Abstract
The resurrection plant (Craterostigma plantagineum) is desiccation tolerant. However, callus derived from this plant, when propagated in vitro, requires exogenously applied abscisic acid (ABA) in order to survive desiccation. Treatment of callus tissue with ABA induces most of the genes that are induced by dehydration in the whole plant. This property has been exploited for the isolation of mutants that show dominant phenotypes resulting from the ectopic expression of endogenous genes induced by the insertion of a foreign promoter. Here we describe new T-DNA tagged Craterostigma desiccation-tolerant (cdt) mutants with different molecular and physiological characteristics, suggesting that different pathways of desiccation tolerance are affected. One of the mutants, cdt-2, constitutively expresses known osmoprotective Lea genes in callus and leaf tissue. Further analysis of this mutant revealed that the tagged locus is similar to a previously characterised gene, CDT-1, which codes for a signalling molecule that confers desiccation tolerance. The nature of the T-DNA insertion provides insight into the mechanism by which the CDT-1/2 gene family functions in ABA signal transduction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Desiccation tolerance in vegetative tissues of higher plants has been extensively studied in resurrection plants, which possess the unique ability to revive from an air-dried state (Gaff 1971). Such plants are often poikilohydrous and their water content correlates with fluctuations in the relative humidity of the local environment. Desiccation tolerance shows a wide taxonomic distribution, and appears to have evolved independently several times as an adaptation to extreme variations in water availability (Oliver et al. 2000).
Using the resurrection plant Craterostigma plantagineum as a model system, the acquisition of desiccation tolerance has been shown to be mediated by abscisic acid (ABA). This conclusion is supported by two observations. Firstly, a comparison of dried and fully hydrated tissues reveals that a group of transcripts and proteins that accumulate during drying also appear when ABA is applied to non-stressed tissues (Bartels et al. 1990). Secondly, in contrast to differentiated tissues, callus tissue requires exposure to ABA in order to acquire desiccation tolerance; dried but otherwise untreated callus is not desiccation tolerant (Chandler et al. 1997). In this experimental system, both ABA-treated and untreated callus tissue lose 90% of their initial fresh weight during dehydration (Furini et al. 1997). This observation suggests that a number of ABA-mediated pathways that lead to the acquisition of desiccation tolerance are not activated by dehydration alone in the callus under experimental conditions.
Progress in understanding the role of ABA in plant development and during dehydration has come from the characterisation of mutants that differ in their responses to ABA. Such mutants show no reduction in endogenous ABA content and their phenotypes cannot be reverted to wild-type by exogenous supply of ABA. Multiple loci have now been characterised that either increase or decrease ABA sensitivity in Arabidopsis (Koornneef et al. 1984; Finkelstein 1994; Cutler et al. 1996). In some of these cases, the identities of the mutated genes and the function of the wild-type gene products are known (Leung and Giraudat 1998; Nambara and McCourt 1999). Many of the mutations that affect ABA sensitivity result in loss of gene function, and the genes affected code for signalling molecules that regulate processes such as phosphorylation and transcription (Finkelstein et al. 2002).
A molecular genetic approach has been used to identify intermediates in the ABA signal transduction pathway that leads to desiccation tolerance in C. plantagineum. T-DNA activation tagging experiments have permitted the isolation of a gene (CDT-1) which codes for a signalling molecule in the ABA transduction pathway (Furini et al. 1997). Constitutive expression of CDT-1 led to desiccation tolerance in the absence of ABA, and to the constitutive expression of a subset of dehydration- and ABA-responsive transcripts. CDT-1 has features of a short interspersed element (SINE) retrotransposon and is unique to C. plantgineum. However, the mechanism by which CDT-1 expression results in desiccation tolerance is unclear, since the mRNA cannot code for a polypeptide of more than 22 amino acids in length. Given that the transcript lacks a longer ORF, an alternative hypothesis is that CDT-1 may perform its function at the RNA level, not as a protein (Furini et al. 1997; Furini et al., unpublished results).
The aim of the present study was to generate and characterise additional gain-of-function mutations that confer on callus the ability to survive desiccation without prior exposure to ABA. For this, T-DNA mediated stable transformation of C. plantagineum was used in combination with activation tagging vectors (Furini et al. 1997; Weigel et al. 2000). Craterostigma desiccation-tolerant (cdt) mutants that are able to survive desiccation of callus without pretreatment with ABA were isolated. Analysis of one of the tagged loci identified a gene that shows similarity to the CDT-1 gene, which provides further insight into a novel gene family whose products influence the ABA signalling pathway.
