Abstract
Brassinosteroids (BR) promote the elongation of cotton fibers and may be a factor in determining their final length. To begin to understand the role of BR-mediated responses in the development of cotton fibers we have characterized the BIN2 genes of cotton. BIN2 is a member of the shaggy-like protein kinase family that has been identified as a negative regulator of BR signaling in Arabidopsis. Sequence analyses indicate that the tetraploid cotton genome includes four genes with strong sequence similarity to BIN2. These genes fall into two distinct subclasses based on sequence and expression patterns. Sequence comparisons with corresponding genes from cotton species that have the diploid A and D genomes, respectively, shows that each pair of genes comprises homeologs derived from the A and D sub-genomes. Transgenic Arabidopsis plants that express these cotton BIN2 cDNAs show reduced growth and similar phenotypes to the semi-dominant bin2 mutant plants. These results indicate that the cotton BIN2 genes encode functional BIN2 isoforms that can inhibit BR signaling. Further analyses of the function of BIN2 genes and their possible roles in determining fiber yield and quality are underway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
More than 14 billion kilogram of cotton fiber are produced annually world-wide. Cotton fibers are seed trichomes that undergo rapid and extensive elongation and can typically reach lengths of 30 mm. Phytohormones, including brassinosteroids (BR), are critical regulators of cotton fiber development. Analysis of fiber development on cotton ovules grown in vitro in liquid medium indicates that exogenous auxin and gibberellic acid (GA) are required for maximal fiber elongation (Beasley and Ting 1974). Likewise, BR treatments increase fiber growth both in cultured ovules and in planta (Ashcraft 1996; Kasukabe et al. 2000). Furthermore, the brassinosteroid biosynthesis inhibitor brassinazole (Asami et al. 2000; Sekimata et al. 2001) strongly inhibits fiber development in ovule culture (Sun et al. 2004). These results indicate that BR plays an important role in promoting cotton fiber initiation and elongation. Therefore, to better understand the role of BR in cotton development, we have undertaken an analysis of the BR signaling pathway in this species.
Brassinosteroids are the only steroid-like growth regulators found throughout the plant kingdom that are required for normal plant development. These molecules have wide ranging effects on plants, which are similar to those of auxin in many respects. Like auxin, BR promotes growth of shoots and inhibits root elongation (Mandava 1988). BR also promotes seed germination and pollen development. Recent transcriptional profiling studies have shown that BR and auxin affect the expression of overlapping sets of genes, but genes that respond specifically to BR or auxin were also identified (Goda et al. 2004). In addition, mutant plants with lesions in genes that disrupt the BR signaling pathway generally remain responsive to auxin and vice versa (Goda et al. 2002).
Several components of the BR signal transduction pathway have recently been identified. Li and Chory (1997) described a gene from Arabidopsis, BRASSINOSTEROID INSENSITIVE 1 (BRI1), which encodes a putative membrane-associated BR receptor. BRI1 codes for a leucine-rich-repeat receptor-like kinase (LRR-RLK) that is localized in the plasma membrane (Friedrichsen et al. 2000; Oh et al. 2000). BRI1 ASSOCIATED RECEPTOR KINASE1 (BAK1) encodes an LRR-RLK distinct from BRI1 that is a component of the BR signaling pathway and may interact with BRI1 (Li et al. 2002). BRASSINOSTEROID INSENSITIVE 2 (BIN2) was identified upon re-examination of BR insensitive dwarf mutants of Arabidopsis. bin2-1 represents a semidominant gain-of-function mutation that causes a bri1-like dwarf phenotype (Li et al. 2001). Therefore, BIN2 was proposed to encode a negative regulator of BR signaling. The protein encoded by the gene BRASSINAZOLE-RESISTANT 1 (BZR1) is a positive regulator of BR signaling that acts downstream of BRI1 and BIN2. BZR1 was found to be stabilized and accumulates in the nucleus in response to treatment with BR, and in plants that carry the dominant allele bzr1-1D (Wang et al. 2002). BRI1-EMS-SUPPRESSOR 1 (BES1) encodes a BZR1-like protein that also accumulates in the nucleus in response to BR, and its accumulation is negatively regulated by BIN2. BES1 has been shown to interact directly with BIN2 in both yeast two-hybrid and GST pull down assays, and BIN2 efficiently phosphorylates BES1 in vitro (Yin et al. 2002). BRASSINOSTEROID SUPPRESSOR1 (BSU1) was recently found to encode a nuclear protein phosphatase that counteracts the action of BIN2 by dephosphorylating BES1 and stabilizing it (Mora-Garcia et al. 2004).
