Abstract
The Aspergillus nidulans homologue of Neurospora crassa cot-1, cotA, encoding a member of the NDR protein kinase family, has been cloned and expressed under the control of the conditional alcA promoter. Depletion of CotA by repression of the alcA promoter led to a severe growth defect accompanied by loss of polarity. Germlings show greatly enlarged volume of the spores and hyphae, accompanied by an increase in number of nuclei per compartment, though the nucleus/volume ratio is not significantly altered. The depleted CotA phenotype was not suppressed by a nuclear migration mutation nudA2. Double mutants showed an additive, defective phenotype, unlike the suppression of the cot-1 ts mutation by ropy mutations seen in N. crassa, suggesting a different relationship between nuclear migration and the cot signalling pathway in A. nidulans. A functional CotA–GFP fusion protein was found in punctate regions of fluorescence similar to the distribution reported for human NDR2, and as a cap at the hyphal tip.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Filamentous fungi show a characteristic mode of growth, in which hyphal cells in the vegetative mycelial state elongate by polar growth through tip extension, without cell separation. The mycelial growth of the model filamentous ascomycete Aspergillus nidulans has been defined in terms of a duplication cycle (Fiddy and Trinci 1976; Trinci 1979), in which nuclei in the tip compartment undergo parasynchronous mitoses. This is followed by multiple septation events in the rear half of the compartment, giving rise to subapical compartments containing several nuclei, and halving the length of the apical compartment. Continued linear apical extension of this compartment, in the absence of mitosis, restores the length of the compartment, and at a certain length or volume mitosis is triggered again. The formation of a second polar axis results in lateral branching which occurs mainly in the subapical compartments, giving approximately one branch per compartment and the new branch eventually establishes a new duplication cycle. A link between mitosis and septation was demonstrated by Harris et al. (1994), who showed the first septum to be formed during or following the third nuclear division. This was further defined to include the importance of cell volume as not only a trigger for septation (Wolkow et al. 1996) but also for regulation of the initiation of mitosis (Fiddy and Trinci 1976; Clutterbuck 1969; Wolkow et al. 1996). Polar growth of hyphal tips and septation are analogous to budding and cell division in the unicellular cell cycle such as seen in the budding and fission yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Since growth and division in these yeasts are much better understood than hyphal growth and septation in filamentous fungi, they provide useful starting points for understanding the genetic control of these processes in filamentous fungi. However, many aspects of filamentous fungal growth, such as the production of multiple axes of polarity when branching, imply different mechanisms from those of yeasts. Genes may be used in different ways, and additional genes may be required where homologues have not been found in the yeasts (Gatherar et al. 2004; Pearson et al. 2004) (for reviews see Momany 2002; Harris and Momany 2004; Harris et al. 2005).
Mutational analysis has been used to isolate temperature sensitive mutants of A. nidulans defective in septa formation, and some of the genes have been identified (Morris 1975; Harris et al. 1994, 1997). Genetic screens have also been used to identify genes involved in polar growth and hyphal branch formation in A. nidulans (Kaminsky and Hamer 1998; Momany et al. 1999; Gatherar et al. 2004) and Neurospora crassa (Seiler and Plamann 2003). A polarity controlling gene of N. crassa, cot-1, has been investigated. cot-1 is a temperature sensitive mutation which results in reduced hyphal growth rate and reduced mean hyphal extension rate at the restrictive temperature due to a cessation of hyphal tip extension (Mitchell and Mitchell 1954; Steele and Trinci 1977; Collinge et al. 1978; Yarden et al. 1992; Lauter et al. 1998). It belongs to a family of conserved genes in eukaryotic cells and homologues have been found in a wide variety of other species. Members include Colletotrichum trifolii TB3 (Buhr et al. 1996; Hanks et al. 1988), S. pombe orb6 (Verde et al. 1998), S. cerevisae CBK1 (Racki et al. 2000; Bidlingmaier et al. 2001), Caenorhabditis elegans sax1 (Zallen et al. 2000), Drosophila Warts (Justice et al. 1995) and the human NDR (Millward et al. 1995). Their gene products are related to Rho kinase, although they lack Rho binding motifs, and have been shown to play a role in the regulation of cell shape. In S. pombe orb6 is required to maintain cell polarity and the protein is seen at the growing tips during interphase, switching to the site of septation during mitosis and cytokinesis (Verde et al. 1998). Similarly, in S. cerevisiae CBK1 is required for proper polarised growth, as well as bipolar bud site selection in diploid cells, and localizes to both sides of the bud neck in late anaphase (Bidlingmaier et al. 2001). In contrast COT1 does not appear to localise specifically to sites of cell growth. It was detected dispersed throughout the cytoplasm and also associated with the cell membrane, but not with the septa. The membrane localised isoform was lost when grown in restrictive conditions (Gorovits et al. 1999, 2000). Further analysis revealed that mutations in three genes encoding subunits of cytoplasmic dynein and dynactin act as partial suppressors of cot-1 (Plamann et al. 1994; Bruno et al. 1996).
As part of the investigation into the genetic control of hyphal branching in A. nidulans we have isolated a putative cot-1 homologue of A. nidulans. This gene, named cotA, was placed under the control of a regulatable promoter in order to further investigate its function, while incorporation of a GFP tag allowed cellular localisation of CotA to be visualised within the hyphae.