Materials and methods
Plant material
Craterostigma plantagineum Hochst. plants were grown in a Conviron growth chamber at 60% humidity, on a photoperiod of 14 h light (60,000 lux) and a day/night temperature regime of 24/20°C. Plants or callus growing in vitro were maintained and propagated in MS basal medium in a growth chamber at 24±1°C on a 16-h day (fluorescent light providing 200 μE/s per m2) under sterile conditions. Dehydration and ABA treatments were performed as described by Bartels et al. (1990).
Nucleic acid analysis
The procedures used for isolation of poly(A)+ RNA and RNA gel-blot analysis have been described previously (Bartels et al. 1990). Genomic DNA was extracted using the Genomic DNA Purification Kit (Qiagen). Hybridisations were performed at high stringency according to Frank et al. (1998).
Plant transformation vectors
Two different vectors (pPCVTAc1 and pPCVRN4) were chosen in order to create a population of T-DNA activation-tagged C. plantagineum plants (Koncz et al. 1994). The reason for using both binary vectors is that transcriptional activation resulting from the insertion of the pPCVTAc1 vector may result in more efficient gene activation, since it contains, in addition to the CaMV35S minimal promoter, four copies of the CaMV 35S transcriptional enhancer. The pPCVTAc1 vector contains the CaMV 35S promoter (Odell et al. 1985) carrying four repeats of the −90 to −440 enhancer domain positioned at the right border of the T-DNA. pPCVRN4 is identical to pPCVTAc1, but does not contain the +1 to −90 CaMV 35S promoter region.
Agrobacterium-mediated transformation of C. plantagineum
The A. tumefaciens strain GV3101 carrying the helper plasmid pMP90RK was used as the host strain. Young leaves were wounded, immersed in a suspension of A. tumefaciens in MSAR medium for 20 min in darkness and co-cultivated for 2 days on solid callus medium without antibiotics (Furini et al. 1994; Koncz et al. 1994).
Plant culture conditions
Plant culture conditions were as described by Furini et al. (1994). After co-cultivation, the infected explants were subcultured for 10 days on MSAR1 (MSAR plus 2 mg/l IAA, 0.5 mg/l 2,4-D, 0.2 mg/l kinetin and 0.2 mg/l 9-iP) plates containing 2.2 g of Gelrite (Kelco), 500 mg/l cefotaxime and 15 mg/l hygromycin. Growing calli were subcultured at 3-week intervals on selective MSAR1. At each subculture step, the cefotaxime concentration was reduced by 100 mg/l. Hygromycin was maintained at 15 mg/l until the plants were transferred to soil. To induce shoot differentiation, embryogenic calli were subcultured on MSAR1a (MSAR plus 0.5 mg/l BAP and 0.1 mg/l NAA). Shoots were rooted on MS medium.
Identification of Craterostigma desiccation-tolerant (cdt) calli
Calli were selected on hygromycin-containing medium and maintained for about 3–4 months. Calli were then transferred to sterile filter paper and air-dried for 24 h at room temperature. During this time, calli lost 94–96% of their initial fresh weight. Untransformed wild-type and calli previously treated with ABA were used as negative and positive controls, respectively. Dried calli were rehydrated for 2 days on callus medium. Antibiotic selection was applied to the transgenic callus population during the rehydration phase. Dried calli were then transferred to callus medium and screened for viability using a dissecting microscope after 2 days.
Cloning of T-DNA flanking regions: Long Range inverse PCR (LR-iPCR)
Genomic DNA isolated from cdt-2 was digested with EcoRI and religated. Ligated DNA was then digested with SmaI in order to linearise the circular DNA structures. LR-iPCR was performed using 500 ng of linearised DNA as a template and an oligonucleotide primer pair (5′-GAGAGGACCTCGAGCTGCAGAATTAC-3′ and 5′-TCCTGCGGGTAAATAGCTGCGCCGATGG-3′) designed to amplify the region of plant DNA flanking the T-DNA right-border sequence and the region downstream of the CaMV 35S promoter. Reactions were performed using the LA PCR (Takara Shuzo) Long-Range PCR kit as recommended by the supplier. The DNA sample was heated to 95°C for 2 min, and PCR was carried out for 35 cycles of 94°C for 30 s, 65°C for 30 s and 68°C for 8 min, followed by an elongation step at 68°C for 10 min. A single 1.7-kb PCR product was obtained and cloned using the pGEM-T-Easy vector system (Promega).