The BR signaling cascade is similar in many respects to the Wingless/Wnt signaling pathway of vertebrates and invertebrates (Kim and Kimmel 2000) with BIN2 acting as a central switch. Under steady-state conditions, BIN2 is active and phosphorylates BZR1 and BES1, which then become targets for degradation. Interaction with BR triggers hetero-dimerization and kinase activation of the BRI1/BAK1 receptor complex (Nam and Li 2002). Although BIN2 does not appear to be a direct target of BRI1/BAK1 (Nam and Li 2004), exposure to BR presumably leads to inactivation of this negative regulator. Reduced BIN2 activity allows hypo-phosphorylated forms of BZR1 and BES1 to accumulate and translocate to the nucleus, where they affect BR-responsive gene expression (He et al. 2002; Zhao et al. 2002).
The Arabidopsis BIN2 (AtBIN2) gene encodes a member of the glycogen synthase kinase-3/SHAGGY-like kinase (GSK3) family, a highly conserved family of serine/threonine kinases found in many eukaryotes (Li and Nam 2002). Embi et al. (1980) first identified GSK3 as an enzyme that phosphorylates glycogen synthase. Members of this family often function as negative regulators of signal transduction pathways (Kim and Kimmel 2000). GSK3 was recently found to be an important and versatile switch in target discrimination and pathway insulation (Kim and Kimmel 2000). The mutant form of AtBIN2 encoded by bin2-1 was shown to have increased kinase activity when expressed in Escherichia coli. This protein kinase apparently blocks BR signal transduction by hyperphosphorylation of BRZ1/BES1, leading to a pronounced BR-insensitive dwarf phenotype. On the other hand, reduced expression of BIN2 partially rescues the phenotype of the weak bri1-5 mutant (Li and Nam 2002). Therefore, BIN2 appears to be a negative regulator of BR signaling in plants, which is constitutively active in the absence of BR and phosphorylates other BR signaling regulators, targeting them for degradation (Clouse 2002).
BRI1 homologs have been identified in rice (Yamamuro et al. 2000), tomato (Montoya et al. 2002), pea (Nomura et al. 2003) and barley (Chono et al. 2003), and we recently characterized two BRI1 genes from cotton (Gossypium hirsutum; Sun et al. 2004). Here we report the functional analysis of four putative BIN2 genes from cotton. Overexpression of each of the cotton BIN2 genes in wild-type Arabidopsis plants resulted in severe stunting and phenotypes similar to those of strong BR-insensitive mutants, indicating they can indeed function as negative regulators of BR signal transduction. We anticipate that further dissection of the BR signaling pathway in cotton will reveal the role of BR in regulating cotton fiber development.
Materials and methods
Plant growth condition
Cotton plants [Gossypium hirsutum cv. Coker 312, G. arboreum (accession A2-347) and G. thurberi (accession D1-4)] were grown in potting soil in a greenhouse. Arabidopsis plants (Arabidopsis thaliana ecotype Ws-2) were grown under continuous fluorescent light in potting soil at room temperature.
Isolation of genomic DNA and total RNA
Cotton and Arabidopsis genomic DNA samples were isolated from leaf tissues using the DNeasy Plant Mini Kit (Qiagen, Santa Clara, CA, USA).
RNA was isolated from leaves, buds and sepals of mature cotton plants and from hypocotyls and roots of seedlings (7–10 days post-imbibition). Flowers were tagged on the day of anthesis, and bolls were harvested at 0 DPA (days post-anthesis), 5 DPA, 10 DPA and 20 DPA. Immature ovules (5–8 DPA), 20-DPA fibers, and 30-DPA fibers were also collected. Samples were ground to a powder in liquid nitrogen and stored at −80°C. Total RNA was extracted using the method of Wan and Wilkins (1994). Arabidopsis RNA was isolated from whole plant tissues using the RNeasy Plant Mini Kit (Qiagen).
Cloning of GhBIN2 cDNAs
Two cotton ESTs (Genbank Accession Nos. AI726755 and AI731919) with high sequence homology to Arabidopsis BIN2 (Li and Nam 2002) were identified in the NCBI GenBank dbEST database using the BLAST program. These ESTs correspond to the 5′-end of the AtBIN2 sequence (96–666 and 1–600) and were used as starting points to amplify GhBIN2 cDNAs from a cotton boll cDNA library. Vector-specific primers that annealed to the lambda ZAP cDNA vector were SK-P (5′-CGCTCTAGAACTAGTGGATC-3′) and T7 (5′-GTAATACGACTCACTATAGGGC-3′). Primers B1 (5′-CTTGATAGCAACCTCGTGCC-3′) and B2 (5′-GAGTTGATGGAAGAAGACCCC-3′), derived from the putative GhBIN2 ESTs were used as BIN2-specific primers. PCR was performed, and the amplified fragments were subcloned into the vector pGEM-T Easy (Cat. No. A1360; Promega, Madison, WI, USA) and sequenced for comparison with AtBIN2. Fragments that shared high sequence similarity were selected for the design of further primers, and PCR amplification was carried out to isolate full length GhBIN2 cDNAs from the cDNA library. Four different GhBIN2 cDNA clones (GhBIN2-B, GhBIN2C, GhBIN2-D, and GhBIN2-E) were sequenced to obtain the four full-length sequences.
