Abstract
Previous studies have shown that heterosis is associated with differential gene expression between hybrids and their parents. In this study, we performed a screen for genes that are differentially expressed between wheat hybrids and their parents in jointing-stage leaves and flag leaves using the differential display technique. Twenty-four differentially expressed cDNA were cloned and sequenced, and their expression patterns were confirmed by reverse-Northern blotting. Sequence analysis and database searches revealed that among the genes that showed differential expression between hybrid and parents were transcription factor genes and genes involved in metabolism, signal transduction, disease resistance, and retrotransposons. These results indicate that hybridization between two parental lines can cause changes in the expression of a variety of genes, and it is concluded that the altered pattern of gene expression in the hybrid may be responsible for the observed heterosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hybrid cultivars have been used commercially in many crop plants and have made significant contributions to the world’s food supply (Duvick 1997). However, the molecular mechanism of hybrid vigor, or heterosis, remains to be revealed. Although the genome in hybrid F1 is derived from its parental inbreds, hybrid performance is often quite different from that of either of the parents, implying that differences in gene expression between hybrids and their parents are responsible for heterosis (Sun et al. 1999).
The differential display technique (Liang and Pardee 1992) has been successfully used to identify genes that are differentially expressed between cereal hybrids and their parents (Xiong et al. 1998; Sun et al. 1999). Previous studies have indeed indicated that the patterns of differential gene expression are correlated with heterosis (Xiong et al. 1998; Wu et al. 2001). Thus, cloning and characterization of genes that are differentially expressed between hybrids and their parents should provide further insight into the molecular mechanisms responsible for heterosis. In wheat, Ni et al (2000) previously identified and cloned a cDNA encoding a novel RNA-binding protein that was specifically expressed in F1 plants but not in the parental lines, and discussed its possible role in wheat heterosis.
In this study, we conducted a differential display analysis of 20 F1 hybrids and their parents. Twenty-four cDNAs derived from genes that are differentially expressed between hybrids and their parents in flag leaves were cloned and sequenced, and their possible roles in heterosis are discussed.
Materials and methods
Wheat materials and identification of true hybrid plants
Four female lines and five male lines were crossed inter se according to the NCII design to form a diallel set of 20 crosses. Forty seedlings per cross were planted in two-row plots with three replications. True hybrid plants were confirmed using simple-sequence repeat (SSR) markers.
RNA extraction
Fully expanded leaves at the jointing stage, and flag leaves, were harvested in the field from the F1 hybrids and their parents. Total RNA was prepared from each sample using the RNeasy kit (Sangon, Shanghai).
Reverse transcription
Equal aliquots (2 μg) of RNA were transcribed into cDNA in 20-μl reactions containing 50 mM TRIS-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 50 μM dNTPs, 200 U of MMLV reverse transcriptase (Promega, Madison, Wis.) and 50 pmol of the anchor oligonucleotide HT11A, HT11C, or HT11G. Reverse transcription was performed for 60 min at 37°C with a final denaturation step at 95°C for 5 min.
PCR amplification of cDNA
The following primers were synthesized according to von der Kammer et al. (1999). As 3′ end anchored primers, the oligonucleotides HT11A (5′-AAGCTTTTTTTTTTTA-3′), HT11C (5′-AAGCTTTTTTTTTTTC-3′) and HT11G (5′-AAGCTTTTTTTTTTTG-3′) were used. DD10 (5′-TGCCGAAGCTTTGGTAGC-3′), DD18 (5′-TGCCGAAGCTTTGGTCAC-3′), DD19 (5′-TGCCGAAGCTTTGGTCAG-3′), DD20 (5′-TGCCGAAGCTTTGGTCAT-3′), DD23 (5′-TGCCGAAGCTTGATTCCG-3′), DD32 (5′-TGCCGAAGCTTGGAGCTT-3′), DD34 (5′-TGCCGAAGCTTTGGTGAC-3′), DD38 (5′-TGCCGAAGCTTGATTGCC-3′), DD46 (5′-TGCCGAAGCTTTGGTGTC-3′), DD54 (5′-TGCCGAAGCTTTGGTTCC-3′), and DD60 (5′-TGCCGAAGCTTCGACTGT-3′) served as 5′ end primers.
In order to improve the reproducibility, the improved differential display protocol was used in our study (von der Kammer et al. 1999). Aliquots (2 μl) of the cDNAs obtained were subjected to PCR employing the corresponding anchor oligonucleotide together with one of the DD (differential display) random primers, 1.5 mM MgCl2, 0.20 mM dNTP and 1 U of Taq polymerase in a 20-μl reaction volume. PCRs were performed as follows: one cycle of 94°C for 1 min, 40°C for 4 min; 72°C for 1 min; followed by 40 cycles of 94°C for 45 s; 60°C for 2 min and 72°C for 1 min. One final step at 72°C for 5 min was added to the last cycle.