DNA sequence analysis
DNA sequences were determined by the ADIS Unit (MPIZ) and analysed using the BLAST network service (Altschul et al. 1990) and the Genetics Computer Group software package (Devereux et al. 1984) version 9.0.
Results
Wild-type C. plantagineum callus requires pre-treatment with ABA in order to acquire desiccation tolerance.
Desiccation tolerance in callus was assessed by comparing the survival rate of ABA treated and untreated callus. After 4 days of exposure to ABA, flavonoid production became evident, as revealed by the reddish colouration of the callus. After desiccation of both untreated and ABA-treated callus, the calli were hydrated and assayed for their ability to recover. Calli that had not been treated with ABA and desiccated did not respond to rehydration. In contrast, the pre-treated callus showed signs of recovery after 24 h of rehydration. Full recovery of 58% of the initial callus clusters was observed 20 days after rehydration, with the callus regaining its normal morphology and proliferation rate. These results represent the average of four independent experiments and indicate that the desiccation/rehydration treatment provides a rigorous screening method.
Generation of a T-DNA tagged population of C. plantagineum
Encouraged by the isolation of the cdt-1 gain-of-function mutant, we extended the screening process to look for further signalling intermediates using two different T-DNA activation tagging vectors. Following the strategy described by Furini et al. (1994), a series of A. tumefaciens-mediated transformations of C. plantagineum was performed. In this way, engineered T-DNA with the CaMV 35S promoter located in the right border region was inserted into the plant genome (see Materials and methods).
The transformation protocol initially involved wounding of young leaves and co-cultivation of the wounded explants with the appropriate A. tumefaciens strain. Approximately 9 weeks after leaf infection, proliferation of hygromycin-resistant callus was observed in at least three different locations on each leaf explant. The tissue exhibited a light-green appearance, and was compact with a slightly nodular surface. The individual callus clusters were subcultured in order to maintain each potential transformation event. Three months after transformation the callus clusters became friable and could not be distinguished from wild-type callus.
After approximately 6 months of continuous subculture, a total of approximately 6,400 independent hygromycin-resistant callus clusters were obtained from equal numbers of transformations using the pPCVTAc1 and pCRVRN4 vectors (see Materials and methods). The T-DNA-tagged population was allowed to proliferate for 1 month before the first desiccation screen was performed. In order to confirm that the callus clusters were indeed transformed, eight randomly picked samples were selected and subjected to a PCR test using primers that were designed to amplify the 1.5-kb hygromycin resistance gene and a 0.5-kb fragment of the carbenicillin resistance gene. PCR products of the predicted size were amplified from seven out of the eight callus clusters tested (data not shown).
Isolation of desiccation-tolerant mutants
Desiccation-tolerant transgenic calli were selected on the basis of their ability to survive a 24-h dehydration treatment without prior application of ABA. The screening process was performed as follows. Tissue was dehydrated overnight under sterile conditions without prior exposure to ABA, and surviving calli were selected following rehydration on MSAR1 medium. Two of the callus clusters survived and had a similar appearance. The selected Craterostigma desiccation-tolerant callus lines (cdt-2 and cdt-3) obtained from the first screen were kept for proliferation. The nomenclature follows that of Furini et al. (1997), who described the first Craterostigma desiccation-tolerant mutant (cdt-1).
Regeneration of plants carrying T-DNA insertions
In order to fully characterise the transgenic C. plantagineum lines, plants were regenerated from the selected callus clusters. Dehydration-resistant callus lines were subcultured for approximately 3 months. Then roughly half of the callus tissue from each line was either transferred to shoot induction media or allowed to proliferate further in the presence of hygromycin. During the second subculture callus tissue became more friable and a large number of embryogenic structures proliferated on the surface of the cluster. Shoots proliferated continuously and did not display any obvious differences from wild-type shoots regenerated on the same medium in the absence of hygromycin.
