Abstract
Meiosis is initiated from the G1 phase of the mitotic cell cycle, and consists of pre-meiotic S-phase followed by two successive nuclear divisions. Here we show that control of gene expression during pre-meiotic S-phase in the fission yeast Schizosaccharomyces pombe is mediated by a DNA synthesis control-like transcription factor complex (DSC1), which acts upon M lu 1 cell cycle box (MCB) promoter motifs. Several genes, including rec8 +, rec11 +, cdc18 +, and cdc22 +, which contain MCB motifs in their promoter regions, are found to be co-ordinately regulated during pre-meiotic S-phase. Both synthetic and native MCB motifs are shown to confer meiotic-specific transcription on a heterologous reporter gene. A DSC1-like transcription factor complex that binds to MCB motifs was also identified in meiotic cells. The effect of mutating and over-expressing individual components of DSC1 (cdc10 +, res1 +, res2 +, rep1 + and rep2 +) on the transcription of cdc22 +, rec8 + and rec11 + during meiosis was examined. We found that cdc10 +, res2 +, rep1 + and rep2 + are required for correct meiotic transcription, while res1 + is not required for this process. This work demonstrates a role for MCB motifs and a DSC1-like transcription factor complex in controlling transcription during meiosis in fission yeast, and suggests a mechanism for how this specific expression occurs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The switch between vegetative mitotic growth and sexual meiotic division is a fundamental process in cell biology. This developmental process has been extensively studied over the past few years in a variety of organisms, and a number of key regulatory components have been discovered.
The fission yeast Schizosaccharomyces pombe is a good model organism in which to study meiosis, as the process is easily manipulated in the laboratory. Meiosis occurs after two haploid cells, arrested in G1 by nutrient depletion, conjugate to form a transient diploid. The result is the production of four haploid spores in an ascus. Crucially, in S. pombe, meiosis is fundamentally similar to that in higher eukaryotes: the pre-meiotic S-phase is followed by two meiotic divisions, the first reductional and the second equational.
A number of molecules and mechanisms have been identified in fission yeast that control the transition into, and passage through, meiosis (Yamamoto 1996; Egel 2000). An important component of this control is the regulation of gene expression. For example, the specific expression of ste11 + regulates the transcription of mei2 +, mat1-Pc , mat1-Mc and other genes, which are all essential for meiotic progression (Sugimoto et al. 1991).
Mitotic cell cycle-regulated transcription has also been well studied in fission yeast. Here, a group of at least ten genes (cdc22 +, cdc18 +, cig2 +, cdt1 +, rad21 +, suc22 +, rad11 +, ste9 +, mik1 + and cdt2 +) are transiently expressed at the beginning of S-phase, and their products are required, either directly or indirectly, for progression through S-phase (Fernandez-Sarabia et al. 1993; Kelly et al. 1993; Connolly and Beach 1994; Hofmann and Beach 1994; Birkenbihl and Subramani 1995; Harris et al. 1996; Parker et al. 1997; Tournier and Millar 2000; Ayte et al. 2001; Ng et al. 2001; Maqbool et al. 2003; Yoshida et al. 2003). The molecular components that co-ordinate mitotic G1-S cell cycle transcription in fission yeast have been uncovered, and comprise a transcription factor complex that binds to common repeated DNA sequences in the promoters of the S-phase genes. The transcription factor has been named DSC1 (also called MBF) and contains the products of the genes cdc10 +, res1 +, res2 + and rep2 + (Lowndes et al. 1992; Caligiuri and Beach 1993; Miyamoto et al. 1994; Sugiyama et al. 1994; Zhu et al. 1994; Nakashima et al. 1995; White et al. 2001). DSC1 has been shown to bind so-called Mlu I cell cycle box (MCB) motifs (Lowndes et al. 1992; Ayte et al. 2001; Maqbool et al. 2003); coincidentally, the MCB DNA sequence ACGCGT is the same as the sequence recognised by the restriction enzyme Mlu I.
In mitotic cells DSC1 contains Cdc10p, Res1p, Res2p and Rep2p (Lowndes et al. 1992; Baum et al. 1997; Whitehall et al. 1999; White et al. 2001). It is believed that DSC1 possesses both stimulatory and repressive functions which together mediate G1-S specific transcription (McInerny et al. 1995; Baum et al. 1997; Whitehall et al. 1999).
Additional roles for cdc10 +, res2 + and another gene, rep1 +, in meiosis have been suggested by the finding that mutations in these genes affect meiotic progression (Beach et al. 1985; Sugiyama et al. 1991; Zhu et al. 1994; Li and Smith 1997; Ding and Smith 1998); and by the observation that ectopic over-expression of res2 + enhances entry into meiosis (Ayte et al. 1997). This has led to the suggestion that a meiotic form of DSC1, containing Cdc10p, Res2p and Rep1p, controls meiotic-specific expression in fission yeast (Sugiyama et al. 1991; Zhu et al. 1994; Sturm and Okayama 1996; Ayte et al. 1997; Zhu et al. 1997), and that it is the change in the composition of DSC1 that results in meiotic G1-S specific transcription in this organism.
In this paper we examine the transcription pattern of a group of fission yeast genes that are expressed during pre-meiotic S-phase. These genes fall into two classes: those that are also transcribed during mitosis and are required for DNA synthesis, such as cdc22 + and cdc18 + (Watanabe et al. 2001); and those specific for meiosis, the rec + genes, whose products are involved in meiotic recombination (Fox and Smith 1998; Davis and Smith 2001). We show that the members of this large group of genes are transcribed simultaneously, during pre-meiotic S-phase, and that this transcription is co-ordinately regulated by a system related to mitotic MCB motifs and DSC1. Thus, the MCB-DSC1 system not only regulates mitotic G1-S cell cycle transcription in fission yeast, but also controls expression of genes during meiosis in this organism.
Materials and methods
Strains and media
General molecular procedures were performed as described by Sambrook et al. (1989); the media used for the propagation of S. pombe have been described by Moreno et al. (1991). The standard genetic procedures of Gutz et al. (1974) and Kohli et al. (1977) were followed. The strains used in this study are listed in Table 1. For all physiological experiments cells were grown in minimal medium (EMM), with shaking, at 25°C or 36°C.