Materials and methods
Strains, media and fungal techniques
The following A. nidulans strains were used in this study; 3K21, yA2; argB2/argB; pantoB2; hhoA::SGFP-TYG (Ramon et al. 2000); G191, fwA1, pyrG89; pabaA1, uaY9 (Ballance and Turner 1985); R153, wA3; pyroA4 (Armitt et al. 1976); SO451, wA3; pyrG89; argB2; pyroA4, nkuA::argB; sE15 (Nayak et al. 2006); WG355, biA1; argB2; galO (Brakhage et al. 1992); XX5, pyrG89; wA3; nudA2, chaA1 (Xiang et al. 1995) while the following strains were constructed in this study: C10, pabaA1, pyrG8; pyr4 alcA(p)::cotA; fwA; CRK17, wA3; pyrG89; argB2; pyroA4, nkuA::argB; sE15; cotA::RFP pyrG; NF1-1, yA2, pabaA1, pyrG89; pyr4 alcA(p)::cotA; PKX23, biA1; argB2; galO; argB alcA(p)::cotA.
L-Broth (1% tryptone, 1% NaCl, 0.5% yeast extract) was used as a bacterial growth media. The routine propagation of A. nidulans strains was carried out on complete medium (CM) and minimal medium (MM) (Pontecorvo et al. 1953). biA1 mutant strains were supplemented with 0.02 μg/ml of biotin while pyrG89 mutant strains were supplemented with 20 mM uracil and 10 mM uridine. Different carbon sources were added in place of glucose depending on the level of induction or repression of the alcA promoter required. YEPD (2% yeast extract, 0.1% peptone, 2% glucose) was also used to repress the alcA promoter. Mycelia were grown at 37°C unless otherwise stated.
Sexual crosses and heterokaryon construction were carried out by standard techniques (Pontecorvo et al. 1953). Fungal protoplasts of the appropriate strain were transformed as described by Ballance and Turner (1985) with 2 μg of plasmid DNA. The transformation plates were incubated at the appropriate temperature for 48 h. Total fungal DNA was extracted using the Promega Wizard Genomic DNA Purification Kit, based on the accompanying protocol for the isolation of genomic DNA from plant tissue.
Microscopy
A Leica DMRB fluorescence microscope, with objectives 10×, 40× and 100× (oil immersion), was used to analyse A. nidulans colonies based on methods described by Harris et al. (1994). Images obtained were captured using video equipment from Princeton Instruments and Openlab 2.0 software on an Apple Mackintosh computer. For each strain a suspension of 106 spores in 20 ml of liquid YEPD was prepared in a sterile universal tube. This was then gently poured into a sterile glass Petri dish, the bottom of which was lined with seven to eight glass coverslips. Conidia settle to the bottom of the Petri dish and adhere to the coverslips. After appropriate incubation the coverslips were removed from the medium and incubated at room temperature for 45 min in fixative solution (3.7% formaldehyde, 50 mM phosphate buffer pH 7.0, 0.2% Triton X-100). The coverslips were rinsed in sterile distilled water prior to staining. A solution of 10 μg of calcofluor/ml dH2O was made fresh for each session. The coverslips were incubated at room temperature with 12 μl of solution for 5 min, and then rinsed in dH2O. They were placed upside down on a glass slide and sealed with clear nail varnish to prevent drying out.
Fluorescence microscopy of strain CRK17 was performed with a Delta Vision Spectris 4.0 microscope running SoftworxTM version 3.2.2 (Applied Precision Instruments, Seattle, USA). The standard FITC filter set was used for GFP and the standard RD-TR-PE filter set was used for RFP. Z-stack images were collected with step sizes of 0.2 μm and deconvolved using SoftworxTM. Images were exported as tif files and image size was cropped using Jasc Paint Shop Pro Version 7.04.
Gene isolation and sequencing
Molecular biology techniques were according to Maniatis et al. (1989) unless otherwise stated. Polymerase chain reaction (PCR) was carried out on total genomic DNA of A. nidulans. Long template PCR for amplifications over 1 kb was carried out using the Expand® Long Template PCR system from Boehringer Mannheim. Amplifications were performed using a DNA Thermocycler from MWG-Biotech Ltd. PCR products were separated from the reaction mixture by passage through a Qiagen Clean-up column, and cloned into the Invitrogen TA-cloning vector pCR2.1. Large-scale preparation of plasmid DNA was achieved using the Promega Wizard® Plus Midipreps DNA Purification System. Sequencing was performed using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction kit from with the standard primers M13 forward and reverse. Colony hybridisation was carried out as described by Maniatis et al. (1989). Sequence comparisons were carried out using software programs available at the NCBI and EBI web sites www.ncbi.nlm.nih.gov and www.ebi.ac.uk, respectively. Primer sequences used are shown in Table 1.
Reverse transcriptase-PCR
RNA was extracted using a procedure based on Yu et al. (2004), and 800 ng RNA was used as template. Reverse transcriptase-PCR (RT-PCR) was performed using the Reverse-iTTM kit from ABgene according to manufacturer’s instructions.
