leaves appearing first near the tip of seedling leaves and then extending down the leaf
Inhibition of reporter gene expression by co-bombardment with Rp1-Kr1N. The X and Y axis indicates the species analyzed and the number (mean of 4 replications) of GUS (ß-glucuronidase) expressing cells respectively. All treatments include the GUS expressing construct co-bombarded with a second plasmid. Dark gray bars denote the Rp1-Kr1N gene with (GUS: Kr1N) with no promoter; light gray bars indicate the Rp1-D gene under the control of the ubiquitin promoter (GUS: Ubi-Rp1-D); black bars denote the -Rp1-Kr1N gene under the control of the ubiquitin promoter (GUS: Ubi-Kr1N), all co-bombarded with GUS. An asterisk denotes a significant difference (P < 0.0001) in the number of GUS expressing cells between the bombardments with the Ubi-Kr1N and the controls. Mz maize, BBS big blue stem, Rc rice, Sg sorghum, Wh wheat
No significant differences were observed in the number of GUS expressing cells in the experimental and control treatments in these species. Stable transgenic maize plants expressing the HRp1-Kr1N transgene have not yet been recovered. In addition, a very high frequency of callus tissue death was encountered during tissue culture, which was probably caused by the Rp1-Kr1N gene.
Identification of the recombinant Rp1-MD*19 gene
The Rp1-MD*19 variant was previously recovered from an rp1-D*19/Rp1-M x rp1/rp1 testcross population (Hu et al. 1996). The rp1-D*19 variant was originally identified as a susceptible recombinant from an Rp1-D/Rp1-D x rp1/rp1 testcross population (Pryor 1987). A recombination event between Rp1-D and the rp1-dp5 paralog gave rise to the rp1-D*19 recombinant (Sun et al. 2001). The first 330 bp of the 5′ coding region was identical to rp1-dp5, while the remainder of the gene was from Rp1-D. It was therefore postulated that this gene was involved in the recombination event that gave rise to Rp1-MD*19, since the gene re-acquired the Rp1-D specificity. If the putative recombinant gene in HRp1-MD*19 is oriented with the 3′ end closest to the telomere, like other Rp1 genes examined (Sun et al. 2001), the flanking marker constitution of the recombinant indicates the gene would probably have the distal (3′) end of the rp1-D*19 gene and the 5′ end of a gene from the Rp1-M parent haplotype. To amplify the putative recombinant gene, the Rp1-D specific reverse primers DR4 and DR5 were coupled with a nonspecific forward primer P6 from the 5′ region of the gene, and used to PCR amplify the gene using DNA from Rp1-MD*19 as template. After cloning and sequencing the amplification product, the sequence data indicated that the recombinant Rp1-MD*19 allele consists of the N-terminus of an rp1 member from the M haplotype and C-terminus of the Rp1-D (from rp1-D*19) gene (Fig. 6). In the recombinant gene product, the entire LRR-encoding region is identical to the Rp1-D gene.
Recombination events, that gave rise to Rp1-MD*19 spontaneous necrotic variant. A recombination event between Rp1-D and rp1-dp5 paralogs gave rise to the rp1-D*19 recombinant, with no detectable resistance phenotype (Sun et al. 2001). A second recombination between rp1-D*19 and a gene from the Rp1-M haplotype, gave rise to Rp1-MD*19 recombinant gene, which confers a spontaneous necrotic phenotype. Arrows indicate recombination exchange points. R indicates haplotype confers a resistance phenotype. S indicates haplotype confers a susceptible phenotype. SHR indicates haplotype confers a spontaneous hypersensitive response
Construction of recombinant genes in vitro
To determine if we could reproduce the phenotype of the Rp1-D*21 and Rp1-MD*19 gene by making similar exchanges in vitro, two recombinant genes were made by swapping parts of the LRR region of the rp1-dp2 gene with fragments of the Rp1-D gene LRR (Fig. 1). The Rp1-dp2-D-1 recombinant construct codes for the amino terminal 495 amino acids of the rp1-dp2 gene, including the NBS-encoding region, and the Rp1-D 760 amino acid LRRI-LRRII region. The Rp1-dp2-D-2 construct codes for the amino terminal 864 amino acids of the rp1-dp2 gene, including the regions coding for the NBS and LRRI regions, and the C-terminal 293 amino acids coding for the Rp1-D LRRII region. Therefore these constructs differ by how much of the Rp1-D LRR they retain. Comparatively, Rp1-D*21 has the rp1-dp2 NBS domain and first 280 amino acids of the LRR region. The DNA coding for the remaining 480 amino acids (LRRI-LRRII region) are from Rp1-D. The Rp1-MD*19 recombinant has the NBS-encoding domain and the first 151 amino acids of the LRR region from a HRp1-M gene. The remaining 609 amino acids of the LRR domain is derived from Rp1-D.