To allow rooting and further development, green shoots were transferred to MS medium without hormones but supplemented with hygromycin. Hygromycin selection was strict, in that non-transformed lines were unable to produce roots in the presence of hygromycin. More than 50% of the transferred shoots gave rise to plantlets with roots, and the regenerated plants showed no obvious phenotypic differences from wild-type, in vitro cultured C. plantagineum plants grown without hygromycin, except that the cdt-2 mutant exhibited a slight yellow colouration (Fig. 1a–c).
C. plantagineum T-DNA insertion lines during development. Wild-type regenerated plants (a) were compared to the cdt-2 (b) and cdt-3 (c) mutant lines after approximately 5 months of in vitro culture. After transfer from in vitro conditions to a growth chamber, the T-DNA insertion line cdt-3 has an elongated stem, with additional leaf rosettes (e). By comparison a wild-type plant displays a more compact rosette structure (d). Both plants were grown under the same conditions and were approximately the same age (8–9 months)
Plants were obtained from both of the mutant lines. An altered phenotype was observed in the cdt-3 plants after 2 weeks of growth (see Fig. 1d, e): the transgenic plants had elongated stems, with additional leaf rosettes. This increase in internode length has not been observed in transgenic lines from a large set of experiments. Apart from the phenotype of the cdt-3 line, the T-DNA insertions did not affect other processes of organ differentiation. Seed formation was induced in these plants to allow segregation analysis. However, both cdt-2 and cdt-3 mutant lines displayed reduced fertility; indeed none of the tested plants produced seeds, even after pollination with wild-type pollen. This contrasts with non-transformed plants derived from tissue culture, which exhibit a high degree of fertility. This phenomenon was also observed in the case of the previously characterised cdt-1 mutant (Furini et al. 1997).
Secondary screening of the putative desiccation-tolerant callus lines
To confirm that the regenerated transgenic plants originated from the desiccation-tolerant callus clusters, a second round of desiccation tolerance screening was performed on callus derived from the leaves of each regenerated plant. Several (10–20) leaflets were taken from the shoots and subcultured for approximately 3 months until callus clusters had formed. A desiccation screen was performed and both mutant lines were found to give rise to desiccation-tolerant callus in the absence of prior treatment with ABA. cdt-2 (transformed with pPCVTAc1) and cdt-3 (transformed with pPCVRN4) were subjected to further molecular analysis. A summary of the characteristics of the selected desiccation-tolerant lines is reported in Table 1.
Molecular characterisation of the T-DNA tagged lines cdt-2 and cdt-3
To confirm the presence and number of T-DNA insertions in the tagged lines, genomic DNA was extracted from cdt-2 and cdt-3 plants, digested with restriction enzymes, and hybridised with either the 300-bp CaMV 35S transcriptional enhancer region or a 1.5-kb DNA fragment corresponding to the hygromycin phosphotransferase (HPT) gene. DNA was digested with EcoRI, BamHI and HindIII. The enzyme EcoRI cleaves at two sites within the T-DNA in both of the binary vectors, releasing a fragment of 1.5 kb that corresponds to the HPT gene. BamHI cleaves at one restriction site in the pPCVTAc1 T-DNA, whereas no restriction sites for HindIII are present in either of the binary vectors used. A band of 1.5 kb was observed in both of the selected mutant lines when the HPT fragment was used as a probe (Fig. 2a), demonstrating that the mutant lines had indeed been transformed. Bands of different sizes were observed in all of the selected mutant lines when the CaMV 35S promoter transcriptional enhancer was used as a probe (Fig. 2b). The numbers of T-DNA insertions in cdt-2 and cdt-3 were 1 and 2, respectively. As expected, fragments larger than 6.6 kb (T-DNA size) were noted in samples digested with HindIII and BamHI. This observation suggests that the T-DNA was not rearranged after insertion into the C. plantagineum genome. These results are also summarised in Table 1.
DNA gel-blot analysis of the T-DNA insertions in cdt-2 and cdt-3. Aliquots (30 μg) of genomic DNA isolated from the T-DNA tagged lines cdt-2 and cdt-3 was digested with EcoRI (lanes 1 and 4), BamHI (lanes 2 and 5) and HindIII (lanes 3 and 6), fractionated in an agarose gel and transferred onto a nitrocellulose membrane. Blots were individually hybridised with a 32P-labelled (a) hygromycin phosphotransferase (HPT) gene and (b) the CaMV 35S enhancer fragment
In order to determine whether or not the tagged loci affected ABA/desiccation-mediated gene expression, transcript analysis was performed. Gel blots bearing RNA extracted from wild-type and transgenic leaf and callus tissue were hybridised with probes for two previously characterised dehydration-responsive cDNAs, pcC27-45 and pcC6-19 (Piatkowski et al. 1990). The transcripts of these genes are only detected during the desiccation process in wild-type C. plantagineum leaves and in ABA-treated callus.