Electrophoresis
PCR products were separated on 4% denaturing polyacrylamide sequencing gels (0.4 mm thick) in a temperature-regulated Bio-Rad Sequencing System (Bio-Rad, Fullerton, Calif.) at 50°C. Gels were stained with silver, and photographed.
Cloning, sequencing and reverse-Northern blot analysis
Bands that showed qualitative differences between hybrid F1 (Nongda3338 × Jingdong6) and its parents were excised from the gel and reamplified using the following PCR conditions: 1 min at 94°C; followed by 40 cycles of 45 s at 94°C, 2 min at 60°C and 1 min at 72°C. One final step at 72°C for 5 min was added to the last cycle. To ensure that there was no DNA contamination in RNA samples, a negative control was prepared without reverse transcriptase. The differentially expressed cDNAs (DEs) were ligated into the pGEM-T Easy vector (Promega) and sequenced. Reverse-Northern analysis was performed according to manufacturer’s instructions using the ECL kit (Amersham, Little Chalfont, Bucks., UK) with minor modifications. Each fragment was reamplified, electrophoresed on a 1.0% agarose gel and transferred to a nylon membrane. Total RNA was labeled using the ECL kit, and hybridized to a Hybond N+ membrane (Amersham) according to the manufacturer’s recommendations.
Results
Patterns of differentially expressed fragments
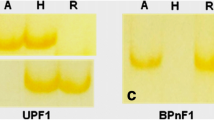
A total of 3066 cDNA fragments were amplified from the leaves harvested at the jointing stage by using 33 primer combinations comprising one of three one-base anchored primers and each of eleven different 5′ end oligonucleotide primers, and 2950 cDNA fragments were amplified from flag leaves by using 27 primer combinations (one of three one-base anchored primers and each of nine 5′ end oligonucleotide primers). The differentially displayed cDNAs showed both quantitative and qualitative differences. Since quantitative differences could not be accurately measured, we only analyzed bands that showed qualitative differences. Such differences can be classified into one of four categories (Fig. 1): (1) bands observed in both parents but not in the F1 (BPnF1, Fig. 1A), (2) bands occurring in one parent but not in the F1 or the other parent (UPnF1, Fig. 1B and C), (3) bands detected in the F1 but in neither of the parents (F1nBP, Fig. 1D), and (4) bands present in one parent and F1 but absent in the other parent (UPF1, Fig. 1E and F). When analyzed across the 20 hybrids, the BPnF1 pattern accounts for 5.99% and 2.87% of the total number of bands resolved in the samples from jointing-stage leaves and flag leaves, respectively. The UPnF1 pattern makes up 8.44% and 8.95%, the F1nBP pattern 3.54% and 4.10%, and the UPF1 pattern for 12.07% and 10.69% of the bands, respectively.
Patterns of differential gene expression between wheat hybrids and their parents. A Bands observed in both patterns but not in the F1 (BPnF1). B Bands occurring in only female parent but not in the F1 and the male (UPnF1). C Bands occurring in only male but not in the F1 and the female parent (UPnF1). D Bands detected in only the F1 but neither of the parents (F1nBP). E Bands present in female parent and F1 but absent in male (UPF1). F Bands present in male and F1 but absent in female parent (UPF1)
The percentages of bands that fall into each of the four categories of differentially displayed cDNA sequences are listed in Table 1. Obviously, the UPF1 pattern is the most prominent class of differentially displayed cDNA fragments at both stages. It is also clear from Table 1 that there is considerable variation in the four categories among the twenty crosses at two stages, indicating that the differences in patterns of gene expression among hybrids may lead to different degrees of heterosis among different hybrids.
Cloning, confirmation and sequencing of differentially expressed cDNAs
Twenty-four cDNA fragments that showed differential expression between the heterotic wheat hybrid F1 Nongda3338×Jingdong6 and its parental inbreds, as detected in DD (differential display), were cloned and sequenced (Table 2). The expression patterns of the twenty-four cDNA fragments were confirmed by reverse Northern blot analysis (Fig. 2). Among the 24 differentially expressed cDNA fragments, nine (DE1–5, DE9, DE12, DE18 and 19) showed reduced expression in the hybrid F1; two (DE6 and 7) were expressed in both parents but not in the F1; four (DE8, 11, 13 and 17) were expressed in one parent, but not in the F1 or the other parent; two (DE10 and 14) showed enhanced expression in the F1; six (DE15, 16, DE20–22 and 24) were present in one parent and the F1, but absent in the other parent; and one (DE23) was expressed in the F1 and in both parents, but the level of expression in the F1 was similar to that in the female parent (Table 2).