Quantitative real-time PCR
Reverse transcription PCR (RT-PCR) assays were carried out on total RNA samples using TaqMan Reverse Transcription Reagents (Cat. No. N808-0234; Applied Biosystems, Foster City, CA, USA). Cotton RNA samples were quantified using a Nanodrop spectrophotometer. All cotton RNA samples were diluted to 100 ng/μl. A total of 0.5 μg of RNA was used for each 50-μl RT-PCR. Random hexamers were used for first-strand cDNA synthesis. Levels of the GhBIN2 mRNAs in total RNA samples from different cotton tissues and transgenic Arabidopsis plants were quantified with an ABI (Applied Biosystems) PRISM 7000 Sequence Detection System. The GhBIN2-specific FAM-labeled TaqMan probes and forward and reverse primers were derived from the 3′ ends of the coding sequences, which are sufficiently divergent to distinguish between the four GhBIN2 sequences. They were designed using ABI Primer Express 2.0 and synthesized by Applied Biosystems (their sequences are listed in Table 1). An 18S rRNA-specific VIC-labeled TaqMan probe and 18S rRNA forward and reverse primers were used for RNA normalization. The TaqMan PCR Core Reagent Kit (N808-0228; Applied Biosystems) was used for quantitative real-time PCR. Control reactions without reverse transcriptase were included to check each RNA sample for possible amplification of contaminating genomic DNA. Each of the four GhBIN2 FAM-labeled probes (first probe), forward and reverse primers, the 18S rRNA VIC-labeled probe (second probe), 18S rRNA forward and reverse primers, AmpErase UNG (uracil-it N-glycosylase), AmpliTaq Gold DNA polymerase and other components were used in the same tube for multiplex PCR. Each RNA sample was assayed three times.
The relative expression levels of all the samples were calculated according to the method recommended in User Bulletin No. 2 for the ABI PRISM 7700 Sequence Detection System. In brief, the threshold cycle (Ct) values for GhBIN2 mRNA and 18S RNA in different samples were obtained by quantitative real-time PCR. The Ct value for the normalizer 18S RNA was subtracted from the GhBIN2 Ct value to obtain the dCt value of the sample. The dCt value of the calibrator sample (the sample with the highest dCt value) was subtracted from every other sample to produce the ddCt value. The relative expression levels are equivalent to 2−ddCt for each sample.
Functional analysis of GhBIN2 cDNAs in transgenic Arabidopsis
The four GhBIN2 cDNAs were separately inserted into pBI121 in place of the uidA gene. The plant gene expression cassettes were constructed as follows. The pGEM-T plasmids containing GhBIN2-B and GhBIN2-D were cleaved with SacII and the ends were blunted by a 3′ fill-in reaction using T4 DNA polymerase, then digested with SacI and the resulting fragments were ligated into pBI121 DNA that had been digested with SmaI and SacI. The pGEM-T plasmids containing GhBIN2-C and GhBIN2-E were digested with SacII and StuI, and the ends blunted by 3′ fill-in reaction using T4 DNA polymerase. The resulting fragments were then ligated into pBI121 DNA that had been digested with SmaI and SacI, and blunted by 3′ fill-in using T4 DNA polymerase. The ligated plasmids were transformed into E. coli. pBI121-GhBIN2 plasmids were recovered from bacterial transformants, and transformed into Agrobacterium tumefaciens GV3101 (C58). A. tumefaciens cells containing pBI121-GhBIN2 constructs were selected on LB plates containing kanamycin (50 μg/ml), gentamycin (25 μg/ml) and rifampicin (100 μg/ml). The plasmids were isolated and the inserts were sequenced to confirm the presence and accuracy of the GhBIN2 ORFs.
The GhBIN2 transgenes were introduced into Ws-2 Arabidopsis plants, using the Agrobacterium-mediated-flower infiltration transformation method (Clough and Bent 1998). The plants in pots were placed in trays and covered with plastic wrap for 16–24 h to maintain high humidity. Plants were watered from the bottom of the tray and watering was reduced as seeds matured. Mature seeds were harvested from individual plant lines.
Arabidopsis seeds were sterilized with 75% ethanol for 1 min and 50% bleach solution for 10 min. The seeds were then rinsed with sterile water (three to five times) and plated. Transformed seedlings were selected on antibiotic-containing MS plates (250 μg/ml carbenicillin, 250 μg/ml cefotaxime, 50 μg/ml kanamycin). The PCR analysis using specific primers designed from the CaMV 35S promoter and the GhBIN2 coding sequences were performed to confirm the insertion of the GhBIN2 transgenes.