The Rp1-dp2-D-1 and Rp1-dp2-D-2 recombinant constructs were tested in stable transgenic maize plants (HiII). Four transgenic maize lines were regenerated from callus carrying the Rp1-dp2-D-1 construct, and all four generated a fragment of expected size when amplified by RT-PCR with primers specific for the gene indicating it was transcribed in these lines. None of the four stable transgenic maize lines expressing the Rp1-dp2-D-1 construct conferred the HRp1-D*21 spontaneous necrotic phenotype (Fig. 4b) and all were highly susceptible to rust isolate IN2, which is avirulent on lines expressing Rp1-D. In contrast, only one of four lines carrying the Rp1-dp2-D-2 construct expressed the transgene as determined by an RT-PCR assay. The three lines not expressing the transgene appeared normal, while all 14 plants regenerated from the callus genotype expressing the transgene showed a very severe spontaneous necrotic phenotype without inoculation (Fig. 4b). All 14 plants died before making viable pollen or seed. A second attempt at producing maize lines expressing the Rp1-dp2-D-2 transgene resulted in six maize lines carrying the transgene, but transcripts could not be detected in any of the six and all had normal phenotypes.
Rp1-D contributes to necrotic spotting phenotypes
Some Rp1 haplotypes with apparently normal race-specific effects have been observed to control mild chlorotic or necrotic spotting phenotypes. This was first observed for an Rp1-DJ haplotype which carries both Rp1-D and Rp1-J in addition to other rp1 homologs in the complex (Hu et al. 1997). This haplotype was associated with chlorotic spotting and partial nonspecific resistance to both common rust (P. sorghi) and southern rust (P. polysora). Additional crossing with this, and other haplotypes indicated these effects were dependent on the genetic background of the cross. In a field experiment planted on 14 April 2000 in Manhattan Kansas, two replications of 25-plant rows were planted for lines A188, H95 and four lines carrying Rp1-A and Rp1-D in the A188 and H95 backgrounds. Each of the genes had been backcrossed at least three times to the A188 and H95 parents. Plants in two rows carrying the Rp1-D haplotype in the A188 background (backcross three to A188) showed necrotic spots at the adult plant stage. When scored during pollen shed, all the plants were scored either as a three (heavy necrotic spotting on leaves below the ear) or a 4 (heavy spots on nearly all leaves). The average rating was 3.50. In contrast, little if any spotting was observed on A188 plants (rp1-A188 haplotype) or plants homozygous for the HRp1-A haplotype in the A188 background. Most plants were rated 0 (no spotting) and several were rated 1 and had few chlorotic spots. Plants homozygous for the HRp1-A or HRp1-D haplotypes in the H95 inbred background were indistinguishable from the H95 parent (Hrp1-H95 haplotype) and had no noticeable spotting phenotypes. The same trend was observed in other years in additional generations with these genes and with other haplotypes that carry the Rp1-D gene. The spotting phenotype with the Rp1-D gene in the A188 background seemed particularly consistent in early spring plantings (data not shown).
Observations made with Rp1-D transgenics indicate this gene, and not some other aspect of the HRp1-D haplotype, contributes to the spotting phenotype. After the original transgenics (HiII background; Ayliffe et al. 2004) were backcrossed twice to A188, a sample of the progeny were grown to maturity in the greenhouse. Five of ten plants that showed the Rp1-D rust resistance (heterozygotes) showed mild spotting phenotypes on the lower leaves at maturity (pollen shedding stage); Three were scored as 2 reactions and two as 1 reactions. The five plants not expressing the Rp1-D resistance did not show noticeable spotting (0 reactions).