Synchronisation of meiotic cells was achieved using the temperature-sensitive mutation pat1-114 as described by Murakami and Nurse (1999). In brief, cells were grown in EMM to mid-exponential phase at 25°C. Subsequent transfer to nitrogen-free EMM for 16 h at 25°C resulted in G1 arrest, and the cells were then shifted to 36°C to induce meiosis. Samples were subsequently removed at regular intervals for RNA extraction, and determination of DNA content by flow cytometry (FACS) analysis.
Cell density in liquid culture was determined from samples fixed in a 0.1% formaldehyde/0.1% NaCl solution. Following sonication, cells were counted electronically with a Z2 Coulter Counter. Flow cytometry analysis (FACS) was performed as previously described (Moreno et al. 1991), using the FACScan system and the Cell Quest analysis program (Becton Dickinson); 10,000 cells were analysed per time point.
To over-express genes using the pREP1 vector (Maundrell 1993) during a synchronous meiosis, cells were grown in EMM supplemented with 5 μg/μl thiamine (nmt1 + promoter “off”) to the mid-exponential phase of growth at 25°C. Cells were washed three times in thiamine-free EMM, and then grown for 16 h in EMM without thiamine (nmt1 + promoter “on”) or nitrogen at the same temperature, before thermal induction of meiosis.
DNA constructs
SPIpMELβ2 (GB 169) was constructed by inserting the S. pombe ura4 + gene (Li et al. 1997) into the Apa I/NcoI sites of URA3 in YIpMELβ2 (Melcher et al. 2000). Integration of the vector into the ura4 locus in fission yeast was facilitated by digestion with Avr II, which linearised the vector within the ura4 + gene.
DNA fragments containing the MCB2 cluster from the cdc22 + promoter (Maqbool et al. 2003), and the synthetic triple MCB sequence (3MCB; Lowndes et al. 1992), were cloned into SPIpMELβ2. For MCB2 the DNA was amplified using oligonucleotides containing Xho I restriction sites to allow cloning into both vectors. The oligos used were GO 42 (5′-GCGCCTCGAGGGTGGTAAATACCGGGAA-3′; reverse, positions –22 to –39 relative to the ATG) and GO 44 (5′-GCGCCTCGAGCATTGATCAACATGACTTAAAG-3′; forward, positions –135 to –114), to create SPIpMELβ2.MCB2 (GB 307). For 3MCB the DNA was made by self-hybridising the oligo GO 549 (5′-TCGATACGCGTAGATCTACGCGTAGATCTACGCGTA-3′), to create SPIpMELβ2.3MCB (GB 303). pSPΔ178.3 M and pSPΔ178.MCB2 have been described previously (Lowndes et al. 1992; Maqbool et al. 2003).
The plasmids pREP1: res1 + (GB 182) and pREP1: res2 + (GB 200) were made by amplifying the ORFs of res1 + and res2 + from cDNA by PCR. The primers used provided an Nde I restriction site at the ATG and a Bam HI site 3′ to the stop codon for each gene, which allowed the genes to be inserted in frame with the nmt1 + promoter in the pREP1 series of plasmids (Maundrell 1993). The oligos used were (the Nde I and Bam HI sites are underlined): for res1 +, GO 170 (5′-CGTACATATGTATAACGACCAAATACATAAAATC-3′) and GO 171 (5′-GGCCAGGATCCTTAAGATCCACTTTGATCTGTATTAATCGT-3′); for res2 +, GO 137 (5′-CGTACATATGGCTCCACGTTCTTCCGCAGT-3′) and GO 138 (5′-GGCCAGGATCCTCATTTTTCTCGGGTTAATGC-3′).
The plasmids pREP1: cdc10 +, pREP1: rep1 + and pREP1: rep2 + have been described previously (McInerny et al. 1995; White et al. 2001). All amplified DNAs were checked by sequencing.
RNA manipulations
S. pombe total RNA was prepared (McInerny et al. 1995) using a Ribolyser (Hybaid) and northern blot analysis carried out using GeneScreen membrane (NEN), following the manufacturer’s suggested protocol. Northern blots were hybridised with DNA probes made by PCR corresponding in each case to ~1 kb of the ORF of each gene. DNA probes were labelled with [α-32P]dCTP using the random hexa-nucleotide labelling procedure of Feinberg and Volgelstein (1983). Transcript profiles were quantified using NIH Image software.
Gel retardation analysis
Whole cell protein extracts were generated from fission yeast cells, and gel retardation analysis was performed as previously described (Ng et al. 2001).
DNA fragments containing MCB motifs from the promoters of G1-S specific genes used as labelled substrates were amplified by PCR with the following oligos. The cdc22 + MCB1 (Maqbool et al. 2003) was obtained using the primer pair GO 36 (5′-GTAGTTCAATCTCATAGA-3′; forward, positions –532 to –515 relative to the ATG) and GO 37 (5′-CTCTGTTTACGACTGAATG-3′; reverse, positions –401 to –419). The rec8 + MCB DNA was made by hybridising the oligo GO 552 (5′-TTTGACGCGTTTAATAAGCTATCTGGTGAACTAACGCGTTCCT-3′; forward, positions –53 to –10 relative to the ATG) with GO 553 (5′-AGGAACGCGTTAGTTCACCAGATAGCTTATTAAACGCGTCAAA-3′).
Results
Co-ordinate gene transcription during pre-meiotic S-phase in fission yeast
Previous reports have shown that a number of genes in fission yeast are transcribed during the early stages of meiosis. These meiotic genes include a group that are also expressed in the G1-S interval during mitosis (e.g. cdc22 + and cdc18 +) and another group that is specific to meiosis and includes the rec + genes (Li and Smith 1997; Watanabe et al. 1999; Mata et al. 2002).
To begin to understand how transcription of these genes is regulated during meiosis, we confirmed and extended previous experiments by measuring transcript abundance for several genes during a synchronous meiosis. This allowed us to map more precisely the incidence and profile of their expression. Meiosis was induced by temperature shift in strains carrying the pat1-114 allele, which encodes a temperature-sensitive repressor of meiosis (Iino and Yamamoto 1985; Nurse 1985), and is widely used to achieve synchronous meioses in fission yeast (Bahler et al. 1991). Diploid pat1-114 cells were thermally induced to enter a synchronous meiosis from G1. We measured mRNA abundance by Northern analysis in samples taken every 15 min, and correlated this with the onset of pre-meiotic S-phase, as determined by flow cytometry analysis. These data are shown in Fig. 1.