Results
Characterization of GhBIN2 cDNAs
The NCBI dbEST database was screened for cotton ESTs that show sequence similarity to BIN2 from A. thaliana (Li and Nam 2002), using the BLAST program. Two cotton ESTs (Genbank Accession Nos. AI726755 and AI731919) from G. hirsutum with high sequence identity to AtBIN2 were identified. These ESTs, which correspond to the 5′-end of the AtBIN2 coding sequence, were used as starting points to amplify GhBIN2 sequences from cotton.
Primers derived from the putative GhBIN2 ESTs were used, together with 5′ and 3′ primers based on the lambda ZAP vector, to amplify corresponding cDNAs from a cotton cDNA library. The DNA fragments obtained by PCR were subcloned, sequenced and compared to AtBIN2. DNA fragments that shared high sequence identity with AtBIN2 were then used for the next amplification step. New primers were designed on the basis of the elongated DNA sequence and, after several such steps, primers were designed to amplify the complete GhBIN2 coding sequences from the cotton cDNA library. Finally, full-length cDNAs were amplified. The amplified cDNA fragments were cloned and twelve representative cDNAs were sequenced. Each of these cDNAs includes a 1143-bp ORF that encodes a derived polypeptide of 381 amino acids—one amino acid longer than AtBIN2.
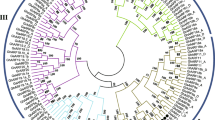
Comparisons of BIN2 cDNA sequences from cotton and Arabidopsis using the Vector NTI Align X program showed that the ORFs of the GhBIN2 cDNAs share between 77.7 and 79.4% nucleotide sequence identity with AtBIN2. Comparisons of the derived amino acid sequences of GhBIN2 proteins with AtBIN2 showed 87.4–90.6% sequence identity (Fig.1). From these alignments, it became clear that the GhBIN2 cDNAs fell into four distinct sequence classes. Representative cDNAs for each class were named GhBIN2-B, GhBIN2-C, GhBIN2-D, and GhBIN2-E. GhBIN2-B and GhBIN2-D share 96.6% nucleotide sequence identity (including the 3′ untranslated region) and their products are 97.4% identical. Although GhBIN2-C and GhBIN2-E share only 93.4% nucleotide sequence identity, their derived amino acid sequences are 99.7% identical, differing only by a single conservative substitution in the 381-amino acid polypeptide. One of the ESTs (AI726755) used as starting points for the amplification of these cDNAs corresponds to GhBIN2-D, while the other (AI731919) corresponds to GhBIN2-E.
BLAST analyses of the four GhBIN2 sequences indicated that they are all very similar to GSK3/SHAGGY-like kinase sequences. The alignment scores (bits) are as high as 600–700. Sequence comparison with mammalian GSK3β (Strausberg 2002) and the Drosophila SHAGGY kinase (Bourouis et al. 1990) showed that derived GhBIN2 sequences share many conservative regions with these two proteins (data not shown). Based on the extensive identity between the GhBIN2 sequences and AtBIN2, we predicted that these cDNAs encode functional homologs of AtBIN2, and are likely to play important roles in the BR signal transduction pathway in cotton.
Expression of GhBIN2 genes
Quantitative real-time PCR assays were performed to compare the relative expression levels of GhBIN2 genes in cotton plants. Total RNAs isolated from a variety of tissues from cotton plants including leaves, buds, hypocotyls, roots, sepals, 5-day to 8-day ovules, and samples from fibers at different developmental stages were used for this analysis. As shown in Fig.1, GhBIN2-B and GhBIN2-D fall into one sequence type, while GhBIN2-C and GhBIN2-E can also be grouped together. Interestingly, gene expression analyses also show that the GhBIN2 genes can be classified into two groups. GhBIN2-C and GhBIN2-E mRNAs are expressed in similar patterns throughout the cotton plant. Highest expression levels were seen in hypocotyls and in buds, with lower levels in sepals. In cotton bolls, these mRNAs were expressed at higher levels in 5-DPA and 10-DPA bolls than in 0-DPA bolls. They were expressed at high levels in 5–8 DPA ovules, and at lower levels in 30-DPA fibers. The expression of GhBIN2-E in 30-DPA fiber was used as the standard for comparative expression analyses. In general, GhBIN2-C and GhBIN2-E are expressed in patterns that are similar to that of GhBRI1 (Sun et al. 2004). Expression of these mRNAs is higher in tissues that are undergoing rapid cell expansion and/or vascular development (Fig.2, lower panels). Conversely, mRNAs corresponding to GhBIN2-B and GhBIN2-D are expressed constitutively at relatively high levels in all of the cotton tissues tested (Fig.2, upper panels). With the exception of the 30-DPA fiber sample, the levels of both GhBIN2-B and GhBIN2-D mRNAs were not significantly different from tissue to tissue and the expression levels of these mRNAs were between 3-fold and 20-fold higher than that of GhBIN2-C or GhBIN2-E in corresponding RNA samples.