Validation by reverse-Northern analysis of the gene expression alterations detected by DDRT. P, paternal parent, F1, hybrid, M, maternal parent. Each of the DDRT fragments was amplified by PCR, electrophoresed on an agarose gel, and transferred to a nylon membrane. Triplicate membranes were prepared and each membrane was hybridized to ECL-labeled total RNA from Nongda3338 (M), Jingdong6 (P), and their hybrid (F1), respectively. Lanes 1–24 correspond to DE1–24, respectively
A BLASTX search in GenBank showed that 15 of the 24 transcripts (DE1 and 2, DE4–6, DE8–10, DE12 and 13, DE15, DE17, DE19, DE21 and 22) showed no homology to any known genes; two transcripts (DE3 and DE24) displayed similarity to genes involved in signal transduction. The predicted product of DE3 is similar to a CaM binding protein, while DE24 is similar to the EDR1 (enhanced disease resistance 1) protein. DE11 is similar to the transcription factor RAPB (Rice HAP B subunit), two transcripts (DE14 and DE23) code for proteins that show high similarity to α-glucan phosphorylase and NADH dehydrogenase (ubiquinone), respectively. One transcript (DE7) encodes a product with a high degree of similarity to NBS-LRR type resistance proteins; two transcripts (DE16, DE18) showed high similarity to retroelement sequences, and one (DE20) codes for a protein with high similarity to twitchin, a myosin-like contractile protein (Table 2).
Discussion
Differential gene expression between hybrids and their parents
The genetic architecture of the hybrid F1s studied is contributed by each of its inbred parents. No novel genes are expected to arise in the hybrid F1 that were not present in either parent. However, the hybrid phenotype is often quite different from that of either of its parents, i.e., the hybrid exhibits heterosis. It is reasonable to speculate that changes in genes expression are the primary determinant of heterosis. In this study, we used an improved differential display protocol (von der Kammer et al. 1999) to analyze changes in gene expression that occur in a hybrid relative to its parents in a common diallel cross between wheat inbreds. The differential expression patterns detected in this study fall into four categories, namely, BPnF1, UPnF1, F1nBP and UPF1, as described above. About 3.8% of the genes expressed at two stages were specifically expressed in the hybrid but not in either of parents, corresponding to overdominant expression or activation of genes in the hybrid. The UPF1 class comprises those genes expressed in the hybrid and in either of the parents, corresponding to dominant expression. Up to 11.4% of the total bands detected at the two stages fall into this category.
There has been much debate about the reproducibility of DDRT-PCR. However, it has been demonstrated that a considerable portion (>90%) of positive bands truly reveal differential expression, if the conditions for PCR are optimized and primers are carefully designed (von der Kammer et al. 1999). In this study, changes in gene expression between hybrid and parents were analyzed using the improved DD procedure described by von der Kammer (1999), in which longer primers and optimized conditions for PCR were used. With this procedure, a total of 1241 cDNA fragments were amplified from wheat roots, and 79% of the amplified bands were reproducible in duplicate PCRs (our unpublished data).
We do not yet know anything about the regulatory mechanisms that underlie these differential expression patterns. We speculate that epigenetic control, such as DNA methylation (Siegfried et al. 1999), paramutation (Hollick et al. 2000; Hollick and Chandler 2001), or even remodelling of chromatin structure (Hoekenga et al. 2000; Chandler et al. 2001), might be involved in the regulation. In fact, differential methylation in CpG or CNG islands has been demonstrated between hybrid and parents in rice and maize (Tsaftaris 1995; Xiong et al. 1999). Hollick et al (2000) noted that a paramutable allele, pl , shows overdominance in gene activity in heterozygotes, and proposed that allele-dependent mechanisms of gene regulation, such as paramutation, could contribute to heterosis. Uniparental silencing of genes in interspecific hybrids has observed for nucleolar dominance (Chen and Pikaard 1997; Chen et al. 1998; Frieman et al. 1999; Pikaard 2000), which may partially explain the UPnF1 pattern. Epigenetic controls have been reported to be responsible for changes in expression between allopolyploids and their parents (Comai 2000). However, which epigenetic mechanisms were involved in the differential expression patterns between hybrids and their parents are still an area that requires further investigation.