Discussion
Three recombinant Rp1 haplotypes have been characterized that confer severe necrotic reactions (Sun et al. 2001, Hu et al. 1996). One of these (HRp1-Kr1N) conferred a diffuse necrosis whose progression was not affected by rust inoculation and the other two (HRp1-D*21 and HRp1-MD*19) conferred necrotic spots. The spots typically occur on fully expanded seedling leaves as they become fully expanded even in the absence of rust but can also be induced by rust infection, similar to the normal hypersensitive reaction, under the appropriate conditions. All three recombinants were associated with novel recombinant Rp1 genes derived from crossovers in their LRR regions.
The recombinant genes in the HRp1-D*21 and HRp1-MD*19 haplotypes both carry part of their 3’, LRR-encoding regions from the Rp1-D gene. The Rp1-D gene itself can also cause a weak necrotic spotting phenotype on the mature leaves of adult plants in some genetic backgrounds. Individual Rp1 genes can therefore have both race-specific effects and nonspecific effects associated with necrotic spotting and partial resistance. The LRR region of most Rp1 genes consists of 14 very degenerate LRRs (LRR I region) followed by an approximately 100 amino acid region with no noticeable LRR homology and another 12 degenerate LRRs at the carboxy terminus (LRR II region). Rp1-D*21 codes for a protein with the last two LRRs of LRR1 and the LRR2 region from the Rp1-D gene and does not confer the Rp1-D specificity since it does not confer any more resistance to rust isolates that are avirulent on Rp1-D than it does to Rp1-D-virulent isolates. In contrast, Rp1-MD*19 does have the Rp1-D specificity in addition to the nonspecific necrotic reaction. The entire LRR-encoding region of this gene is identical to Rp1-D but the N terminal and NBS-encoding regions come from a gene from the Hrp1-M haplotype. The rp1-dp1-D-1 gene was synthesized to carry the whole Rp1-D LRR but the N-terminus of rp1-dp2, the same N-terminus as the Rp1-D*21 gene. This gene conferred no spontaneous necrosis or resistance when expressed in transgenic plants. The other gene synthesized in vitro (Rp1-dp2-D-2) carried LRR2 from Rp1-D and the N-terminus and LRR1 from rp1-dp2. This appeared to confer a very severe spontaneous necrotic phenotype similar to the Rp1-D*21 gene but only a single transformation event was regenerated and every seedling from this event died before race specificity could be examined. Thus it appears the nonspecific spontaneous necrotic phenotype can be caused by a recombinant LRR-encoding region, or exchanging the whole Rp1-D LRR region onto a different N-terminus.
The LRR domains have been proposed to be responsible for the specific recognition functions in the NBS-LRR class of disease resistance genes and thus determine resistance specificities (Baker et al. 1997; Ellis et al. 1999; Dodds and Schwechheiner 2002, Ellis et al. 2007). The results with the Rp1 recombinants are consistent with this idea. The recombination event that gave rise to Rp1-MD*19 restored an Rp1-D specificity and the recombinant gene in this haplotype included an intact Rp1-D LRR. Apparently, this LRR, and recombinant LRR with only the C-terminal half of it, are also able to confer a nonspecific spontaneous necrotic phenotype when combined with the appropriate N-terminus. The effect may be small, as with the Rp1-D gene whose nonspecific reactions are only noticeable in certain genetic backgrounds in adult plants, or they may be very strong. It is not clear if they function by actually interacting with a specific protein or compound made by the fungus or the fungal interaction because they can cause HR in the absence of the fungus. It is possible their ability to respond to fungal infection under the appropriate conditions is caused by priming cells towards defense reactions which then occurs when the cells are challenged by a pathogen.
HRp1-Kr1N also confers a spontaneous necrotic phenotype, but does not exhibit a classic lesion mimic, spotting phenotype. It is also unique in that the necrosis is not inducible by rust inoculation, but appears to be developmentally controlled (Fig. 4c). Rust resistance has also been assayed on HRp1-Kr1N seedlings prior to necrosis, and it is clear that the HRp1-Kr1N haplotype still confers the Rp1-Kr1 parental resistance specificity (Hu et al 1996). Consequently, the recombinant gene in this haplotype is probably different from that causing the Rp1-Kr1 specificity. The function of the Rp1-Kr1N recombinant gene was tested in transient and stable transformation experiments. Transient transformation experiments demonstrated that delivery of the gene into single cells interfered with cell viability sufficiently to inhibit reporter gene expression (Fig. 5). Therefore, the Rp1-Kr1N recombinant gene kills cells spontaneously and confers the necrotic phenotype in maize supporting the postulate that the change in the distal half of LRRII domain of the Kr1N recombinant gene is responsible for the spontaneous necrotic phenotype. Moreover, the inability of the two control constructs to kill cells demonstrated that the harmful effect on cell viability was due to the expression of the recombinant Kr1N gene and not the expression of any rp1 resistance gene. There were no stable transgenic maize plants expressing the Rp1-Kr1N transgene recovered. The high frequency of callus tissue death during tissue culture were likely due to expression of the Kr1N gene. These two factors indicated that the recombinant confers a lethal phenotype in stable transgenic plants, making it difficult to recover transgenics expressing this gene. It is plausible that the change in the LRR domain of Kr1N allows it to inappropriately interact with plant signaling components to trigger activation of the defense response pathway in the absence of a pathogen.