pcC6-19 transcripts were detected at a relatively high level in hydrated callus from cdt-2 and also, albeit to a lesser degree, in the cdt-3 line. The level of the pcC6-19 transcript in callus from cdt-2 was slightly lower than that found in ABA-treated callus (Fig. 3a). Constitutive expression of pcC27-45 was detected in callus originating from the cdt-2 line and in dehydrated wild-type tissue, but not in callus originating from cdt-3 or in fully hydrated wild-type tissue. In contrast, the pcC6-19 transcript was detected in leaves originating from the cdt-2 line, but not in leaves from cdt-3. Moreover, pcC27-45 was not detectable in leaves from either of the mutant lines (Fig. 3b and Table 1).
RNA gel-blot experiments showing the steady-state level of transcripts of the dehydration-responsive genes pcC27-45 and pcC6-19 in wild-type and mutant lines of C. plantagineum. (a) Poly(A)+ RNA (3 μg per lane) from fully hydrated callus derived from the cdt-3 (lane 1) and cdt-2 (lane 2) mutant lines. Samples from fully hydrated wild-type callus and from callus pre-treated with ABA for 4 days were loaded in lanes 3 and 4, respectively. (b) Poly(A)+ RNA (3 μg per lane) from fully hydrated leaves from the mutant lines cdt-3 (lane 5) and cdt-2 (lane 6). RNA samples from fully hydrated wild-type plants and from plants that had been dehydrated for 72 h were loaded in lanes 7 and 8, respectively. Samples were fractionated in a denaturing gel and transferred to a nylon membrane. Two different Lea gene fragments, pcC27-45 and pcC6-19, were used as 32P-labelled probes. The ribosomal cDNA fragment pTA 71 (Gerlach and Bedbrook, 1979) was used to monitor the loading of the RNA
CDT-2 shares sequence homology with CDT-1
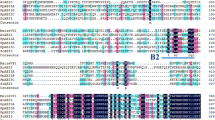
The cdt-2 mutant was further studied to determine the nature of the single T-DNA insertion event. The downstream sequence immediately adjacent to the CaMV35S minimal promoter and CaMV 35S transcriptional enhancer regions was isolated by long-range inverse PCR. A 1.7-kb genomic sequence was obtained and found to share sequence motifs with the 3′ region of the CDT-1 gene (Fig. 4a, b). The region of sequence identity corresponds to a relatively thymidine-rich (~37%) segment (positions 511–921), which ends with a polyA-stretch at the 3′ end, but does not include the 5′ region of CDT-1 that has been suggested to encode a short peptide (Furini et al. 1997) (see Fig. 4b). Database searches revealed that no other regions of the rescued genomic T-DNA flanking sequence shared homology with previously reported genes.
Structure of the 1.7-kb T-DNA flanking region rescued from the cdt-2 mutant. (a) Four copies of the CaMV 35S enhancer sequence (E, dark grey boxes) and the minimal CaMV 35S promoter (p, light grey box) originating from the activation tagging vector (pPCVTAc1) are located upstream of a transcribed region with similarity to the previously reported CDT-1 gene. The region of high homology is highlighted (stippled) and the positions of the oligoA stretches are indicated (A). (b) Alignment of the T-DNA flanking region rescued from the cdt-2 mutant with the CDT-1 cDNA and the consensus sequence. The ORF identified by Furini et al. (1997) in the 5′ region of CDT-1 is underlined. The conserved thymidine-rich 3′ region is boxed. C RNA gel-blot analysis of mutant and wild-type samples reveals a cdt-2 specific transcript. Poly(A)+ RNA from the following sources were fractionated in a denaturing gel: fully hydrated cdt-2 callus (lane 1); fully hydrated wild-type callus tissue (lane 2); callus tissue pre-treated with ABA for 4 days (lane 3); fully hydrated leaves (lane 4); leaves dehydrated for 72 h (lane 5), fully hydrated roots (lane 6), roots dehydrated for 72 h (lane 7). The samples were transferred to a nylon membrane and hybridised with a 32P-labelled probe that corresponded to the rescued genomic T-DNA flanking region originating from the cdt-2 mutant. The ribosomal cDNA fragment pTA71 was used to monitor the loading of the RNA
To investigate whether the T-DNA insertion leads to altered expression of genes present in the isolated flanking regions, RNA gel blot analysis was performed (see Fig. 4c). On RNA blots that included RNA from wild-type callus and leaf tissues, and from transgenic callus tissue, the T-DNA flanking region originating from the cdt-2 mutant hybridised to two transcripts. One transcript, approximately 1 kb in length, accumulated to broadly similar levels in callus from the cdt-2 mutant, untreated and ABA-treated wild-type callus, and in hydrated leaves and roots. Barely detectable levels of the transcript were observed in dehydrated leaves and roots. A slightly smaller transcript was found only in the mutant callus line, which is likely to have resulted from ectopic transcriptional activation mediated by the upstream CaMV 35S promoter.