GhBIN2s expression levels in cotton plants. The levels of GhBIN2 mRNAs in total RNA samples were assayed by a duplex TaqMan quantitative real-time PCR assay using 18S rRNA as a reference. The sample with the lowest ratio of GhBIN2-E mRNA to 18S RNA (30 DPA fiber) was set to 1 and the values for the other samples are expressed relative to that value. Values are means (±SD) of three independent assays
Identification of the two homeologous GhBIN2 loci in the tetraploid cotton genome
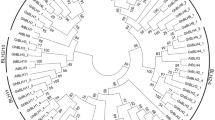
Cultivated cotton varieties (G. hirsutum and G. barbadense) are allotetraploids (2 n = 4 x = 52, AADD) that originated from diploid cotton ancestors (Wendel et al. 1992). The two extant diploid cotton species with the A-genome (2 n = 2 x = 26, AA), G. arboreum and G. herbaceum, are thought to be closely related to the A sub-genome of the tetraploid cottons. G. thurberi and G. raimondii are among several extant D-genome diploid cotton species (2 n = 2 x = 26, DD) and G. raimondii is considered to be most closely related to the D sub-genome of the tetraploid cotton species (Iqbal et al. 2001). As shown in Fig.1, GhBIN2-B and GhBIN2-D fall into one sequence type, while GhBIN2-C and GhBIN2-E represent a distinct type, and gene expression data further support the grouping of these genes (see Fig.2). Based on these results, it is apparent that these cDNAs represent four GhBIN2 genes that fall into two distinct structural and functional classes. Therefore, we speculated that one member of each pair of GhBIN2 cDNAs is derived from the A sub-genome while the second originates from the D sub-genome. If this were the case, we would expect diploid cottons to contain two BIN2 loci; one locus corresponding to the GhBIN2-B/D class of genes and a second corresponding to the GhBIN2-C/E genes. To test this hypothesis, BIN2 cDNA fragments were amplified from both G. arboreum (GaBIN2) and G. thurberi (GtBIN2), using specific primers corresponding to each of the GhBIN2 cDNA classes. The resulting cDNA amplicons were sequenced and compared to the GhBIN2 cDNAs. As predicted, two distinct BIN2 sequences were identified from G. arboreum (GaBIN2-1 and GaBIN2-2) and from G. thurberi (GtBIN2-1 and GtBIN2-2). Nucleotide sequence comparisons showed that GaBIN2-1 is most closely related to GhBIN2-B (99.3% identity) and GtBIN2-1 is nearly identical to GhBIN2-D (99.4% identity). Likewise, GtBIN2-2 shares 99.8% identity with GhBIN2-C and GaBIN2-2 is 99.8% identical to GhBIN2-E. A phylogenetic tree based on AlignX analyses of these eight sequences, together with the coding sequences of 10 Arabidopsis shaggy-like protein kinase genes, is shown in Fig.3. AtSK21 corresponds to AtBIN2 while AtSK22 and AtSK23 are the most closely related shaggy-like protein kinase genes. This analysis places GhBIN2-B and GhBIN2-D within a clade together with GaBIN2-1 and GtBIN2-1. GhBIN2-C and GhBIN2-E form a second clade with GaBIN2-2 and GtBIN2-2 (Fig.3). We interpret these data to indicate that GhBIN2-B and GhBIN2-E represent orthologous gene loci derived from the A sub-genome ancestor of cotton, while GhBIN2-D and GhBIN2-C represent the D sub-genome homeologues.
Phylogenetic tree based on the alignment of amino acid sequences derived from BIN2-encoding cDNAs from G. hirsutum (GhBIN2-B, GhBIN2-D, GhBIN2-C, and GhBIN2-E), and from the A-genome diploid species G. arboreum (GaBIN2-1 and GaBIN2-2) and the D-genome species G. thurberi (GtBIN2-1 and GtBIN2-2). Sequences derived from 10 Arabidopsis genes for shaggy-like kinases were also included in the analysis, which was performed using AlignX (Vector NTI)
Functional analysis of GhBIN2 gene products in Arabidopsis
To investigate the functional characteristics of these GhBIN2 genes, transgenic Arabidopsis plants were developed that express each of the four GhBIN2 cDNAs under the control of the strong, constitutive CaMV 35S promoter. Since overexpression of AtBIN2 in otherwise wild-type Arabidopsis was sufficient to produce a strong BR-insensitive phenotype (Li and Nam 2002), we predicted that, if functional, expression of the GhBIN2 cDNAs should also produce a dwarf and bin2 mutant phenotype.