Delivery of Rp1-Kr1N into cells of rice, big blue stem, sorghum and wheat had no noticeable effect on cell viability, as assayed by reporter gene expression. Therefore, the Rp1-Kr1N gene does not appear to function in these distantly related cereal species. Similarly, stable transgenic wheat plants expressing the Rp1-D*21 gene showed no noticeable resistance or spontaneous necrosis. This may be due to a lack of necessary complementary components of a protein complex or signaling component required for activation of the defense response pathway and Rp1-Kr1N expression. Alternatively, Ayliffe et al. (2004) found many of the Rp1-D transcripts in stable wheat and barley transgenics were truncated, raising the possibility that other cereal species were not making Rp1 proteins properly.
Characterization of the three Rp1 variants provides evidence that some spontaneous necrotic variants are the result of an alteration of actual disease resistance genes. The hypothesis that similar types of resistance genes, or other genes involved in defense responses, are affected in other spontaneous necrotic variants is also supported by the analysis of spontaneous necrotic variants in several different species (Walbot et al. 1983; Dietrich et al. 1994; Johal et al. 1995; Lorrain et al. 2003). One example is the Arabidopsis dll1 (disease-like lesions1) spontaneous lesion variant. This variant produces lesions spontaneously that mimics bacterial speak disease and showed accumulation of genes related to defense. These spontaneous or ‘autoactive’ alleles have been best characterized at the L locus in flax (Howles et al. 2005). Several mutant and recombinant variants were created with alterations in the NBS and LRR regions that caused a gene dosage-dependent dwarf phenotype and constitutive expression of plant defense genes in the absence of the pathogen. Similarly, the autoactive Rp1 genes described here were also created by alterations (intragenic recombination) in the LRR region. It is therefore plausible that regions of Rp1 proteins function as negative regulators of defense signaling that are mediated by other regions of the R-protein. Once these interactions are hindered by recombination in the LRR region, constitutive signaling can occur activating plant defenses in the absence of the pathogen or the AVR protein.
One of the most interesting aspects of the Rp1 alleles conferring spontaneous necrotic phenotypes is that their necrotic reaction was induced by inoculation with any rust biotype, and every rust species that was tested (Hu et al. 1996). Therefore, the race-nonspecific resistance of these genes has the greatest potential for durable disease control. The one disadvantage of using resistance genes to control disease is the fact that their resistance is race-specific and therefore not durable due to shifts in pathogen populations. Quantitatively inherited disease resistances are usually considered more durable, but they are very difficult to manipulate genetically (Hooker 1967). Race-nonspecific resistance that can be manipulated as single loci would be invaluable as a method for controlling disease. The variant Rp1 genes provide evidence that a race-specific rust resistance gene can be engineered to confer race-nonspecific resistance and provides some insight as to the information needed for designing the types of changes that might be made to resistance genes to create genes with the desired nonspecific effects.