Isolation of the cotA gene
Amplification of the cotA gene used the degenerate primers named MSF and MSR, which were designed from an alignment of N. crassa COT1 (Yarden et al. 1992), C. trifolii TB3 (Buhr et al. 1996) and a S. cerevisiae homologue, subsequently named CBK1 (Nasr et al. 1996; Dorland et al. 2000; Racki et al. 2000). The PCR product of 280 bp, obtained after 20 cycles using the optimum conditions of 56°C annealing temperature, 2.5 mM MgCl2, 100 pM primer, 0.1 μg genomic DNA, was ligated into the vector pCR2.1, resulting in plasmid pAM1. Isolation of the entire cotA gene fragment was carried out by probing the cosmid-based genomic library of A. nidulans (Brody et al. 1991), obtained from the Fungal Genetics Stock Centre, with the cotA fragment excised from pAM1. Cosmids were checked by PCR using the internal primers SFF and SFR resulting in a product of approximately 220 bp, and comparisons of restriction digest patterns using BamHI and EcoRI. Cosmids pSW03H10, pSL11A05 and pSL09H05, were identified as containing the cotA gene at this time and the restriction band patterns suggested that all three cosmids were related. Physical mapping had shown that the inserts in these cosmids were derived from linkage group V (Prade et al. 1997). The cotA gene was subcloned by inserting a 4.3 kb XbaI restriction fragment of cosmid pSW03H10 into the vector pBS SK to give the plasmid pMS1, and verified by sequencing of the insert (GenBank Accession No. AY620243). Intron positions were identified by RT-PCR, while the start codon was assigned by alignment of cotA with the A. fumigatus homologue (Fig. 1). The putative A. nidulans homologue of cot-1, cotA, corresponds to gene locus number AN5529.2 in the subsequently released A. nidulans genome database (http://www.broad.mit.edu/annotation/fungi/aspergillus/index.html).
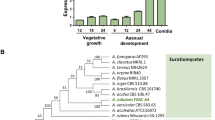
Phylogenetic analysis of the COT1 family. Aligned sequences were edited to remove large gaps and unaligned regions prior to tree construction using the Minimum Evolution method in Mega3 (Kumar et al. 2004). Bootstrap values are given for 1,000 replicates
Promoter exchange
Promoter exchange was achieved using the pAL3 vector (Waring et al. 1989) which carries the alcA promoter of A. nidulans and the N. crassa pyr-4 gene as a selectable marker for fungal transformation. 1.39 kb of the 5′ region of cotA, from nt −47 to nt 1,342 relative to the start codon, was amplified from pMS1 using the oligonucleotides ALF2 and ALAR1 and inserted as a KpnI/XbaI fragment into pAL3 to give pMS3 (Fig. 2a). The fusion of the alcA promoter to cotA was verified by sequencing using the primers Ig61 and Ig63. The circular plasmid was then used to transform the A. nidulans strain G191 in order to replace the wild type cotA promoter with alcA. Integration was verified by Southern blot using a BamHI/EcoRI fragment of cotA (data not shown). The resulting strain, named C10, was crossed with strain 3K21 in which the gene encoding GFP had been fused to nuclear localisation signals, removing the need to stain with DAPI (Ramón et al. 2000), and a double mutant strain NF1-1 recovered. A second alcA(p)–cotA strain was constructed by transforming WG355 with a linear fragment derived from plasmid pKO2. It was generated by removing an EcoRI/BglII fragment from pMS3 containing the alcA promoter and a 5′ section of cotA gene and inserting it into pBluescriptKS digested with EcoRI/BamHI. This resulted in the removal of the BamHI site from the vector sequence, allowing the use of this enzyme for inserting the argB selectable marker. A 1-kb fragment of sequence upstream of the cotA start codon was amplified from pMS1, inserting an XbaI restriction site at the start. This was cloned into the vector using EcoRI restriction digest. The resulting plasmid was then subjected to restriction digest using XbaI, which removed a cassette containing the 5′ upstream region of cotA, the argB selectable marker, the alcA promoter and the N-terminus of cotA (Fig. 2b).
a–b Genomic organization of the cotA gene and the strategies used for promoter exchange. a Schematic view of the organization of the plasmid pMS3, bearing the alcA(p)–cotA fusion, and its integration into the genome of A. nidulans. The left-to-right hatched arrow (cotA) indicates the first 1,342 and 47 bp upstream from the start codon of the cotA gene, while the shaded box (alcA) depicts the alcA promoter. Once integrated into the genome at the cotA locus, indicated by right-to-left hatching (cotA gene), the wild type cotA promoter is replaced by the alcA promoter and a portion of the 5′ of cotA gene sequence from the plasmid. b Representation of the pK02 plasmid and linear cassette used to create a gene replacement event into the A. nidulans genome. The striped arrow (5′ cotA) indicates 5′ untranslated region, while other shading remains the same. The fusion between alcA promoter and cotA gene sequences is also shown in greater detail
GFP-tagging of CotA
For 3′ tagging of CotA, RFP and GFP tagging cassettes were constructed using vectors pFN03 and pXDRFP4 (Yang et al. 2004; Zarrin et al. 2005). Primers are listed in Table 1. PCR was performed using the Expand High Fidelity PCR kit from Roche according to manufacturer’s recommendations. All PCR products were purified using a GFX kit (Amersham–Pharmacia), and strain S0451 was transformed using the final linear PCR product.