The expression of genes of diverse categories is altered in hybrids relative to their parents
In this study, we found that genes that are differentially expressed between hybrids and their parents include several that are involved in signal transduction, metabolism, disease resistance, transcriptional control, and retrotransposon activity. The predicted products of DE3 and DE24 show high similarity to a CaM binding protein and EDR1 (enhanced disease resistance 1) from rice, respectively. The plant CaM binding protein (CBP) is involved in Ca2+/CaM-mediated signaling pathways related to morphogenesis, cell division, cell elongation, ion transport, gene regulation, cytoskeletal organization, cytoplasmic streaming, pollen function, and stress tolerance (Reddy et al. 2002), and EDR1 forms part of a signaling transduction pathway activated by plant hormones (Frye et al. 2001). It would be premature to speculate how changes in signaling transduction in hybrids might affect heterosis, but alterations in signaling systems may be important for other modifications in patterns of gene expression in hybrids that affect heterosis in their turn. The product of DE11 shows high similarity to RAPB (rice HAP B subunit), a transcription factor which binds to CCAAT boxes present in many genes (such as genes regulated by light) to regulate their expression. Transcription factors can negatively regulate expression of some genes by binding to their CCAAT box, Therefore, silencing of DE11 in the hybrid may enhance the expression of genes related to photosynthesis and/or disease resistance, which may in turn contribute to heterosis. The DE14 and DE23 proteins are homologous to starch phosphorylase and NADH dehydrogenase, respectively. Starch phosphorylase is involved in starch metabolism, and the gene encoding starch phosphorylase is expressed at a higher level in hybrid than in either of its parents, indicating that the hybrid has a greater ability to convert the starch stored in leaves into sucrose for transport into grains, which may contribute to the heterosis for kernel weight. DE7 showed high similarity to NBS-LRR type resistance proteins, which may be related to the enhanced resistance of the hybrid to infection stress.
DE16 is present in the male parent and the F1, but absent in the female parent (UPF1), while DE18 was observed in both parents and in the hybrid F1, but the amount of expression in hybrid F1 was lower than that in either of its parents (HL). These two cDNAs showed high similarity to transcripts of retrotransposons. Retrotransposons are ubiquitous in plants and play a major role in plant gene and genome evolution (Kumar and Bennetzen 1999). Retrotransposons can generate mutations by inserting near or within genes, and these elements may provide regulatory sequences for gene expression and alter the expression of adjacent genes (Fedoroff 2000; Kashkush et al. 2003). These studies indicated that the changes in retrotransposon activity might cause alterations in the expression patterns of other genes. In this study, the change in the expression profiles of DE16 and DE18 in the hybrid may contribute to modifications in the expression of other genes in the hybrid.
In this study, the expression patterns of 24 sequenced cDNA fragments were confirmed in only one of the 20 hybrids, the highly heterotic hybrid Nongda3338 × Jingdong6 and its parents. The 20 hybrids showed considerable variation in both hybrid performance and heterosis in different agronomic traits. We have shown that fragments that occurred only in the F1 but in neither of the parents (F1nBP) can be positively correlated with heterosis for some agronomic traits, and fragments observed in both parents but not in the F1 (BPnF1) are negatively correlated with heterosis for some agronomic traits (our unpublished data). We expect that a similar heterosis-linked expression pattern of the very same genes will also be found in other heterotic F1 hybrids, but might not be found in F1 hybrids that show low levels of heterosis.
Polyploidy has played an important role in the evolution of higher plants, and 50–70% of all angiosperm species are of polyploid origin (Masterson 1994). During the last two decades, molecular data have provided new insights into the mechanism and evolutionary aspects of polyploidy (Wendel 2000). To establish themselves as successful species, the genomic structure and gene expression patterns of newly formed allopolyploids often change due to rearrangements in noncoding genomic DNA, through epigenetic changes and regulation of gene expression (Kashkush et al. 2002). Kashkush et al (2003) found that a variety of different genes (rRNA genes and genes involved in metabolism, disease resistance, and cell cycle regulation and retroelements) showed alteration in expression between a synthetic wheat allotetraploid F1 and its two diploid progenitors, and they also found that retrotransposons activated in the allotetraploid F1 alter the expression of adjacent genes (Kashkush et al. 2003). In this study, we also found that similar types of genes (such as genes involved in metabolism, disease resistance, and retroelements) are altered in expression in hybrids relative to their parents. These results indicate that similar genetic mechanisms may be responsible for both the evolution of polyploidy and heterosis.