References
Altpeter F, Vasil V, Srivastava V, Stoger E, Vasil IK (1996) Accelerated production of transgenic wheat (Triticum aestivum L.) plants. Plant Cell Rep 16:12–17
Armstrong CL, Green CE (1985) Establishment and maintenance of friable embryonic maize callus and the involvement of l-proline. Planta 164:207–217
Ayliffe MA, Steinau M, Park RF, Rooke L, Pacheco MG, Hulbert SH, Trick HN, Pryor AJ (2004) Aberrant mRNA processing of the maize Rp1-D rust resistance gene in wheat and barley. Mol Plant Microbe Interact 17:853–864
Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar SP (1997) Signaling in plant–microbe interactions. Science 276:726–733
Belkhadir Y, Nimchuk Z, Hubert DA, Mackey D, Dangl JL (2004a) Arabidopsis RIN4 negatively regulate disease resistance mediated by RPS2 and RPM1 downstream or independent of NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRp12 or AvrRpm1. Plant Cell 16:2822–2835
Belkhadir Y, Subramaniam R, Dangl JL (2004b) Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr Opin Plant Biol 7:391–399
Bendahmane A, Farnham G, Moffett P, Baulcombe DC (2002) Constitutive gain-of- function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J 32:195–204
Bozkurt O, Hakki EE, Akkaya MS (2007) Isolation and sequence analysis of wheat NBS-LRR type disease resistance gene analogs using degenerate PCR primers. Biochem Genet 45:469–486
Brueggeman R, Rostoks N, Kudrna D, Kilian A, Han F, Chen J, Druka A, Steffenson B, Keinhofs A (2002) The barley stem rust-resistance gene Rpg1 is a novel disease-resistance gene with homology to receptor kinases. Proc Natl Acad Sci 99:9328–9333
Chern M, Fitzgerald HA, Canlas PE, Navarre D, Ronald PC (2005) Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol Plant Microbe Interact 18:511–520
Christensen AH, Sharrock RA, Quail PH (1992) Maize polyubiquitin genes: structure, thermal perturbation of expression and transcription splicing, and promoter activity following transferred protoplasts by electroporation. Plant Mol Biol 18:675–689
Collins N, Drake J, Ayliffe M, Sun Q, Ellis J, Hulbert S, Pryor T (1999) Molecular characterization of the maize Rp1-D rust resistance haplotype and its mutants. Plant Cell 11:1365–1376
Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL (1994) Arabidopsis mutants simulating disease resistance response. Cell 77:565–577
Dodds PN, Schwechheiner C (2002) A breakdown in defense signaling. Plant Cell 14(Suppl):S5–S8
Ellis JG, Lawrence GJ, Luck JE, Dodds PN (1999) Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11:495–506
Ellis JG, Dodds PN, Lawrence GJ (2007) Flax rust resistance gene specificity is based on direct resistance-avirulence protein interactions. Annu Rev Phytopathol 45:289–306
Finer JJ, McMullen MD (1991) Transformation of soybean via particle bombardment of embryogenic suspension culture tissue. In Vitro Cell Dev Biol 27P:175–182
Fromm ME, Morrish F, Armstrong C, Williams R, Thomas J, Klein TM (1990) Inheritance and expression of chimeric genes in the progeny of transgenic maize plants. Bio/Tech 8:833–839
Frost D, Way H, Howles P, Luck J, Manners J, Harham A, Finnegan J, Ellis J (2004) Tobacco transgenic for the flax rust resistance gene L expresses allele-specific activation of defense responses. Mol Plant Microbe Interact 17:224–232
Fu D, Uauy C, Distelfeld A, Blechl A, Epstein L, Chen X, Sela H, Fahima T, Dubcovsky J (2009) A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323:1357–1360
Greenberg J, Guo A, Klessig DF, Ausubel FM (1994) Programmed cell death in plants: a pathogen-triggered response activated coordinately with multiple defense functions. Cell 77:551–563
Hooker AL (1967) The genetics and expression of resistance in plants to rusts of the genus Puccinia. Annu Rev Phytopathol 5:163–182
Howles P, Lawrence G, Finnegan J, McFadden H, Ayliffe M, Dodds P, Ellis J (2005) Autoactive alleles of the flax L6 rust resistance gene induce non-race-specific rust resistance associated with the hypersensitive response. Mol Plant Microbe Interact 18(18):570–582
Hu G, Richter T, Hulbert S, Pryor A (1996) Disease lesion mimicry caused by mutations at the rust resistance gene Rp1. Plant Cell 8:1367–1376
Hu G, Webb CA, Hulbert SH (1997) Adult plant phenotype of the Rp1-DJ compound rust resistance gene in maize. Phytopathology 87:236–241
Hwang C-F, Williamson VM (2003) Leucine-rich repeat-mediated intramolecular interactions in nematode recognition and cell death signaling by the tomato resistance protein Mi. Plant J 34:585–593