The GhBIN2 transgenes were transformed into wild-type Ws-2 plants via floral inoculation, and T1 seedlings were selected on kanamycin-containing medium. Although many of the kanamycin-resistant plants had near-normal phenotypes, some of the GhBIN2-overexpressing plants were stunted to various degrees (Fig.4). The most severely affected had phenotypes similar to those of bin2 mutant plants. These plants grew as small, tightly compact rosettes with curled leaves that tended to be darker in color than wild-type plants. The shoot heights of these transgenic Arabidopsis plants were measured after 5 weeks of growth and compared to that of the wild-type Ws-2 plants. PCR analyses using specific primers designed from CaMV 35S promoter and GhBIN2 coding sequences were used to confirm the presence of GhBIN2 transgenes, and quantitative real-time PCR was used to assay the expression levels of the four GhBIN2s in individual transgenic plants.
Comparison of a wild-type Arabidopsis plant (Ws-2) with selected transgenic plants that express GhBIN2 gene constructs. Plants representing a series of bin2-like phenotypes are shown. The phenotypes range from mild growth inhibition to severe stunting. Photographs were taken 25 days after sowing. Scale bar 4 mm
Expression of each of the four GhBIN2 transgenes in Arabidopsis plants resulted in phenotypes ranging from mild to severe dwarfism and bin2 phenotypes. Quantitative real-time PCR analysis showed the level of GhBIN2 expression was negatively correlated with the height of the transgenic plants. As shown in Fig.5 (upper panels), GhBIN2-B and GhBin2-D expression levels were 4-fold to 6-fold higher in severely dwarfed transgenic plants than in moderately dwarfed plants. In addition, as also shown in Fig.5 (lower panels), GhBIN2-C and GhBin2-E expression levels were 3-fold to 4-fold higher in severely dwarfed plants than in plants with moderate growth inhibition. Correlation analysis showed a strong inverse correlation between GhBIN2 expression levels and the heights of the transgenic plants (mean correlation, r = −0.90). These results show that expression of each of the GhBIN2 genes in Arabidopsis results in a phenotype that is indistinguishable from that of the bin2-1 mutant or AtBIN2-overexpressing transgenic plants, indicating that all four GhBIN2 genes encode functional proteins that can act as negative regulators of the BR signal transduction pathway.
GhBIN2 expression levels are inversely correlated with the height of transgenic Arabidopsis plants. The mean height of wild-type plants (Ws-2) is shown. Several independent T0 GhBIN2 transgenic plants of similar heights were measured, then assayed for transgene expression. Plant heights are shown as shaded bars and GhBIN2 expression levels are shown as open bars. The error bars represent standard deviation (SD)
Discussion
Four cotton cDNAs were identified that display strong sequence identity to BIN2, a negative regulator of the BR signaling pathway in Arabidopsis. These four cDNAs encode putative proteins that fall into two sequence classes; two (GhBIN2-B and GhBIN2-D) are more than 97% identical to each other, and the members of the second pair (GhBIN2-C and GhBIN2-E) differ from each other by only a single amino acid. The identification of these subgroups is further supported by differences in their expression profiles.
There are at least 10 genes for GSK3/SHAGGY-like protein kinases (AtSK) in Arabidopsis (Dornelas et al. 1998) and Charrier et al. (2002) divided the entire AtSK gene family into four subgroups based on sequence similarity. In this scheme, AtSK22 and AtSK23 are most closely related to AtBIN2 (AtSK21). Our phylogenetic tree analysis showed that the GhBIN2 genes are more closely related to AtBIN2 than to AtSK22, AtSK23 or other AtSKs (Fig.3), which supports the hypothesis that the GhBIN2s are bona fide BIN2 homologs.
GSK3/SHAGGY-like protein kinases such as BIN2 are a group of conserved serine/threonine kinases that are involved in a number of signaling pathways (Li and Nam 2002). These enzymes are reported to control metabolism, cell fate determination, and tissue patterning in many organisms. Catalytic domains of the GhBIN2 family share 70% sequence identity to mammalian GSK3β (Strausberg 2002) and Drosophila SHAGGY kinase (Bourouis et al. 1990), and they also share many conservative regions including the eight subdomains found in other serine/threonine kinases (Hanks et al. 1988).