Results
cotA sequence
The coding region of the cotA gene of A. nidulans contains three introns which coincide with introns 2–4 of N. crassa. The A. fumigatus homologue Afu4g11890 appears to have a conserved exon–intron structure relative to A. nidulans. As previously observed with this sub-family of putative protein kinases, the greatest variation is seen in the N-terminal region. In many species, this region has glutamine-rich stretches, particularly extensive in S. cerevisiae, Candida albicans, Eremothecium gossypii, C. trifolii and Ustilago maydis homologues, though not so marked in A. nidulans and N. crassa. This N-terminal region also shows substantial length variation between species, being short in S. pombe and human homologues. It is not clear to what extent this N-terminal variation reflects actual functional differences in these kinases. They have been assigned to the same sub-family on the basis of sequence similarity over the kinase domains region, and a tree based on alignment of this conserved region is shown in Fig. 1. The tree reflects the phylogeny of the species from which these putative orthologues originate. For example, Colletotrichum, Gibberella, Claviceps and Neurospora (Sordariomycetes) are closely grouped.
Promoter exchange
Attempts were made to disrupt the cotA gene but no homologous integrants could be detected in heterokaryons. Either the gene is essential or the disrupted strain cannot form a balanced heterokaryon. As an alternative methodology to evaluate the effect of cotA gene expression on A. nidulans growth the wild type promoter was exchanged for the alcA promoter. The initial strain produced by this method, using a circular plasmid (Fig. 2a), NF1-1, proved to be unstable when grown for the long periods required for a sexual cross. Therefore strain PKX23 was produced using a linear cassette (Fig. 2b). In order to verify that a single integration event had taken place a Southern blot was probed with a 4.4 kb XbaI fragment containing the cotA gene along with the surrounding 5′ and 3′ regions from plasmid pMS1. Detection of a restriction fragment of 7.5 kb indicated a single integration event at the site of the the cotA locus in strain PKX23 (data not shown). Stable cotA growth was demonstrated by growth on the repressive YEPD medium for 4 days.
RT-PCR was used to confirm absence of alcA(p)–cotA transcript in PKX23 under repressing conditions. MM containing 100 mM glycerol and 100 mM threonine was used to induce alcA(p), MM with glucose was used to partially repress alcA(p) and YEPD to obtain maximum repression. Mycelia were grown overnight in liquid media at 37°C with shaking.
A transcript was detected from wild type cells under all conditions (Fig. 3). No transcript was detected from PKX23 grown on YEPD, while transcript was observed in mycelia grown on MM with glucose, and in inducing medium. This data indicates that alcA(p) is being regulated as expected in PKX23.
Repression of cotA transcription. Transcripts were analysed by RT-PCR in strains PKX23 and WG355 under inducing and repressing conditions for alcA(p). Primers AL84 and AL85 (Table 1) amplify a 3′ section of the transcript producing a band of 1,111 bp (Fig. 5a). As a control a 600 bp section of the actin transcript was also amplified from each sample using primers PSM36 and PSM37 (Table 1; Fig. 5b). Lane 1 WG355 inducing medium, lane 2 WG355 YEPD, lane 3 PKX23 inducing medium, lane 4 PKX23 MM with glucose, lane 5 PKX23 YEPD. Each lane contains 10 μl of a 50 μl RT-PCR reaction
Morphological comparison of CotA-depleted and normal germlings
A microscopic study was undertaken in order to characterise the effects of cotA repression, (Fig. 4). Strains NF1-1 and 3K21 were grown in YEPD for 12–19 h at 37°C to achieve similar morphological ages and calcofluor was used to stain cell walls. The number of nuclei, together with length, width, and volume measurements of germinating spores, were determined from a sample of 100 germlings for each genotype. The two-sample Student t test was used to make statistical comparisons between the mutant and wild type data, and these results are shown in Table 2. The most visually striking difference is the severely swollen spore of cotA, which has significantly larger volume (t=19.40, df=103, P<0.001), approximately four times the size of the wild type spore (Fig. 5a). This was accompanied by an increased number of nuclei present within the spore (Fig. 5b). However, despite the higher numbers (mean of 17.00/cotA spore, compared to 4.38/wt spore), there was no significant increase in the nuclei/volume ratio (t=1.31, df=169, P>0.05) (Fig. 4c). Irregularities in hyphal diameter and shape between different cotA-depleted germlings resembled the morphology seen in cot-1 of N. crassa grown at the restrictive temperature (Collinge et al. 1978) and it was not possible to determine the volume of these irregular-shaped compartments. In addition, a large number of branches formed in the growing tip, before septa were laid down.
The growth defect of CotA depleted strain is not suppressed by nuclear migration mutation nudA2
The isolation of suppressor mutations is a useful approach for discovery of gene interactions, and could be used to find genes with functions related to that of cotA/cot-1. As a result of attempts to identify substrates or processes regulated by the Cot-1 kinase by isolation of suppressor mutants, ropy mutants were obtained, and shown to be defective in nuclear migration (Plamann et al. 1994). This approach was later adopted as a large scale screen for nuclear distribution mutants (Bruno et al. 1996). ropy mutations partially suppress the colonial phenotype of cot-1, showing a curled hyphal growth phenotype, though the mechanism(s) causing this suppression has not been explained. We were interested to know whether any future screens for cotA suppressor mutations in A. nidulans might result in the re-isolation of nuclear migration mutants, as seen in N. crassa. Nuclear migration has been studied intensively in A. nidulans in parallel with the studies in N. crassa (Xiang and Fischer 2004). A series of mutants defective in nuclear migration, nudA,C,F,G, were originally isolated as temperature sensitive mutants in A. nidulans, and nudA, which encodes the heavy chain of cytoplasmic dynein, is the homologue of N. crassa ro-1 (Xiang et al. 1994; Plamann et al. 1994). We were therefore interested to find out whether the temperature sensitive allele nudA2 could suppress the cotA depletion phenotype.