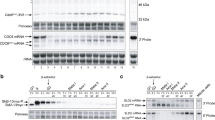
Regulation of gene expression during pre-meiotic S-phase in fission yeast. A population of diploid pat1-114 cells (GG 208) was thermally induced to enter a synchronous meiosis from G1, and samples were taken every 15 min for Northern blot analysis and at 60-min intervals for flow cytometry. The Northern blot was successively hybridised with radioactive probes for the indicated genes, which are displayed in three groups. An asynchronous wild-type mitotic control sample (M) is included in the last lane. Equal loading of RNA samples was confirmed by ethidium bromide staining of rRNA. The plots show the level of each transcript relative to that of rRNA at each timepoint
In agreement with previous work (Li and Smith 1997; Mata et al. 2002), most of the genes we analysed were induced during meiosis. Furthermore, these data show that the two groups of genes are induced together, with transcripts first appearing at ~80 min, and reaching high levels at ~120 min. For the cdc22 + group of genes, very low levels of transcript were present at earlier time points (0–60 min) during meiosis. Flow cytometry indicated that pre-meiotic S-phase started at between 1 and 2 h after the temperature shift, and was completed by 3 h—and was therefore coincident with the initiation of gene expression. Such co-ordinate transcription suggests that the same molecular processes may regulate the expression of both groups of genes.
We also examined the transcription profile of the genes that encode components of DSC1 in the same experiment (see below): cdc10 + and res1 + were found to be constitutively expressed during meiosis, whereas rep1 + and res2 + were induced. Low levels of rep1 + mRNA were detectable as early as 15 min after the temperature shift, i.e. before the onset of meiotic transcription of the cdc22 + and rec + groups of genes. The rep1 + gene was not transcribed during mitosis (Fig. 1; “M”, last lane) and is the earliest gene among this group to be induced during meiosis. This is consistent with the suggestion that Rep1p has a primary role in regulating meiotic expression (Li and Smith 1997; Ding and Smith 1998). res2 + mRNA appeared at the same time as the transcripts of the cdc22 + group of genes. Interestingly, res2 + contains one MCB-like sequence in its promoter at position –34 relative to the ATG (Fig. 2A, see next section), suggesting that its transcription may also be under the control of DSC1. rep2 + transcripts were absent during meiosis (data not shown).
Distribution and functional analysis of MCB motifs associated with genes expressed during pre-meiotic S-phase. A Presence of MCB motifs in the promoter regions of genes transcribed at pre-meiotic S-phase in fission yeast. The genes are placed in two classes: those required for DNA synthesis, which are expressed in both mitosis and meiosis; and those involved in recombination, expressed exclusively during meiosis. In each case the positions of the MCB motifs are numbered relative to the translation start codon (ATG) of each gene. B Synthetic MCB sequences confer meiotic-specific transcription in fission yeast. Haploid pat1-114 cells containing pSPΔ178.3 M (GG 664) were thermally induced to enter a synchronous meiosis from G1, and samples were taken every 15 min for Northern analysis, and at 60-min intervals for flow cytometry. The Northern blot was hybridised with radioactive probes for cdc22 +, rec8 + and lacZ. Equal loading of RNA samples was confirmed by ethidium bromide staining of rRNA. The level of each transcript, relative to rRNA, is plotted against time. C cdc22 + MCB motifs confer meiotic-specific transcription in fission yeast. Haploid pat1-114 cells containing SPIpMELβ2.MCB2 integrated in single copy (GG 675) were thermally induced to enter a synchronous meiosis from G1, and samples were taken every 15 min for Northern analysis and at 60-min intervals for flow cytometry. The Northern blot was successively hybridised with radioactive probes for cdc22 +, rec8 + and lacZ. Equal loading of RNA samples was confirmed by ethidium bromide staining of rRNA. Quantification of each transcript against rRNA is shown as in B
MCB sequences confer meiotic transcription in fission yeast
The molecular components that regulate mitotic cell cycle transcription of the cdc22 + group of genes have been identified; transcriptional specificity is mediated by the binding of the transcription factor complex DSC1 to a particular DNA motif (the MCB) present in the promoters of all of these genes.
It has been noted previously (Ding and Smith 1998; Mata et al. 2002) that the promoters of the rec + genes, which are expressed exclusively during meiosis, also contain sequences that resemble MCB motifs (Fig. 2A), suggesting that these motifs may also have a role in meiotic transcription in fission yeast. It is interesting to note that there appears to be correlation between the number and complexity of MCB motifs and the transcriptional profile of a given gene, with meiotic-specific genes having relatively simple arrays, and the mitotic/meiotic genes having more complex patterns (Fig. 2A).
To test if MCB motifs have a role in meiotic transcription we exploited two UAS reporter constructs, pSPΔ178 and SPIpMELβ2. pSPΔ178 is a multi-copy plasmid containing the budding yeast cytochrome c (CYC1) minimal promoter (Lowndes et al. 1992), and SPIpMELβ2 is an integration vector containing the minimal budding yeast MEL1 promoter which we adapted for use in fission yeast (see Materials and methods). Into both plasmids were inserted DNA fragments containing either three adjacent synthetic MCB motifs (“3MCB”; Lowndes et al. 1992) or an MCB cluster from the cdc22 + promoter named “MCB2” (Maqbool et al. 2003).
pSPΔ178.3MCB was transformed, and SPIpMELβ2.MCB2 was integrated in single copy, into haploid pat1-114 cells. Each strain was then induced to undergo synchronous meiosis from G1 arrest. Northern analysis of RNA obtained from the two different strains revealed specific induction of the lacZ reporter gene, coincident with the appearance of the endogenous cdc22 + and rec8 + transcripts (Fig. 2B, C). These experiments demonstrated that MCB motifs, both synthetic and derived from a natural promoter, confer meiotic transcription in fission yeast. These results confirm that MCB sequences present in the promoters of G1-S specific genes regulate their transcription during meiosis. Essentially identical results were also obtained using a promoter fragment from the rec15 + gene containing MCB motifs (data not shown).