Discussion
Activation tagging as a tool for dissecting gene function
Activation tagging is effective in organisms with functional gene redundancy, such as the polyploid C. plantagineum (n=28). Functional redundancy has become more apparent as sequencing of eukaryotic genomes has revealed the existence of many duplicated genes that are very similar both in their coding regions and their non-coding (nc), regulatory regions (Weigel et al. 2000). Thus, screening of other T-DNA populations created using activation-tagging vectors has led to the identification of a histidine kinase gene from A. thaliana, whose overexpression can bypass the requirement for cytokinin in the regeneration of shoots (Kakimoto 1996), and recently to the identification of A. thaliana genes for transcription factors such as LEAFY PETIOLE, which encodes a protein with a domain that is similar to the DNA binding domains of members of the AP2/EREBP family that is thought to function in either cell division or leaf patterning (Van der Graaf et al. 2000). Other examples include a MYB gene that regulates phenylpropanoid biosynthesis and enhances the accumulation of lignin, hydroxycinnamic acid, esters and flavonoids, including various anthocyanins that give rise to a purple leaf colouration (Borevitz et al. 2000), and the ORCA3 gene from Catharanthus roseus, which codes for a jasmonate-responsive transcriptional regulator with an AP2-domain which is thought to regulate primary and secondary metabolism (Van der Fits and Memelink 2000).
The resurrection plant C. plantagineum is desiccation tolerant, but callus derived from this plant and propagated in vitro requires exogenously applied ABA in order to survive severe dehydration. ABA treatment of callus induces most of the genes that are induced by dehydration in the whole plant (Bartels et al. 1990). This property has been exploited for the isolation of dominant mutants that have acquired the capacity for desiccation tolerance without exposure to ABA—which may result from ectopic expression of endogenous genes activated by the insertion of a foreign promoter.
An experimental system designed to screen for a gain-of-function phenotype in T-DNA-transformed C. plantagineum cells identified two mutant lines that displayed desiccation tolerance in dedifferentiated callus tissues. The callus resulting from the regenerated plants also exhibited desiccation tolerance and was characterized by the expression of desiccation-inducible transcripts in the absence of exogenous ABA. The mutated callus lines, cdt-2 and cdt-3, were characterised further and the tagged region was isolated from the cdt-2 mutant.
Function of the tagged loci
Considerable evidence now exists to suggest that ABA plays a central regulatory role in the expression of the osmoprotective Lea genes (Himmelbach et al. 2003). This is based on the observation that seedlings homozygous for mutations in genes for ABA biosynthesis produce low levels of ABA, and this is correlated with the inability to express multiple classes of Lea genes (Pla et al. 1989, 1991; Giordani et al. 1999). Conversely, ectopic expression of the ABI-3 transcriptional activator gene allows the ABA-mediated activation of Lea genes in vegetative tissues of A. thaliana (Parcy et al. 1994). As previously mentioned, the CDT-1 gene is predicted to encode a molecule that interacts with ABA signalling networks, based on the ectopic expression of Lea genes seen in the dominant cdt-1 mutant (Furini et al. 1997). Here we show that the cdt-2 mutant resembles cdt-1 in showing constitutive, high-level expression of known C. plantagineum osmoprotective Lea genes (Piatkowski et al. 1990). Therefore it is likely that the CDT-2 gene also plays a regulatory role in ABA signal transduction.