References
Chandler VL, Vaucheret H (2001) Gene activation and gene silencing. Plant Physiol 125:145–148
Chen ZF, Pikaard CS (1997) Transcriptional analysis of nucleolar dominance in polyploid plants: biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proc Natl Acad Sci USA 94:3442–3447
Chen ZF, Comai L, Pikaard CS (1998) Gene dosage and stochastic effects determine the severity and direction of uniparental ribosomal RNA gene silencing (nucleolar dominance) in Arabidopsis allopolyploids. Proc Natl Acad Sci USA 95:14891–14896
Comai L (2000) Genetic and epigenetic interactions in allopolyploid plants. Plant Mol Biol 43:387–399
Duvick DN (1997) Heterosis: feeding people and protecting natural resources. In: Coors JG, Pandey S (eds) Genetics and exploitation of heterosis in crops. American Society of Agronomy, Madison, Wis. pp 19–29
Fedoroff N (2000) Transposons and genome evolution in plants. Proc Natl Acad Sci USA 97:7002–7007
Frieman M, Chen ZF, Saez-Vasquez J, Shen LA, Pikaard CS (1999) RNA polymerase I transcription in a Brassica interspecific hybrid and its progenitors: tests of transcription factor involvement in nucleolar dominance. Genetics 152:451–460
Frye CA, Tang D, Innes RW (2001) Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc Natl Acad Sci USA 1:373–378
Hoekenga O, Muszynski MG, Cone KC (2000) Developmental patterns of chromatin structure and DNA methylation responsible for epigenetic expression of a maize regulatory gene. Genetics 155:1889–1902
Hollick JB, Chandler VL (2001) Genetic factors required to maintain repression of a paramutagenic maize pl allele. Genetics 157:369–378
Hollick JB, Patterson GI, Asmundsson IM, Chandler VL (2000) Paramutation alters regulatory control of the maize pl locus. Genetics 154:1827–1838
Kashkush K, Feldman M, Levy AA (2002) Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160:1651–1659
Kashkush K, Feldman M, Levy AA (2003) Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nature Genet 33:102–106
Kumar A, Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33:479–532
Liang P, Pardee AB (1992) Differential display of eukaryotic messager RNA by means of the polymerase chain reaction. Science 257:967–971
Masterson J (1994) Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264:421–423
Ni Z, Q Sun, Z Liu, L Wu, X Wang (2000) Identification of a hybrid-specific expressed gene encoding novel RNA-binding protein in wheat seedling leaves using differential display of mRNA. Mol Gen Genet 263:934–938
Pikaard CS (2000) Nucleolar dominance: uniparental gene silencing on a multi-megabase scale in generic hybrids. Plant Mol Biol 43:163–177
Reddy ASN, Day IS, Narasimhulu SB, Safadi F, Reddy VS, Golovkin M, Harnly MJ (2002) Isolation and Characterization of a novel calmodulin-binding protein from potato. J Biol Chem 6:4206–4214
Siegfried Z, Eden S, Mendelsoln M, Feng X, Tsuberi B and Cedar H (1999) DNA methylation represses transcription in vivo. Nature Genet 22:203–206
Sun QX, Ni ZF, Liu ZY (1999) Differential gene expression between wheat hybrids and their parental inbreds in seedling leaves. Euphytica 106:117–123
Tsaftaris SA (1995) Molecular aspects of heterosis in plants. Physiologia Plantarum 94:362–370
Von der Kammer H, Albrecht C, Mayhaus M, Hoffmann B, Stanke G, Mnitsch R (1999) Identification of genes regulated by muscarinic acetylcholine receptors: application of an improved and statistically comprehensive mRNA differential display technique. Nucleic Acids Res 27:2211–2218
Wendel JF (2000) Genome evolution in polyploids. Plant Mol Biol 42:225–249
Wu LM, Ni ZF, Wang ZK, Lin Z, Sun QX (2001) Relationship between differential gene expression patterns of multigene families and heterosis in a wheat diallel crosses. Acta Genetica Sinica 28:256–266
Xiong LZ, Yang GP, Xu CG, Zhang QF, Saghai Maroof MA (1998) Relationships of differential gene expression in leaves with heterosis and heterozygosity in a rice diallel cross. Mol Breeding 4:129–136
Xiong LZ, Xu CG, Saghai Maroof MA, Zhang Qifa (1999) Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol Gen Genet 261:439–446
Acknowledgements
This work was financially supported by the State Key Basic Research and Development Plan of China (2001CB1088), the National Science Fund for Distinguished Young Scholars (39925026) and National Natural Science Foundation of China (30270824)
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Hagemann
Rights and permissions
About this article
Cite this article
Wu, L.M., Ni, Z.F., Meng, F.R. et al. Cloning and characterization of leaf cDNAs that are differentially expressed between wheat hybrids and their parents. Mol Genet Genomics 270, 281–286 (2003). https://doi.org/10.1007/s00438-003-0919-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-003-0919-y