A sexual cross between alcA(p)::cotA strain PKX23, and temperature sensitive nudA2 strain XX5 readily produced cleistothecia on glucose-containing medium, despite repression of cotA expression. Colonies grown from ascospores on MM with glycerol at 26°C were randomly picked off onto master plates of similar medium and in similar conditions. Progeny were tested for the presence or absence of alcA(p)::cotA and nudA by replication onto medium containing high levels of glucose and grown at 30°C, or minimal media with glycerol grown at the restrictive temperature 44°C. These conditions were chosen for scoring of the progeny phenotypes because they simply demonstrate either alcA(p)::cotA restricted growth on the glucose, or temperature sensitive nudA growth at 44°C. From comparison between the two plates, progeny could be allocated as either parental or recombinant.
Four different phenotypes were observed in the progeny (Fig. 6), parental alcA(p)::cotA and nudA, wild type recombinants, and a putative double mutant phenotype. This mutant grew as small densely packed colonies on glucose, demonstrating both the colonial morphology of alcA(p)::cotA and the curly disordered hyphae of nudA. Rather than nudA2 suppressing the depleted alcA(p)::cotA phenotype, the mutations are additive. On glycerol the alcA promoter has a certain degree of control over the morphology of the double mutant. Colonies became somewhat less dense in that individual hyphae are more discernible and more light penetrates through the mycelium. The nudA2 strain still has aberrant growth even at 30°C, and lowering the temperature did not alter the appearance of the double mutant mycelium (data not shown). Spontaneous suppressor mutations of the alcA(p)::cotA allele on YEPD were not observed, though low frequency suppressor mutations at the creA locus, leading to de-repression of alcA(p), have been obtained after mutagenesis (Gatherar 2004).
Phenotypic comparison of progeny produced in the sexual cross between strains PKX23 (alcA(p)::cotA) and XX5 (nudA2); a grown on YEPD at 30°C for alcA repression, b grown on MM with glycerol at 44°C for 48 h for temperature sensitivity, c grown in YEPD at 44°C for 15 h. (i) alcA(p)::cotA; (ii) nudA; (iii) wild type; (iv) alcA(p)::cotA, nudA. Scale bars 20 μm
GFP tagging of CotA
To locate the CotA protein, 3′ GFP and RFP tagged strains were constructed. Tagging cassettes were created and 11 transformants from each of the resulting transformations were selected for further analysis. Nine of the 11 RFP transformants were shown to be single-copy homologous integrants by PCR screen and Southern blot, as were all of the GFP transformants. No morphological differences were seen compared to the SO451 recipient, indicating normal function of the CotA fusions. Data is given for strain CRK17 (CotA-RFP), though other tested transformants showed a similar phenotype.
Microscopic localisation of CotA
Fluorescence microscopy showed the presence of bright patches throughout the cytoplasm in swollen spores, young germlings and older hyphae (Fig. 7) in most samples examined. A small proportion of hyphae (15 h) showed a distinct cap at the growing tip, and cytoplasmic patches were less obvious. An N-terminal GFP fusion was also constructed under the control of the alcA promoter (data not shown). Similar cytoplasmic patches were also observed, but no cap was seen at the tip.
Localisation of CotA-RFP in CRK17. The presence of bright patches of CotA throughout the cytoplasm is seen in both germinating spores (a) and older hyphae ranging from 15 to 20 h (b, c). Occasionally a cap of CotA can be seen at the growing tip and in these cells cytoplasmic patches are less obvious (d)
Discussion
cot-1 like genes have been identified in all eukaryotic species examined, but it is not clear to what extent these genes are truly orthologous, since functional analysis has revealed some differences. A common theme in filamentous fungi and yeasts is that they are required for maintaining cell polarity. They have been classified by similarities in their kinase domains, and the presence of additional, non-conserved sequence between domains VII and VIII. The greatest variation in the members of this family is in the N-terminal region, which is of variable length, and differs in the presence and length of glutamine rich tracts. The function of these tracts in this family of proteins is unclear, though they are often associated with transcriptional activation (Gerber et al. 1994; Chen and Dickman 2002). N. crassa and other Sordariomycetes have the most closely related genes to the Aspergillus species, but the Aspergillus glutamine tracts are not very extensive compared to the Sordariomycetes. While S. cerevisiae and C. albicans have glutamine-rich N-terminal regions, S. pombe Orb6 and related mammalian kinases NDR1 and NDR2 lack this region entirely.
In N. crassa, the cot-1 phenotype is partially suppressed by mutations in genes affecting nuclear migration, including ro-1, ro-3 and ro-4, which encode subunits of the dynein/dynactin complex (Bruno et al. 1996), though the mechanism of suppression remains unclear. Nevertheless, it provided a very effective method of isolating new nuclear migration mutants in N. crassa. Since cotA appears to be the orthologue of cot-1, and orthologues of ro-1 and other nuclear migration mutants have been investigated extensively in A. nidulans, we were interested to see whether the cotA phenotype of the downregulated strain could be suppressed by a ts allele of nudA, the orthologue of N. crassa ro-1, which encodes for the dynein large subunit. This is a component of the motor protein needed for nuclear migration along the microtubules, and mutations in ro-1 are the most commonly isolated suppressors of cot-1 in N. crassa.