Expression analysis using real-time PCR revealed that these genes, including BIN2, are expressed in all tested Arabidopsis organs, with higher levels being found in flower buds and open flowers than in the other organs (Charrier et al. 2002). We found that each of the GhBIN2 genes is also expressed in all cotton tissues tested. However, GhBIN2-B and GhBIN2-D transcripts are expressed at consistent levels throughout the plants, while the GhBIN2-C and GhBIN2-E mRNAs are differentially expressed—with highest levels occurring in hypocotyls, leaf buds, and in immature bolls and ovules. Generally, GhBIN2-C and GhBIN2-E are expressed in rapidly growing tissues in a pattern similar to that of GhBRI1, the putative BR receptor gene in cotton (Sun et al. 2004). Since our experiments show that all four GhBIN2 genes encode proteins that can negatively regulate BR signaling in transgenic Arabidopsis plants, the significance of the differential expression patterns among these genes is not clear. However, it is important to remember that mRNA levels do not necessarily correlate with levels of functional BIN2 within the cells and, that the expression could be limited to specific tissue or cell types within the organs tested. Therefore, more detailed expression analyses to determine the spatial patterns of GhBIN2 expression should provide a more complete understanding of BIN2 function.
Upland cotton (G. hirsutum) is a tetraploid plant that originated from A-genome and D-genome cotton species (Iqbal et al. 2001). Sequence comparison of the four GhBIN2 genes with those of diploid A-type (G. arboreum) and D-type (G. thurberi) cotton suggests a specific ancestral relationship. Based on this analysis, we predict that GhBIN2B, which is most similar to GaBIN2-1, represents the A sub-genome homeolog of the constitutively expressed GhBIN2 pair. Likewise, GhBIN2E, which most closely matches GaBIN2-2, represents the A sub-genome homeolog of the developmentally regulated GhBIN2 pair. On the other hand, constitutively expressed GhBIN2-D, and developmentally regulated GhBIN2-C, which most closely resemble sequences from G . thurberi (GtBIN2-1, and GtBIN2-2 respectively) represent the corresponding D sub-genome homeologs. If this is the case, the cotton BIN2 gene family was probably derived via a gene duplication event that predates the divergence of the A-genome and D-genome diploid Gossypium species. Based on their distinct expression patterns, these genes may have diverged functionally, as well as structurally. The subsequent polyploidization event that resulted in tetraploid G. hirsutum produced the current gene family consisting of two homeologous pairs of genes. This contrasts with the BRI1 genes: here, single A sub-genome and D sub-genome homeologs were identified (Sun et al. 2004).
Current models of the BR signal transduction pathway indicate that BIN2 negatively regulates BR-mediated responses (Clouse 2002). According to this model BIN2 is constitutively active in the absence of BR and it phosphorylates the positive downstream regulators BZR1 and BES1 (BZR2), targeting them for degradation (Clouse 2002). Li and Nam (2002) reported that about 10% of the kanamycin-resistant plants transformed with a wild-type BIN2 gene construct displayed BR-insensitive phenotypes. These BIN2 transgenic plants formed an allelic series of bin2 phenotypes, ranging from minor reductions in growth to the severe dwarfing characteristic of the semi-dominant bin2 mutant plants. Northern hybridization analysis revealed a correlation between the level of BIN2 expression and severity of the phenotype in these transgenic Arabidopsis plants (Li and Nam 2002). Overexpression of each of the GhBIN2 cDNAs in Arabidopsis gave similar results. About 10% of the transgenic plants showed bin2-like phenotypes, of varying severity (see Fig.4) and quantitative real-time PCR analysis showed that the heights of the transgenic plants were inversely correlated with the GhBIN2 expression levels. We interpret these results to indicate that the GhBIN2 genes encode proteins that are functionally homologous to AtBIN2, and we speculate that these genes may negatively regulate BR signal transduction in cotton.