Identification of a DSC1-like MCB binding complex in meiotic cells
Having established a role for MCB motifs in meiotic transcription we were next interested to see if a transcription factor complex similar to DSC1 regulates their expression. Initially, we sought to identify a DSC1-like DNA binding complex in meiotic cells using gel retardation analysis.
Diploid pat1-114 cells were thermally induced to enter meiosis synchronously from G1, and samples taken every hour to extract protein for gel retardation analysis using a DNA fragment from the cdc22 + promoter as the labelled substrate. No DSC1-like complex was detected at the beginning of the experiment in G1-arrested cells, but 2 h after the temperature shift and entry into meiosis, and coincident with the pre-meiotic S-phase (data not shown), a retarded complex appeared, of similar mobility to the mitotic DSC1 complex (Fig. 3A; lanes 7–11).
A meiotic form of DSC1 that binds to MCB motifs appears during a synchronous meiosis. A Gel retardation assay using a cdc22 + MCB1 DNA promoter fragment as the labelled probe with protein extracts from diploid pat1-114 cells (GG 208) undergoing synchronous meiosis. Two cultures were induced to enter meiosis, one was arrested in G1 by the removal of nitrogen (-N), the other was asynchronous (+N). Samples were taken every hour. The large arrow indicates the DSC1 complex, the small arrow the free probe. Lane F contains free probe; for lane M 20 μg of protein extract from dividing (mitotic) cells was used. B Gel retardation assay using a rec8 + MCB1 DNA promoter fragment as the labelled probe with protein extracts from G1-arrested diploid pat1-114 cells undergoing synchronous meiosis. The large arrow indicates the DSC1 complex, the small arrow free probe. Lane F contains the free probe
In diploid pat1-114 cells that were thermally induced to enter meiosis, but had not been pre-arrested in G1, a DSC1-like complex was detectable throughout the experiment (Fig. 3A; lanes 2–6), similar to that observed previously by others (Caligiuri et al. 1997; Ayte et al. 1997). The DSC1 detected in non G1 arrested meiotic cells is likely to represent the mitotic form of the complex, which under normal conditions would be expected to disappear, as fission yeast cells can only enter meiosis after nitrogen starvation (Yamamoto 1996).
We also examined complexes that could bind to rec + gene promoter fragments containing MCB motifs during a synchronous meiosis that was induced from G1 arrest. A DSC1-like complex was detected that bound to rec8 + and rec7 +, rec11 +, rec15 + MCB DNA, but differed from that formed on the cdc22 + probe in that it was present throughout the meiotic induction (Fig. 3B, and data not shown).
The observation that a DSC1-like complex could bind to rec + gene MCB motifs throughout meiosis in nitrogen-starved, G1-arrested cells, raised the possibility that DSC1 might bind to these sequences in mitotic cells. We tested this hypothesis by examining whether promoter fragments containing MCB motifs from the various rec + genes could bind to DSC1 in mitotic cells. When MCB motifs from the rec8 + and rec7 +, rec11 + and rec15 + genes were used both as labelled substrate and non-labelled competitor DNA, the DNAs bound to mitotic DSC1 (data not shown).
Altering the levels of components of DSC1 affects meiotic transcription and pre-meiotic S-phase
Having identified a DSC1-like complex in meiotic cells in fission yeast, we were next interested to determine whether manipulating components of DSC1 affected meiotic transcription. We took two approaches to examine this possibility. We first tested the effect of mutants defective and deficient for components of DSC1 on meiotic transcription and S-phase progression. Secondly, the effect of over-expressing the same components was studied.
In the first approach we combined mutations in genes for individual components of DSC1 with pat1-114. This was done in separate experiments with haploid cdc10-129, cdc10-V50 , res1 Δ, res2 Δ, rep1 Δ and rep2Δ cells. In each experiment samples were taken to quantify cdc22 +, rec8 + and rec11 + transcript abundance by Northern analysis, and monitor pre-meiotic S-phase progression by flow cytometry. These genes were chosen as representatives of the two groups shown in Fig. 1A.
We also tested the effect of over-expression of individual components of DSC1 on cdc22 +, rec8 + and rec11 + transcript levels during meiosis. To do this we placed cdc10 +, res1 +, res2 +, rep1 + and rep2 + under the control of the regulatable nmt1 + promoter (Maundrell 1993), and separately over-expressed each gene in haploid pat1-114 cells, and then induced synchronous meiosis. In each case, over-expression was confirmed by hybridising Northern blots with the appropriate probe (data not shown).
cdc10 +
In agreement with previous experiments (Beach et al. 1985), cdc10-129 cells were found to be unable to enter pre-meiotic S-phase (Fig. 4). A likely explanation for this observation is the fact that these cells fail to express cdc22 +, rec8 + and rec11 + (Fig. 4), which encode products required for meiotic S-phase and recombination. Very low levels of rec8 + and rec11 + mRNAs were apparent late in meiosis. However, the amounts were significantly lower than the peak levels observed in the wild-type samples on the same Northern blot, taken from the experiment shown in Fig. 1 (Fig. 4; “wt 105 and 180”). Identical results were obtained with another mutant allele of cdc10 +, cdc10-V50 (Fig. 4).
Role of cdc10 + in meiotic transcription and progression. Haploid pat1-114 cdc10-129 (GG 216), pat1-114 cdc10-V50 (GG 307) and pat1-114 cells containing pREP1: cdc10 + (GG 304), were thermally induced to enter a synchronous meiosis from G1, and samples were taken every 15 min for Northern analysis, and at 60-min intervals for flow cytometry. The Northern blots were hybridised with radioactive probes for cdc22 + , rec8 + and rec11 +. Control RNA lanes containing trough (105 min) and peak (180 min) samples from the “wild-type” meiosis shown in Fig. 1, and an asynchronous mitotic wild-type sample (M) are included in the last three lanes. Equal loading of RNA samples was confirmed by ethidium bromide staining of rRNA (data not shown). A wild-type meiotic profile of each transcript is included for direct comparison
Over-expression of wild-type cdc10 + did not affect the time at which cdc22 +, rec8 + and rec11 + transcripts first appeared, but resulted in higher transcript levels than those observed in wild-type cells undergoing meiosis (Fig. 4; compare with “wt 105 and 180”). Combined, these data imply that cdc10 + is not only required for, but has a stimulatory function in controlling meiotic MCB-regulated gene expression.
res1 +
The res1Δ mutation had no effect on the cdc22 +, rec8 + or rec11 + expression profile during meiosis, implying it has no role in this process. Surprisingly, therefore, over-expression of res1 + resulted in higher cdc22 +, rec8 + and rec11 + transcript levels (Fig. 5). All three mRNAs also appeared earlier than in wild-type cells, with cdc22 + coming up especially early, peaking at 45 min. Neither manipulation of res1 + affected pre-meiotic S-phase progression, at least as judged by flow cytometry (Fig. 5).