In the case of A. nidulans, the mutations were additive, suggesting species differences in the relationship between the cotA signalling pathway and nuclear migration. However, this additive phenotype may be specific to the alcA(p)::cotA allele used in this study. Attempts to introduce the amino change present in the N. crassa cot-1 ts mutant were unsuccessful (Gatherar 2004). The mechanism of suppression in N. crassa has not been explained, but Plamann et al. (1994) proposed that the need for COT1 kinase activity is partially bypassed by decreased dynein activity, since the suppression effect is observed with mutations in several different ro genes.
Functional and genetic analysis of the orb6 gene of S. pombe led to a model for a signalling pathway in which Orb6 acts to promote cell polarity, but also to delay mitosis (Verde et al. 1998).
Partial downregulation of orb6 in S. pombe led to shorter cells, while overexpression led to elongated cells, and it was proposed that orb6 might act negatively on cdc2 and delay mitosis, in addition to activating polar growth. In A. nidulans, overexpression of CotA does not lead to any significant change in morphology compared to wild-type strains. Downregulation leads to enlarged germinating spore compartments with an increased number of nuclei. However, the nucleus number/volume ratio did not change significantly. This suggests that the increased nuclear number may be a response to the increase in cell volume which results from the polarity defect, rather than removal of negative regulation of mitosis. Unlike the yeasts, hyphal compartments are multinucleate, with nuclear numbers broadly related to compartment length/volume. Apical hyphal compartments in growing mycelium are typically long, with around 40–80 nuclei. Parasynchronous mitoses followed by multiple septation leads to about four nuclei per sub-apical compartment (Fiddy and Trinci 1976; Trinci 1979). The control circuit which maintains a relationship between cell volume, mitosis and septation is only partly understood (Wolkow et al. 1996), but unlike the S. pombe downregulated orb6 mutant, the relationship of cell volume and mitosis appears to be still intact after depletion of CotA in A. nidulans. Similarly, mutations in hypA and other hyp loci also lead to activated isotropic growth and mitosis in hyphal compartments (Kaminsky and Hamer 1998). hypA encodes a TRS120 homologue, a component of the TRAPP II complex mediating Golgi traffic, and mutation leads to a polarity defect (Shi et al. 2004).
Cbk1p of S. cerevisiae is involved in polar growth, but also needed for cell separation. It has been shown that Cbk1p acts in association with Mob2p on ACE2, which encodes a transcription factor involved in cell separation (O’ Conallain et al. 1999), but the polarity related targets remain to be elucidated.
Mob2 is related to Mob1, a component of the mitotic exit network, and Mob1 is associated with a kinase Dbf2, which resembles Cbk1p. These genes, together with other Ace2p transcribed genes, are required for the M–G1 transition.
NDR1 and NDR2 have also been shown to associate with human homologues of Mob1 and Mob2 (Devroe et al. 2004), and NDR2 autophosphorylation is stimulated by Mob2, though the substrates of these kinases are not yet known. Disruption of NDR2 is found in some murine B-cell lymphomas, implying a role in cell proliferation/tumour progression in mammals.
Both NDR1 and NDR2 are found in the cytoplasm, though NDR1 is also found in the nucleus, while NDR2 seems to be excluded. This difference in localisation is difficult to explain, since there is very little difference in the putative nuclear localisation signal. It was suggested that glutamine-rich N-terminal region of TB3 of C. trifolii is associated with nuclear localisation (Chen and Dickman 2002), though there as yet is no clear correlation between structure/presence of the N-terminal region and localisation in this family. As is the case with CotA, tagged-NDR2 shows a punctate distribution in the cytoplasm, and it was suggested that this might reflect presence of some of this kinase in a subcellular compartment other than the nuclei. In addition, we occasionally observed a cap of CotA at the growing tip, as seen for Orb6 in S. pombe (Verde et al. 1998), which may reflect its role in polar growth. Further work is underway to identify the subcellular compartments in which it is usually found.
The A. nidulans genome contains a gene similar to MOB1 (AN6288.2, 37% amino acid identity), and a weaker hit that might correspond to MOB2 (AN1370.2, 32%). A good candidate for an ACE2 homologue is also present (AN4873.2, 50%). Putative MOB1 homologues appear to be better conserved than the putative MOB2 homologues within the filamentous fungal genomes available to date.
While CBK1, cot-1 and cotA may carry out some similar functions, deletion of CBK1 indicates that it is not essential, whereas deletion/depletion of cot-1 and cotA genes respectively results in a severe growth defect (Yarden et al. 1992; this work). Recently, it was shown that deletion of the Claviceps purpurea homologue is possible, but leads to a severe growth defect and loss of pathogenicity (Scheffer et al. 2005). The sequence differences and functional differences of the genes in this family indicate mechanistic differences for these putative orthologues.