Characterisation of the T-DNA tagged locus in the cdt-2 line reveals a similar gene/genomic region to cdt-1, which is probably part of the larger gene family in the C. plantagineum genome, as suggested by Southern analysis (Furini et al. 1997). Given that the cdt-2 mutant line contains a single T-DNA insertion, and expresses a novel transcript that hybridised specifically to the genomic region downstream of the CaMV 35S promoter, it is not likely that a secondary mutation is responsible for the altered phenotype. The cdt-1 mutant also carries a single T-DNA insertion that activates CDT-1 gene expression; furthermore, the desiccation-tolerant phenotype was found to segregate with the T-DNA insertion, and transformation experiments have confirmed that CDT-1 confers desiccation tolerance on C. plantagineum callus (Furini et al. 1997). The fact that two independently derived mutant loci are homologous was surprising, but provides more evidence that the CDT-1/2 gene family is important for the acquisition of desiccation tolerance in callus. Consequently, our studies strongly support the hypothesis that CDT-1 and CDT-2 have similar functions, since both cdt-1 and cdt-2 mutants display activated Lea gene expression and desiccation-tolerance in callus. However, independent overexpression studies, similar to that performed with CDT-1, are required to further validate the function of CDT-2.
Furini et al. (1997) provided several lines of genetic and biochemical evidence indicating that CDT-1 specifies an intermediate in the ABA signal transduction pathway leading to desiccation tolerance. The data presented in this study support a role for the CDT-1/2 genes as regulatory molecules. Initially it was suggested that CDT-1 could either function via the short peptide it may encode or as a regulatory RNA. However, unravelling the mechanism of action has proven to be difficult. Here we show that a CDT-1 related gene that does not include the short 5′ ORF identified by Furini et al. (1997) in CDT-1 is sufficient to confer desiccation tolerance. This observation lends strong support to the possibility that CDT-1/2 function as regulatory nc RNA molecules. Given the sequence conservation between the CDT-1/2 genes, the functionally important elements are likely to be located in the conserved thymidine-rich 3′ region indicated in Fig. 4b. Non-coding RNAs are thought to play roles that require highly specific nucleic acid recognition without complex catalysis, such as directing post-transcriptional regulation of gene expression or in guiding RNA modifications. The connection between ABA signalling and RNA metabolism is further highlighted by the discovery dysfunctional alleles of the genes abscisic acid hypersensitive 1 (ABH1; Hugouvieux et al. 2001), hyponastic leaves 1 (HYL1; Lu and Fedoroff 2000) and supersensitive to ABA and drought 1 (SAD1; Xiong et al. 2001), which encode RNA binding proteins.
The cdt mutants isolated so far can be distinguished by several phenotypic characteristics, suggesting that different pathways may be affected. Thus, cdt-3 does not appear to dramatically affect the two Lea marker genes and, unlike cdt-1 and cdt-2, may also affect developmental processes that control internode length. These attributes of the cdt-3 mutant plant are interesting: callus desiccation tolerance and the elongated internode, a known response to overproduction of, or sensitivity to gibberellins. The absence of enhanced levels of ABA-dependent transcripts in this mutant also suggests that desiccation tolerance in callus may not always involve ABA-dependent pathways.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bartels D, Schneider K, Terstappen G, Piatkowski D, Salamini F (1990) Molecular cloning of abscisic acid modulated genes which are induced during desiccation of the resurrection plant Craterostigma plantagineum Planta 181:27–34
Borevitz JO, Xia Y, Blount J, Dixon R, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12:2383–2393
Chandler JW, Abrams SR, Bartels D (1997) The effect of ABA analogues on callus viability and gene expression in Craterostigma plantagineum Physiol Plant 99:465–469
Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt PA (1996) Protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis Science 273:1239–1241
Devereux J, Haeberli P, Smithies O (1984) A comprehensive set of sequence analysis programs for the VAX Nucl Acids Res 12:387–395