Hwang C-F, Bhakta AV, Truesdell GM, Pudlo WM, Williamson VM (2000) Evidence for a role of the N terminus and Leucine-rich repeat region of the MI gene product in regulation of localized cell death. Plant Cell 12:1319–1329
Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5:387–405
Johal GS, Hulbert SH, Briggs SP (1995) Disease lesion mimics of maize: a model for cell death in plants. BioEssays 17:685–692
Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFadden H, Bossolini E, Selter LL, Keller B (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323:1360–1363
Lorrain S, Vaillau F, Balague C, Roby D (2003) Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci 8:263–271
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
McHale L, Tan X, Koehl P, Michelmore WR (2006) Plant NBS-LRR proteins: adaptable guards. Genome Biol 7:212.1–212.11
Ohwaki Y, Kawagishi-Kobayashi M, Wakasa K, Fujihara S, Yoneyama T (2005) Induction of class-1 non-symbiotic hemoglobin genes by nitrate, nitrite and nitric oxide in cultured rice cells. Plant Physiol 46:324–331
Oldroyd GED, Staskawicz BJ (1998) Genetically engineered broad-spectrum disease resistance in tomato. Proc Natl Acad Sci USA 95:10300–10305
Pryor AJ (1987) The origin and structure of fungal disease resistance genes in plants. Trends Genet 3:157–161
Pryor A (1993) Transposon tagging of a rust resistance gene in maize. In: Nestler EW, Verma DPS (eds) Advances in molecular genetics of plant-microbe interactions, vol 2. Kluwer Academic, Dordrecht, pp 469–476
Rairdan GJ, Moffett P (2006) Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binging and inhibition of activation. Plant Cell 18:2082–2093
Richter T, Pryor T, Bennetzen J, Hulbert S (1995) New rust resistance specificities associated with recombination in the Rp1 complex in maize. Genetics 141:373–381
Rosewarne G, Singh R, Huerta-Espino J, Rebetzke G (2008) Quantitative trait loci for slow-rusting resistance in wheat to leaf rust and stripe rust identified with multi- environment analysis. Theor Appl Genet 116:1027–1034
Shirano Y, Kachroo P, Shah J, Klessig DF (2002) A gain-of-function mutation in an Arabidopsis toll interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14:3149–3162
Smith SM, Hulbert SH (2005) Recombination events generating a novel Rp1 race specificity. Mol Plant Microbe Interact 18:220–228
Songstad DD, Armstrong CL, Petersen WL, Hairston B, Hinchee MAW (1996) Production of transgenic maize plants and progeny by bombardment of HI-II immature embryos. In Vitro Cell Dev Biol 32:179–183
Sun Q, Collins NC, Ayliffe M, Smith SM, Drake J, Pryor A, Hulbert SH (2001) Recombination between paralogues at the rp1 rust resistance locus in maize. Genetics 158:423–438
Tao Y, Fenghua Y, Leister RT, Ausubel FM, Katagiri F (2000) Mutational analysis of the Arabidopsis nucleotide binding site-leucine-rich repeat resistance gene RPS2. Plant Cell 12:2541–2554
Walbot V, Hoisington DA, Neuffer MG (1983) Disease lesion mimic mutations. In: Kosuge T, Meredith CP, Hollaender A (eds) Genetic engineering of plants. Plenum Publishing Corp, New York, pp 431–442
Young ND (1996) QTL mapping and quantitative disease resistance plants. Ann Rev Phytopathol 43:479–501
Zhang XC, Gassman W (2007) Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol 145:1577–1587
Zhang Y, Dorey S, Swiderski M, Jones JDG (2004) Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1, SGT1 and HSP90. Plant J 40:213–224
Acknowledgments
We thank Julie Essig and Dr. Marcy Main for technical assistance. This work was supported by the US Department of Agriculture–National Research Institute (Grant 2001-35319-10014) and National Science Foundation (grant MCB-0090883). This article is contribution no. 09-293-J from the Kansas Agricultural Experimental Station, Kansas State University, Manhattan, Kansas.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Shirasu.
Nucleotide sequence data reported for Rp1-Kr1N is available in the GenBank database under the accession number GU942722.
Rights and permissions
About this article
Cite this article
Smith, S.M., Steinau, M., Trick, H.N. et al. Recombinant Rp1 genes confer necrotic or nonspecific resistance phenotypes. Mol Genet Genomics 283, 591–602 (2010). https://doi.org/10.1007/s00438-010-0536-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-010-0536-5