Role of res1 + in meiotic transcription and progression. Haploid pat1-114 res1Δ (GG 248) and pat1-114 cells containing pREP1: res1 + (GG 432) were thermally induced to enter a synchronous meiosis from G1, and samples were taken every 15 min for Northern analysis and at 60-min intervals for flow cytometry. The Northern blots were hybridised with radioactive probes for cdc22 + , rec8 + and rec11 +. Control RNA lanes containing trough (105 min) and peak (180 min) samples from the “wild-type” meiosis shown in Fig. 1, and an asynchronous mitotic wild-type sample (M) are included in the last three lanes. Equal loading of RNA samples was confirmed by ethidium bromide staining of rRNA (data not shown). A wild-type meiotic profile of each transcript is included for direct comparison
res2 + and rep1 +
In both res2Δ and rep1Δ cells induction of cdc22 +, rec8 + and rec11 + transcripts was affected during meiosis (Figs. 6 and 7). All three transcripts were absent in rep1Δ cells, but were differently altered in res2Δ, with cdc22 + mRNA being undetectable and rec8 + and rec11 + reduced compared to wild-type. Both of these deletion mutants failed to undergo pre-meiotic S-phase, as reported previously (Sugiyama et al. 1994; Zhu et al. 1994; Li and Smith 1997). These results suggest that res2 + and rep1 + are required for full meiotic transcription of cdc22 +, rec8 + and rec11 +. Over-expression of res2 + had no effect on the levels of cdc22 +, rec8 + and rec11 + transcripts, while over-expressing rep1 + enhanced cdc22 + rec8 + and rec11 + transcription in cells undergoing meiosis. This latter observation implies that Rep1p has a stimulatory function in controlling meiotic gene expression.
Role of res2 + in meiotic transcription and progression. Haploid pat1-114 res2Δ (GG 157) and pat1-114 cells containing pREP1: res2 + (GG 344) were thermally induced to enter a synchronous meiosis from G1, and samples were taken every 15 min for Northern blot analysis and at 60-min intervals for flow cytometry. The Northern blots were hybridised with radioactive probes for cdc22 + , rec8 + and rec11 +. Control RNA lanes containing trough (105 min) and peak (180 min) samples from the “wild-type” meiosis shown in Fig. 1, and an asynchronous mitotic wild-type sample (M) are included in the last three lanes. Equal loading of RNA samples was confirmed by ethidium bromide staining of rRNA (data not shown). A wild-type meiotic profile of each transcript is included for direct comparison
Role of rep1 + in meiotic transcription and progression. Haploid pat1-114 rep1Δ (GG 172) and pat1-114 cells containing pREP1: rep1 + (GG 306) were thermally induced to enter a synchronous meiosis from G1, and samples were taken every 15 min for Northern blot analysis, and at 60-min intervals for flow cytometry. The Northern blots were hybridised with radioactive probes for cdc22 + , rec8 + and rec11 +. Control RNA lanes containing trough (105 min) and peak (180 min) samples from the “wild-type” meiosis shown in Fig. 1, and an asynchronous mitotic wild-type sample (M) are included in the last three lanes. Equal loading of RNA samples was confirmed by ethidium bromide staining of rRNA (data not shown). A wild-type meiotic profile of each transcript is included for direct comparison
rep2 +
The rep2Δ mutation and over-expression of rep2 + had differing and interesting effects on cdc22 +, rec8 + and rec11 + mRNA levels (Fig. 8). In rep2Δ cells, higher levels of all three transcripts were observed during meiosis than were detectable in a wild-type meiosis. Over-expression of rep2 + had no effect on cdc22 + transcription, but repressed that of rec8 + and rec11 +; only extremely low levels of transcript were expressed from the latter two genes.
Role of rep2 + in meiotic transcription and progression. Haploid pat1-114 rep2Δ (GG 303) and pat1-114 cells containing pREP1: rep2 + (GG 305) were thermally induced to enter a synchronous meiosis from G1, and samples taken every 15 min for Northern analysis, and at 60-min intervals for flow cytometry. The Northern blots were hybridised with radioactive probes for cdc22 + , rec8 + and rec11 +. Control RNA lanes containing trough (105 min) and peak (180 min) samples from the “wild-type” meiosis shown in Fig. 1, and an asynchronous mitotic wild-type sample (M) are included in the last three lanes. Equal loading of RNA samples was confirmed by ethidium bromide staining of rRNA (data not shown). A wild-type meiotic profile of each transcript is included for direct comparison
In rep2Δ cells pre-meiotic S-phase occurred at the same time as in wild-type cells, as indicated by flow cytometry. In contrast, pre-meiotic S-phase was delayed by about an hour in cells over-expressing rep2 +.
Discussion
Fission yeast meiotic gene expression
On undertaking the developmental switch between mitotic growth and meiosis, diploid fission yeast cells undergo a programme of molecular changes that result in DNA replication, two meiotic divisions, and the formation of four haploid spores. An essential early stage in this process is pre-meiotic S-phase: the first, reductional, meiotic division will not occur without a preceding S-phase (Watanabe et al. 2001).
The experiments described in this paper analyse the transcription control system that regulates gene expression during pre-meiotic S-phase in fission yeast. We show that a large group of genes are simultaneously transcribed during this early stage of meiosis (Fig. 1), the promoters of which all contain MCB motifs (Fig. 2A). MCB motifs were shown to confer a similar expression pattern on heterologous reporter genes during meiosis (Fig. 2B, C), and so control transcription during pre-meiotic S-phase in fission yeast. Furthermore, we identified a DSC1-like complex that binds to MCB motifs in meiotic fission yeast cells (Fig. 3). Loss of function mutants for, and over-expression of, components of DSC1 affected meiotic transcription and progression through pre-meiotic S-phase. Combined, these data demonstrate a role for the DSC1-MCB transcription control system in controlling meiotic gene expression in fission yeast.