References
Armitt S, McCullough W, Roberts CF (1976) Analysis of acetate non-utilizing (acu) mutants in Aspergillus nidulans. J Gen Mirobiol 92:263–282
Ballance DJ, Turner G (1985) Development of a high-frequency transforming vector for Aspergillus nidulans. Gene 36:321–331
Bidlingmaier S, Weiss EL, Seidel C, Drubin DG, Snyder M (2001) The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol Cell Biol 21:2449–2462
Brakhage AA, Browne P, Turner G (1992) Regulation of Aspergillus nidulans penicillin biosynthesis and penicillin biosynthesis genes acvA and ipnA by glucose. J Bacteriol 174:3789–3799
Brody H, Griffith J, Cuticchia A, Arnold J, Timberlake W (1991) Chromosome-specific recombinant DNA libraries from the fungus Aspergillus nidulans. Nucleic Acids Res 19:3105–3109
Bruno KS, Tinsley JH, Minke PF, Plamann M (1996) Genetic interactions among cytoplasmic dynein, dynactin, and nuclear distribution mutants of Neurospora crassa. Proc Natl Acad Sci USA 93:4775–4780
Buhr TL, Oved S, Truesdell GM, Huang C, Yarden O, Dickman MB (1996) A kinase encoding gene from Colletotrichum trifolii complements a colonial growth mutant of Neurospora crassa. Mol Gen Genet 251:565–572
Chen C, Dickman MB (2002) Colletotrichum trifolii TB3 kinase, a COT1 homolog, is light inducible and becomes localized in the nucleus during hyphal elongation. Eukaryot Cell 1:626–633
Clutterbuck AJ (1969) Cell volume per nucleus in haploid and diploid strains of Aspergillus nidulans. J Gen Microbiol 60:133–135
Collinge AJ, Fletcher MH, Trinci APJ (1978) Physiology and cytology of septation and branching in a temperature sensitive colonial mutant (cot-1) of Neurospora crassa. Trans Br Mycol 77:107–120
Devroe E, Erdjument-Bromage H, Tempst P, Silver PA (2004) Human Mob proteins regulate the NDR1 and NDR2 serine-threonine kinases. J Biol Chem 279:24444–24451
Dorland S, Deegenaars ML, Stillman DJ (2000) Roles for the Saccharomyces cerevisiae SDS3, CBK1 and HYM1 genes in transcriptional repression by SIN3. Genetics 154:573–586
Fiddy C, Trinci APJ (1976) Mitosis, septation, branching, and the duplication cycle in Aspergillus nidulans. J Gen Microbiol 97:169–184
Gatherar IM (2004) The genetic control of hyphal morphology in Aspergillus. PhD Thesis, University of Sheffield
Gatherar IM, Pollerman S, Dunn-Coleman N, Turner G (2004) Identification of a novel gene hbrB required for polarised growth in Aspergillus nidulans. Fungal Genet Biol 41:463–471
Gerber HP, Seipel K, Georgiev O, Hofferer M, Hug M, Rusconi S, Schaffner W (1994) Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science 263:808–811
Gorovits R, Propheta O, Kolot M, Dombradi V, Yarden O (1999) A mutation within the catalytic domain of COT1 kinase confers changes in the presence of two COT1 isoforms and in Ser/Thr protein kinase and phosphatase activities in Neurospora crassa. Fungal Genet Biol 27:264–274
Gorovits R, Sjollema KA, Sietsma JH, Yarden O (2000) Cellular distribution of COT1 kinase in Neurospora crassa. Fungal Genet Biol 30:63–70
Hanks SK, Quinn AM, Hunter T (1988) The protein-kinase family-conserved features and deduced phylogeny of the catalytic domains. Science 241:42–52
Harris SD, Momany M (2004) Polarity in filamentous fungi: moving beyond the yeast paradigm. Fungal Genet Biol 41:391–400
Harris SD, Morrell JL, Hamer JE (1994) Identification and characterisation of Aspergillus nidulans mutants defective in cytokinesis. Genetics 136:517–532
Harris SD, Hamer L, Sharpless KE, Hamer JE (1997) The Aspergillus nidulans sepA gene encodes an FH1/2 protein involved in cytokinesis and the maintenance of cellular polarity. EMBO J 16:3474–3483
Harris SD, Read ND, Roberson RW, Shaw B, Seiler S, Plamann M, Momany M (2005) Polarisome meets spitzenkörper: microscopy, genetics and genomics converge. Eukaryot Cell 4:225–229
Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ (1995) The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev 9:534–546
Kaminsky SGW, Hamer JE (1998) hyp loci control cell pattern formation in the vegetative mycelium of Aspergillus nidulans. Genetics 148:669–680
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Lauter FR, Marchfelder U, Russo VE, Yamashiro CT, Yatzkan E, Yarden O (1998) Photoregulation of cot-1, a kinase-encoding gene involved in hyphal growth in Neurospora crassa. Fungal Genet Biol 23:300–310
Maniatis T, Fritsch EF, Sambrook J (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Millward T, Cron P, Hemmings BA (1995) Molecular cloning and characterization of a conserved nuclear serine(threonine) protein kinase. Proc Natl Acad Sci USA 92:5022–5026
Mitchell MB, Mitchell HK (1954) A partial map of linkage group D in Neurospora crassa. Proc Natl Acad Sci USA 40:436–440
Momany M (2002) Polarity in filamentous fungi: establishment, maintenance and new axes. Curr Opin Microbiol 5:580–585
Momany M, Westfall PJ, Abramowsky G (1999) Aspergillus nidulans swo mutants show defects in polarity establshment, polarity maintenance and hyphal morphogenesis. Genetics 151:557–567