Finkelstein RR (1994) Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations Plant J 5:765–771
Finkelstein RR, Gampalab SSL, Rock CD (2002) Abscisic acid signaling in seeds and seedlings Plant Cell 14:S15–S45
Frank W, Phillips J, Salamini F, Bartels D (1998) Two dehydration-inducible transcripts from the resurrection plant Craterostigma plantagineum encode interacting homeodomain-leucine zipper proteins Plant J 15:413–421
Furini A, Koncz C, Salamini F, Bartels D (1994) Agrobacterium-mediated transformation of the desiccation-tolerant plant Craterostigma plantagineum Plant Cell Rep 14:102–106
Furini A, Koncz C, Salamini F Bartels D (1997) High level transcription of a member of a repeated gene family confers dehydration tolerance to callus tissue of Craterostigma plantagineum EMBO J 16:3599–3608
Gaff DF (1971) Desiccation tolerant flowering plants in Southern Africa. Science 174:1033–1034
Gerlach WL, Bedbrook JR (1979) Cloning and characterisation of ribosomal RNA genes from wheat and barley. Nucl Acids Res 7:1869–1885
Giordani T, Natali L, D-Ercole A, Pugliesi C, Frambrini M, Vernieri P, Vitagliano C, Cavallini A (1999) Expression of a dehydrin gene during embryo development and drought stress in ABA-deficient mutants of sunflower (Helianthus annuus L.) Plant Mol Biol 39:739–748
Himmelbach A, Yang Y, Grill E (2003) Relay and control of abscisic acid signaling Curr Opin Plant Biol 6:470–479
Hugouvieux V, Kwak JM, Schroeder JI (2001) An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis Cell 106:477–487
Kakimoto T (1996) CKI1, a histidine kinase homolog implicated in cytokinin signal transduction Science 274:982–991
Koncz C, Martini N, Szabados L, Hrouda M, Bachmair A, Schell J (1994) Specialised vector for gene tagging and expression studies Plant Mol Biol Manual B2:1–22
Koornneef M, Reuling G, Karssen CM (1984) The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana Physiol Plant 61:377–383
Leung J, Giraudat J (1998) Abscisic acid signal transduction Annu Rev Plant Physiol Plant Mol Biol 49:199–222
Lu C, Fedoroff N (2000) A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin and cytokinin Plant Cell 12:2351–2366
Nambara E, McCourt P (1999) Protein farnesylation in plants: a greasy tale Curr Opin Plant Biol 2:388–392
Odell JT, Nagy F, Chua N-H (1985) Identification of DNA-sequences required for activity of the cauliflower mosaic virus-35S promoter Nature 313:810–812
Oliver MJ, Tuba Z, Mishler BD (2000) The evolution of desiccation tolerance in land plants. Plant Ecol 151:85–100
Piatkowski D, Schneider K, Salamini F, Bartels D (1990) Characterisation of five abscisic acid-responsive cDNA clones isolated from the desiccation-tolerant plant Craterostigma plantagineum and their relationship to other water-stress genes Plant Physiol 94:247–258
Pla M, Goday A, Vilardell J, Gomez J, Pages M (1989) Differential regulation of ABA-induced 23–25 kDa proteins in embryo and vegetative tissues of the viviparous mutants of maize Plant Mol Biol 13:385–394
Pla M, Gomez J, Goday A, Pagès M (1991) Regulation of the abscisic acid-responsive gene rab28 in maize viviparous mutants Mol Gen Genet 230:394–400
Van der Fits L, Memelink J (2000) ORCA3, a Jasmonate-Responsive transcriptional regulator of plant primary and secondary metabolism Science 289:295–299
Van der Graaff E, Den Dulk-Ras A, Hooykaas PJJ, Keller B (2000) Activation tagging of the LEAFY PETIOLE gene affects leaf petiole development in Arabidopsis thaliana Development 127:4971–4980
Weigel D et al (2000) Activation tagging in Arabidopsis Plant Physiol 122:1003–1014
Xiong L, Gong Z, Rock CD, Subramanian S, Guo Y, Xu W, Galbraith D, Zhu J-K (2001) Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis Dev Cell 1:771–781
Acknowledgements
We are grateful to Dr. Csaba Koncz for providing us with the T-DNA tagging vectors and would like to thank B. Eilts and M. Feck for technical assistance. Financial support from an EU Biotech programme is gratefully acknowledged. C. S. was partially supported by CONACyT (Mexico).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Herrmann
Rights and permissions
About this article
Cite this article
Smith-Espinoza, C., Phillips, J., Salamini, F. et al. Identification of further Craterostigma plantagineum cdt mutants affected in abscisic acid mediated desiccation tolerance. Mol Genet Genomics 274, 364–372 (2005). https://doi.org/10.1007/s00438-005-0027-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-005-0027-2