How does the DSC1-MCB system stimulate meiotic-specific expression? It is known that mutations in a number of genes for components of the mitotic DSC1, including cdc10 + and res2 +, prevent or delay passage through meiosis (Beach et al. 1985; Sugiyama et al. 1994; Zhu et al. 1994; Li and Smith 1997; Figs. 4 and 6). Furthermore, ectopic over-expression of res2 + induces meiosis (Ayte et al. 1997). Deletion of rep1 + also reduces rec + gene transcription during meiosis (Li and Smith 1997; Ding and Smith 1998), and prevents induction of res2 + (Sugiyama et al. 1994). Here we show, both by deleting and over-expressing cdc10 +, res2 + and rep1 +, that they are all required, and positively regulate expression of cdc22 +, rec8 + and rec11 +, at pre-meiotic S-phase. Over-expression of cdc10 + caused over-expression of MCB-regulated genes during meiosis (Fig. 4), a phenomenon not seen when this gene is over-expressed in mitosis (Baum et al. 1997, White et al. 2001), implying that Cdc10p-mediated transcription is controlled differently in the two processes. As rep1 + is exclusively expressed during meiosis under normal conditions, and it is the first gene in this group to be induced during pre-meiotic S phase (Fig. 1), it is likely that it has a primary role in controlling meiotic-specific expression of these genes.
Interestingly, res2 + is also induced during meiosis, coincident with the other genes at pre-meiotic S-phase (Fig. 1; Mata et al. 2002). It is likely that this gene is also under DSC1 control, as it contains an MCB motif in its promoter (Fig. 2A), and res2 + is not expressed in meiotic cells mutant for either cdc10 + or rep1 + (data not shown). This observation implies that res2 + regulates its own expression during meiosis, as part of a feedback loop. As res2 + is not under the control of DSC1 during mitosis (Whitehall et al. 1999), this meiotic-specific regulation may contribute to mediating meiotic expression of MCB-controlled genes.
Manipulating rep2 + had interesting effects on meiotic transcription (Fig. 8). Specifically, its deletion resulted in higher than wild-type levels of cdc22 +, rec8 + and rec11 + RNAs during meiosis. This observation suggests Rep2p is required to repress MCB-regulated gene expression during meiosis. Furthermore, over-expressing rep2 + had no effect on cdc22 + expression, but repressed both rec8 + and rec11 +. This latter result is consistent with rep2 + having a repressive role, at least for the rec + genes. rep2 + is not normally expressed during meiosis, but is only transcribed during mitosis (Nakashima et al. 1995)—a property which is highly unusual in fission yeast (Mata et al. 2002). This observation, in combination with the gel retardation data, may explain how DSC1 results in inhibition of rec + gene, but not cdc22 +, transcription in mitotic cells.
Gel retardation analysis led to the provocative observation that MCB motifs in rec + gene promoters have alternative DSC1 binding properties. Like the MCB motifs of the cdc22 + group of genes, they bind to this complex in mitotic cells (Fig. 2). However, significantly, the fact that rec + genes are not expressed during mitosis indicates that DSC1 causes their repression. The simplest explanation for this is that two forms of DSC1 exist in mitotic cells: one is repressive and binds to the MCBs of rec + genes, and the other is stimulatory and binds to the cdc22 + group MCBs. This difference is probably accounted for by the variation in complexity of MCB motifs seen in the promoters of two types of genes (Fig. 2A), with the simpler rec + gene arrays binding to the repressive form of DSC1.
Finally, res1 + appears to have no role in regulating meiotic gene transcription. We show that deleting res1 + has no effect on cdc22 +, rec8 + or rec11 + expression during meiosis (Fig. 6). This observation suggests that res1 + is not required for meiotic transcription, which is consistent with the finding that res1Δ has no effect on mating efficiency (Tanaka et al. 1992). Surprisingly, therefore, over-expression of res1 + resulted in cdc22 +, rec8 + and rec11 + mRNA appearing earlier than in wild-type cells and peaking at higher levels (Fig. 6). However, it is known that over-expression of res1 + during mitosis results in continuous maximal transcription of MCB-regulated genes (Baum et al. 1997). We suggest that res1 + has similar properties when over-expressed in meiosis, but this does not occur under normal conditions, as the gene is only transcribed at low levels (Fig. 1; Ayte et al. 1997). It is likely that it is this property which accounts for the ability of res1 +, when over-expressed, to suppress pat1-114 induced meiosis (Tanaka et al. 1992).
To summarise the conclusions from these data, we propose that during mitosis the mitotic form of DSC1 binds to MCB motifs in both cdc22 + and rec + gene promoters. It is the presence of Rep2p (along with Cdc10p, Res1p and Res2p) in this complex that specifically represses rec + gene expression during mitosis, whilst stimulating cdc22 + transcription. This difference may be accounted for by alternative forms of DSC1 binding to the MCB motifs in the two promoter types, with the relatively simpler rec + gene promoter motifs (Fig. 2A) being specifically repressed by Rep2p under these conditions.
Upon nitrogen starvation and initiation of meiotic S-phase, mitotic DSC1 is replaced by a meiotic form of the transcription factor complex, containing Cdc10p, Res2p and Rep1p, and possibly other components. Rep1p has the primary role in initiating meiotic transcription, as it is the first of these genes to be expressed at this stage. Cdc10p has an important stimulatory role as its over-expression induced MCB-regulated gene transcription. res2 + contains an MCB motif in its own promoter, and is itself regulated by DSC1 in a feedback loop to stimulate its meiotic expression. It is the combination of Rep1p and Res2p, together with Cdc10p, binding to MCB motifs in the promoters of genes that stimulates meiotic-specific transcription in fission yeast.