References
Asami T, Min YK, Nagata N, Yamagishi K, Takatsuto S, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S (2000) Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol 123:93–100
Ashcraft CW (1996) The effect of brassinolide on cotton fiber development. MS Thesis, Texas Tech University
Beasley C, Ting IP (1974) The effect of plant growth substances on in vitro fiber development from unfertilized cotton ovules. Am J Bot 61:188–194
Bourouis M, Moore P, Ruel L, Grau Y, Heitzler P, Simpson P (1990) An early embryonic product of the geneFrequencies of spontaneous base substitutions at the AatII target site in the chloroplast DNA of C. reinhardtii shaggy encodes a serine/threonine protein kinase related to the CDC28/cdc2+ subfamily. EMBO J 9:2877–2884
Charrier B, Champion A, Henry Y, Kreis M (2002) Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol 130:577–590
Chono M, Honda I, Zeniya H, Yoneyama K, Saisho D, Takeda K, Takatsuto S, Hoshino T, Watanabe Y (2003) A semidwarf phenotype of barley uzu results from a nucleotide substitution in the gene encoding a putative brassinosteroid receptor. Plant Physiol 133:1209–1219
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Clouse SD (2002) Brassinosteroid signal transduction: clarifying the pathway from ligand perception to gene expression. Mol Cell 10:973–982
Dornelas MC, Lejeune B, Dron M, Kreis M (1998) The Arabidopsis SHAGGY-related protein kinase (ASK) gene family: structure, organization and evolution. Gene 212:249–257
Embi N, Rylatt DB, Cohen P (1980) Glycogen synthase kinase-3 from rabbit skeletal muscle. Separation from cyclic-AMP-dependent protein kinase and phosphorylase kinase. Eur J Biochem 107:519–527
Friedrichsen DM, Joazeiro CA, Li JM, Hunter T, Chory J (2000) Brassinosteroid-Insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol 123:1247–1255
Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S (2002) Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 130:1319–1334
Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S (2004) Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 134:1–19
Hanks SK, Quinn AM, Hunter T (1988) The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241:42–52
He JX, Gendron JM, Yang YL, Li JM, Wang ZY (2002) The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a posiive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA 99:10185–10190
Iqbal MJ, Reddy OUK, El-Zik KM, Pepper AE (2001) A genetic bottleneck in the ‘evolution under domestication’ of upland cotton Gossypium hirsutum L. examined using DNA fingerprinting. Theor Appl Genet 103:547–554
Kasukabe Y, Iwara I, Maekawa Y (2000) Improvement of cotton fiber properties by genetic engineering. Sen-i Gakkaishi 56:99–103
Kim L, Kimmel AR (2000) GSK3, a master switch regulating cell-fate specification and tumorigenesis. Curr Opin Genet Dev 10:508–514
Li JM, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90:929–938
Li JM, Nam KH (2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295:1299–1301
Li JM, Nam KH, Vafeados D, Chory J (2001) BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol 127:14–22
Li J, Wen JQ, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110:213–222
Mandava NB (1988) Plant growth-promoting brassinosteroids. Annu Rev Plant Physiol Plant Mol Biol 39:23–52
Montoya T, Nomura T, Farrar K, Kaneta T, Yokota T, Bishop GJ (2002) Cloning the tomato Curl3 gene highlights the putative dual role of the leucine-rich repeat receptor kinase tBRI1/SR160 in plant steroid hormone and peptide hormone signaling. Plant Cell 14:3163–3176
Mora-Garcia S, Vert G, Yin Y, Cano-Delgado A, Cheong H., Chory, J (2004) Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Gene Dev 18:448–460
Nam KH, Li JM (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110:203–212
Nam KH, Li JM (2004) The Arabidopsis transthyretin-like protein is a potential substrate of BRASSINOSTEROID-INSENSITIVE 1. Plant Cell 16:2406–2417
Nomura T, Bishop GJ, Kaneta T, Reid JB, Chory J, Yokota T (2003) The LKA gene is a BRASSINOSTEROID INSENSITIVE 1 homolog of pea. Plant J 36:291–300
Oh MH, Ray WK, Huber SC, Asara JM, Gage DA, Clouse SD (2000) Recombinant Brassinosteroid Insensitive 1 receptor-like kinase auto-phosphorylates on serine and threonine residues and phosphorylates a conserved peptide motif in vitro. Plant Physiol 124:751–766
Sekimata K, Kimura T, Kaneko I, Nakano T, Yoneyama K, Takeuchi Y, Yoshida S, Asami T (2001) A specific brassinosteroid biosynthesis inhibitor, Brz2001: evaluation of its effects on Arabidopsis, cress, tobacco, and rice. Planta 213:716–721
Strausberg R (2002) Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA 99:16899–16903
Sun Y, Fokar M, Asami T, Yoshida S, Allen RD (2004) Characterization of the Brassinosteroid insensitive 1 genes of cotton. Plant Mol Biol 54:221–232
Wan CY, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223:7–12
Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410:380–383
Wendel JF, Brubaker CL, Percival AE (1992) Genetic diversity in Gossypium hirsutum and the origin of upland cotton. Am J Bot 97:1291–1310
Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12:1591–1605
Yin YH, Wang ZY, Mora-Garcia S, Li JM, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109:181–191
Zhao J, Peng P, Schmitz RJ, Decker AD, Tax FE, Li JM (2002) Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol 130:1221–1229
Acknowledgements
We wish to thank Ms. Jing Wang for development of the cotton boll cDNA library. We also thank Dr. Jianming Li for providing us with the BIN2 cDNA sequence prior to publication. We also thank Dr. Robert Bradley for assistance with sequence alignments. This research was supported by USDA-NRI Grant No. 2003-35304-13384
Author information
Authors and Affiliations
Corresponding author
Additional information
G. Jürgens
Rights and permissions
About this article
Cite this article
Sun, Y., Allen, R.D. Functional analysis of the BIN2 genes of cotton. Mol Genet Genomics 274, 51–59 (2005). https://doi.org/10.1007/s00438-005-1122-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-005-1122-0