Morris NR (1975) Mitotic mutants of Aspergillus nidulans. Genet Res 26:237–254
Nasr F, Becam AM, Herbert CJ (1996) The sequence of 36.8 kb from the left arm of chromosome XIV reveals 24 complete open reading frames: 18 correspond to new genes, one of which encodes a protein similar to the human myotonic dystrophy kinase. Yeast 12:169–175
Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, Hynes MJ, Osmani SA, Oakley BR (2006) A versatile and efficient gene targeting system for Aspergillus nidulans. Genetics (in press)
O’ Conallain C, Doolin MT, Taggart C, Thornton F, Butler G (1999) Regulated nuclear localisation of the yeast transcription factor Ace2p controls expression of chitinase (CTS1) in Saccharomyces cerevisiae. Mol Gen Genet 262:275–282
Pearson CL, Xu K, Sharpless KE, Harris SD (2004) MesA, a novel fungal protein required for the stabilization of polarity axes in Aspergillus nidulans. Mol Biol Cell 15:3658–3672
Plamann M, Minke PF, Tinsley JH, Bruno KS (1994) Cytoplasmic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J Cell Biol 127:139–149
Pontecorvo G, Roper JA, Hemmons LM, MacDonald K, Bufton AWJ (1953) The genetics of Aspergillus nidulans. Adv Genet 5:141–239
Prade RA, Griffith J, Kochut K, Arnold J, Timberlake WE (1997) In vitro reconstruction of the Aspergillus (=Emericella) nidulans genome. Proc Natl Acad Sci USA 94:14564–14569
Racki WJ, Becam AM, Nasr F, Herbert CJ (2000) Cbk1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. EMBO J 19:4524–4532
Ramon A, MuroPastor MI, Scazzocchio C, Gonzalez R (2000) Deletion of the unique gene encoding a typical histone H1 has no apparent phenotype in Aspergillus nidulans. Mol Microbiol 35:223–233
Scheffer J, Ziv C, Yarden O, Tudzynski P (2005) The COT1 homologue CPCOT1 regulates polar growth and branching and is essential for pathogenicity in Claviceps purpurea. Fungal Genet Biol 42:107–118
Seiler S, Plamann M (2003) The genetic basis of cellular morphogenesis in the filamentous fungus Neurospora crassa. Mol Biol Cell 14:4352–4364
Shi X, Sha Y, Kaminskyj S (2004) Aspergillus nidulans hypA regulates morphogenesis through the secretion pathway. Fungal Genet Biol 41:75–88
Steele GC, Trinci APJ (1977) Effect of temperature and temperature shifts on growth and branching of a wild type and a temperature sensitive colonial mutant (cot-1) of Neurospora crassa. Arch Microbiol 113:43–48
Trinci APJ (1979) The duplication cycle and branching in fungi. In: Burnett JH, Trinci APJ (eds) Fungal walls and hyphal growth. Cambridge University Press, Cambridge, pp 319–358
Verde F, Wiley DJ, Nurse P (1998) Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and co-ordinates cell morphogenesis with the cell cycle. Proc Natl Acad Sci USA 95:7526–7531
Waring RB, May GS, Morris NR (1989) Characterisation of an inducible expression system in Aspergillus nidulans using alcA and tubulin-coding genes. Gene 79:119–130
Wolkow TD, Harris SD, Hamer JE (1996) Cytokinesis in Aspergillus nidulans is controlled by cell size, nuclear positioning and mitosis. J Cell Sci 109:2179–2188
Xiang X, Fischer R (2004) Nuclear migration and positioning in filamentous fungi. Fungal Genet Biol 41:411–419
Xiang X, Beckwith SM, Morris NR (1994) Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc Natl Acad Sci USA 91:2100–2104
Xiang X, Roghi C, Morris NR (1995) Characterisation and localisation of the cytoplasmic dynein heavy chain in A. nidulans. Proc Natl Acad Sci USA 92:9890–9894
Yang L, Ukil L, Osmani A, Nahm F, Davies J, De Souza CP, Dou X, Perez-Balaguer A, Osmani SA (2004) Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot Cell 3:1359–1362
Yarden O, Plamann M, Ebbole DJ, Yanofsky C (1992) cot-1, a gene required for hyphal elongation in Neurospora crassa, encodes a protein kinase. EMBO J 11:2159–2166
Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Dominguez Y, Scazzocchio C (2004) Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol. 41:973–981
Zallen JA, Peckol EL, Tobin DM, Bargmann CI (2000) Neuronal cell shape and neurite initiation are regulated by the Ndr kinase SAX-1, a member of the Orb6/COT-1/warts serine/threonine kinase family. Mol Biol Cell 11:3177–3190
Zarrin M, Leeder AC, Turner G (2005) A rapid method for promoter exchange in Aspergillus nidulans using recombinant PCR. Fungal Genet Biol 42:1–8
Acknowledgements
This work was supported by BBSRC grant 50/G06623 to GT, BBSRC studentships to SJ and ACL and a Ministry of Culture and Higher Education of Iran studentship to MS. We thank Mrs. J. Price and Ms. N. Frima for technical assistance. All genetic manipulation experiments were carried out in accordance with local regulations.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Punt.
Rights and permissions
About this article
Cite this article
Johns, S.A., Leeder, A.C., Safaie, M. et al. Depletion of Aspergillus nidulans cotA causes a severe polarity defect which is not suppressed by the nuclear migration mutation nudA2 . Mol Genet Genomics 275, 593–604 (2006). https://doi.org/10.1007/s00438-006-0113-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-006-0113-0