Meiotic gene expression in budding yeast
The regulation of transcription at early and later stages of meiosis has also been extensively studied in the budding yeast Saccharomyces cerevisiae, and many mechanisms of its control have been elucidated (Mitchell 1994; Kupiec et al. 1997; Clancy 1998; Chu et al. 1998). As in fission yeast, some genes containing MCB motifs in their promoters, which are expressed at G1-S during mitosis, are also induced during meiosis (Johnston et al. 1986). However, no role for MCB sequences in meiotic transcription has been established in this species (Cole and Mortimer 1989). So the cis -acting promoter sequences that confer early meiotic transcription in the two species of yeasts are likely to be different. A related transcription factor complex may be operating in budding yeast, as homologues of cdc10 +, SWI6 and SWI4, have been reported to be transcribed during meiosis (Leem et al. 1998). In addition, deletion of SWI6 reduces expression of RAD51 and RAD54 during meiosis, two genes required for meiotic recombination. However, swi6 Δ cells are not delayed in pre-meiotic S-phase progression (Leem et al. 1998), so there must also be significant differences between the two organisms.
Fission yeast meiotic gene expression and progression
The genes regulated by the DSC1-MCB complex include those required for pre-meiotic DNA synthesis, such as cdc22 + and cdc18 +, and those required for recombination, such as rec8 + and rec11 +. Recent experiments have suggested that the two processes of pre-meiotic S-phase and recombination, both essential for the first meiotic division, are not dependent on each other, and represent two separate pathways (Muramaki and Nurse 2001). We propose that the meiotic form of DSC1, containing Cdc10p, Res2p and Rep1p, initiates these two pathways, as it regulates the transcription of both groups of genes by binding to MCB-sequence motifs present in their promoters. Thus, their specific expression, under the control of DSC1 and MCB motifs, is an important part of meiotic progression.
References
Ayte J, Leis JF, Decaprio JA (1997) The fission yeast protein p73 res2 is an essential component of the mitotic MBF complex and master regulator of meiosis. Mol Cell Biol 17:6246–6254
Ayte J, Schweitzer C, Zarzov P, Nurse P, Decaprio JA (2001) Feedback regulation of the MBF transcription factor by cyclin Cig2. Nat Cell Biol 3:1043–1050
Bahler J, Schuchert P, Grimm C, Kohli J (1991) Synchronized meiosis and recombination in fission yeast: observations with pat1-114 diploid cells. Curr Genet 19:445–451
Baum B, Wuarin J, Nurse P (1997) Control of S-phase periodic transcription in the fission yeast mitotic cycle. EMBO J 16:4676–4688
Beach D, Rodgers L, Gould J (1985) RAN1+ controls the transition from mitotic division to meiosis in fission yeast. Curr Genet 10:297–311
Birkenbihl RP, Subramani S (1995) The rad21 gene product of Schizosaccharomyces pombe is a nuclear, cell cycle regulated phosphoprotein. J Biol Chem 270:7703–7711
Caliguiri M, Beach D (1993) Sct1 functions in partnership with Cdc10 in a transcription complex that activates cell cycle START and inhibits differentiation. Cell 72:607–619
Caligiuri M, Connolly T, Beach D (1997) Ran1 functions to control the Cdc10/Sct1 complex through Puc1. Mol Biol Cell 8:1117–1128
Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I (1998) The transcriptional program of sporulation in budding yeast. Science 282:699–705
Clancy MJ (1998) Meiosis: step-by-step through sporulation. Curr Biol 8:461–463
Cole GM, Mortimer RK (1989) Failure to induce a DNA repair gene, RAD54 , in Saccharomyces cerevisiae does not affect DNA repair or recombination phenotypes. Mol Cell Biol 9:3314–3322
Connolly T, Beach D (1994) Interaction between the Cig1 and Cig2 B-type cyclins in the fission yeast cell cycle. Mol Cell Biol 14:768–776
Davis L, Smith GR (2001) Meiotic recombination and chromosome segregation in Schizosaccharomyces pombe. Proc Natl Acad Sci USA 98:8395–8402
Ding R, Smith GR (1998) Global control of meiotic recombination genes by Schizosaccharomyces pombe rec16 (rep1). Mol Gen Genet 258:663–670
Egel R (2000) Fission yeast on the brink of meiosis. Bioessays 22:854–860
Feinberg AP, Volgelstein B (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 1:6-13
Fernandez-Sarabia MJ, McInerny CJ, Harris P, Gordon CB, Fantes PA (1993) The cell cycle genes cdc22 + and suc22 + of the fission yeast Schizosaccharomyces pombe encode the large and small subunits of ribonucleotide reductase. Mol Gen Genet 238:241–251
Fox ME, Smith GR (1998) Control of meiotic recombination in Schizosaccharomyces pombe. Prog Nucleic Res Mol Biol 61:345–378
Gutz H, Heslot H, Leupold U, Loprieno N (1974) Schizosaccharomyces pombe. In: King RC (ed) Handbook of genetics, vol 1. Plenum, New York, pp 395–446
Harris P, Kersey PJ, McInerny CJ, Fantes PA (1996) Cell cycle, DNA damage and heat shock regulate suc22 + expression in fission yeast. Mol Gen Genet 13:284–291
Hofmann JFX, Beach D (1994) cdt1 is an essential target of the Cdc10/Sct1 transcription factor: requirement for DNA replication and inhibition of mitosis. EMBO J 13:425–434
Iino Y, Yamamoto M (1985) Mutants of Schizosaccharomyces pombe which sporulate in the haploid state. Mol Gen Genet 198:416–421
Johnston LH, Johnson AL, Barker DG (1986) The expression in meiosis of genes which are transcribed periodically in the mitotic cell cycle of budding yeast. Exp Cell Res 165:541–549
Kelly TJ, Martin GS, Forsburg SL, Stephen RJ, Russo A, Nurse P (1993) The fission yeast cdc18 + gene product couples S phase to START and mitosis. Cell 74:371–382
Kohli J, Hottinger H, Munz P, Strauss A, Thuriaux P (1977) Genetic mapping in Schizosaccharomyces pombe by mitotic and meiotic analysis and induced haploidization. Genetics 87:23–471
Kupiec M, Byers B, Esposito RE, Mitchell AP (1997) Meiosis and sporulation in Saccharomyces cerevisiae. In: Pringle JR, Broach JR, Jones EW (eds) The molecular biology of the yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., pp 889–1036
Leem SH, Chung CN, Sunwoo Y, Araki H (1998) Meiotic role of SWI6 in Saccharomyces cerevisiae. Nucleic Acids Res 26:3154–3158
Li YF, Smith GR (1997) The Schizosaccharomyces pombe rec16 gene product regulates multiple meiotic events. Genetics 146:57–67
Li YF, Numata M, Wahls WP, Smith GR (1997) Region-specific meiotic recombination in Schizosaccharomyces pombe: the rec11 gene. Mol Microbiol 23:869–878
Lowndes NF, McInerny CJ, Johnson AL, Fantes PA, Johnston LH (1992) Control of DNA synthesis genes in fission yeast by the cell-cycle gene cdc10 +. Nature 355:449–453
Maqbool Z, Kersey PJ, Fantes PA, McInerny CJ (2003) MCB regulation of cdc22 +cell cycle-specific transcription in fission yeast. Mol Genet Genomics 269:765–775
Maundrell K (1993) Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123:127–130
McInerny CJ, Kersey PJ, Creanor J, Fantes PA (1995) Positive and negative roles for cdc10 in cell cycle gene expression. Nucleic Acids Res 23:4761–4768
Melcher K, Sharma B, Ding WV, Nolden M (2000) Zero background yeast reporter plasmids. Gene 247:53–61
Mata J, Lyne R, Burns G, Bahler J (2002) The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet 32:143-147
Mitchell AP (1994) Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev 58:56–70
Miyamoto M, Tanaka K, Okayama H (1994) res2 +, a new member of the cdc10 +/ SWI4 family, controls the ‘start’ of mitotic and meiotic cycles in fission yeast. EMBO J 13:1873–1880
Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of the fission yeast Schizosaccharomyces pombe. Methods Enzymol 194:795–823
Murakami H, Nurse P (1999) Meiotic DNA replication checkpoint in fission yeast. Genes Dev 13:2581–2593
Murakami H, Nurse P (2001) Regulation of premeiotic S phase and recombination-related double-strand DNA breaks during meiosis in fission yeast. Nat Genet 28:290–293
Nakashima N, Tanaka K, Sturm S, Okayama H (1995) Fission yeast Rep2 is a putative transcriptional activator subunit for the cell cycle ‘start’ function of Res2-Cdc10. EMBO J 14:4794–4802
Ng SS, Anderson M, White S, McInerny CJ (2001) mik1 + G1-S transcription regulates mitotic entry in fission yeast. FEBS Lett 503:131–134
Nurse P (1985) Mutants of the fission yeast Schizosaccharomyces pombe which alter the shift between cell proliferation and sporulation. Mol Gen Genet 198:497–502
Parker A, Clyne R, Carr A, Kelly T (1997) The Schizosaccharomyces pombe rad11 + gene encodes the large sub-unit of replication protein A. Mol Cell Biol 17:2381–2390
Sambrook J, Fritch EF, Maniatis T (1989) Molecular cloning: a laboratory manual (2nd edn). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
Sturm S, Okayama H (1996) Domains determining the functional disitinction of the fission yeast Start molecules Res1 and Res2. Mol Biol Cell 7:1967–1976
Sugimoto A, Iino Y, Maeda T, Watanabe Y, Yamamoto M (1991) Schizosaccharomyces pombe ste11 + encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev 5:1990–1999
Sugiyama A, Tanaka K, Okazaki K, Nojima H, Okayama H (1994) A zinc finger protein controls the onset of premeiotic DNA synthesis of fission yeast in a Mei2-independent cascade. EMBO J 13:1881–1887
Tanaka K, Okazaki K, Okazaki N, Ueda T, Sugiyama A, Noijima H, Okayama H (1992) A new cdc gene required for S-phase entry of Schizosaccharomyces pombe encodes a protein similar to the cdc10 + and SWI4 gene products. EMBO J 11: 4923–4932
Tournier S, Millar JBA (2000) A role for the START gene-specific transcription factor complex in the inactivation of cyclin B and Cut2 destruction. Mol Biol Cell 11:3411–3424
Watanabe Y, Yokobayashi S, Yamamoto M, Nurse P (2001) Pre-meiotic S phase is linked to reductional chromosome segregation and recombination. Nature 409:359–363
White S, Khaliq F, Sotiriou S, McInerny CJ (2001) The role of DSC1 components cdc10 +, rep1 + and rep2 + in MCB gene transcription at the mitotic G1-S boundary in fission yeast. Curr Genet 40: 251–259
Whitehall S, Stacey P, Dawson K, Jones N (1999) Cell cycle-regulated transcription in fission yeast: Cdc10-Res protein interactions during the cell cycle and domains required for regulated transcription. Mol Biol Cell 10:3705–3715
Yamamoto M (1996) The molecular control mechanisms of meiosis in fission yeast. Trends Biochem Sci 21:18–22
Yoshida S, Al-Amodi H, Nakamura T, McInerny CJ, Shimoda C (2003) The Schizosaccharomyces pombe cdt2 + gene, a target of G1-S phase-specific transcription factor complex DSC1, is required for mitotic and premeiotic DNA replication. Genetics 164:881–893
Zhu Y, Takeda T, Nasmyth K, Jones N (1994) pct1 +, which encodes a new DNA-binding partner of p85 cdc10, is required for meiosis in the fission yeast Schizosaccharomyces pombe. Genes Dev 8:885–898
Zhu Y, Takeda T, Whitehall S, Peat N, Jones N (1997) Functional characterisation of the fission yeast Start-specific transcription factor Res2. EMBO J 16:1023–1034
Acknowledgements
We would like to thank Chikashi Shimoda, Viesturs Simanis, Gerry Smith and Kiochi Tanaka for strains and plasmids. We would particularly like to thank Gerry Smith for his encouragement to initiate this research. The work was supported by The Biotechnology and Biological Sciences Research Council, The Royal Society, and a Wellcome Trust Studentship to L.C. Thanks to Tracy Riddell for assistance with FACS analysis, and to other members of the lab for suggestions during the course of this work. The work described in this paper has been performed in compliance with the UK laws covering genetic experimentation
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. P. Hollenberg
Rights and permissions
About this article
Cite this article
Cunliffe, L., White, S. & McInerny, C.J. DSC1-MCB regulation of meiotic transcription in Schizosaccharomyces pombe . Mol Genet Genomics 271, 60–71 (2004). https://doi.org/10.1007/s00438-003-0956-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-003-0956-6