Abstract
Homologous recombination results in the exchange and rearrangement of DNA, and thus generates genetic variation in living organisms. RecA is known to function in all bacteria as the central enzyme catalyzing strand transfer and has functional homologues in eukaryotes. Most of our knowledge of homologous recombination in eukaryotes is limited to processes in the nucleus. The mitochondrial genomes of higher plants contain repeated sequences that are known to undergo frequent rearrangements and recombination events. However, very little is known about the proteins involved or the biochemical mechanisms of DNA recombination in plant mitochondria. We provide here the first report of an Arabidopsis thaliana homologue of Escherichia coli RecA that is targeted to mitochondria. The mtrecA gene has a putative mitochondrial presequence identified from the A. thaliana genome database. This nuclear gene encodes a predicted product that shows highest sequence homology to chloroplast RecA and RecA proteins from proteobacteria. When fused to the GFP coding sequence, the predicted presequence was able to target the fusion protein to isolated mitochondria but not to chloroplasts. The mitochondrion-specific localization of the mtrecA gene product was confirmed by Western analysis using polyclonal antibodies raised against a synthetic peptide from a unique region of the mature mtRecA. The Arabidopsis mtrecA gene partially complemented a recA deletion in E. coli, enhancing survival after exposure to DNA-damaging agents. These results suggest a possible role for mtrecA in homologous recombination and/or repair in Arabidopsis mitochondria.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Homologous DNA recombination is a universal process that occurs in nearly all organisms. In this process two homologous DNA duplexes interact, resulting in an exchange of genetic information and rearrangement of DNA segments. In bacteria, the RecA enzyme catalyzes exchange between two homologous DNA strands. RecA is involved in recombination repair and is capable of catalyzing homologous DNA pairing at double strand breaks. It also serves as a coprotease in Escherichia coli to activate self-digestion of LexA during the SOS response and self-digestion of the lambda repressor during prophage induction (Cox 2000). Accessory proteins are involved in each of these processes.
RecA homologues have been identified in all bacterial species, as well as in many eukaryotes, including plants (Angulo et al. 1985; Ogawa et al. 1993; Sato et al. 1995). In eukaryotes many of the RecA homologues, including the Rad51/Dmc1 group of proteins, have been implicated in meiotic recombination, while some have been associated with DNA repair (Hong et al. 2001). However, almost all current information on homologous recombination in eukaryotes and its associated proteins relates to the process in the nucleus. There is very little information on homologous recombination or related activity in organelles.
Mitochondrial DNA (mtDNA) molecules undergoing recombination have been observed in yeast (Sena et al. 1986). Several proteins involved in recombination and repair have been identified in yeast mitochondria (Foury and Lahaye 1987; Manna et al. 1991; MacAlpine et al. 1998) and a link between recombination junctions and segregation in yeast has been proposed (Lockshon et al. 1995). Studies on a yeast nuclear mutation that abolishes mtDNA recombination show that recombination is required for the stable inheritance of yeast mtDNA (Ling et al. 1995). In humans, sequence rearrangements, deletions and single-base mutations in mtDNA have been associated with several diseases that affect specific tissues or organs and various aging disorders (Larsson and Clayton 1995). A recent report demonstrates a high rate of DNA recombination activity in heart muscle mitochondria (Kajander et al. 2001). Bacterial-like homologous DNA recombination activity in human mitochondria has also been reported (Thyagarajan et al. 1996).
In plants, an Arabidopsis thaliana gene that encodes a Mg2+- and ATP-dependent RecA homologue targeted to chloroplasts has been characterized (Cerutti et al. 1992; Cao et al. 1997). This DNA-damage-responsive protein shares high homology with bacterial RecA and cross-reacts with antibodies raised against E. coli RecA (Cerutti et al. 1992).
In plant mitochondria, homologous recombination appears to be a routine process involved in generating genetic permutations. An example of active mtDNA recombination involves the atp6 gene in Arabidopsis, which is present in two copies that code for preproteins whose presequences differ due to recombination at the junction with the sequence encoding the mature protein (Marienfeld et al. 1996). It was therefore suggested that the atp6 gene might contain sequences that are preferentially involved in homologous recombination. In addition, both large (6.5 and 4.2 kb) direct repeats and numerous smaller (30–560 bp, totaling 144 in number) repeats are present in A. thaliana mtDNA (Unseld et al. 1997). There is some evidence to suggest that recombination across the large repeats occurs often with no loss of genetic information resulting from most rearrangements (Klein et al. 1994), while the small repeats may not be active in frequent recombination (Unseld et al. 1997).
Some alterations in plant mitochondrial genomes result in cytoplasmic male sterility (CMS), a commercially exploited phenotype characterized by the production of infertile pollen (Conley and Hanson 1995; Gutierres et al. 1999). Not all mtDNA rearrangements result in CMS or other obvious phenotypes, and may be due to rearrangements that occur in the abundant non-coding regions (Geiss et al. 1994; Marienfeld et al. 1997; Small and McAlister-Henn 1997). Recombination may also result in selective gene amplification, which has been reported in plant mitochondria (Conley and Hanson 1995; Muise and Hauswirth 1995).
Although a substantial amount of genetic and physical evidence for recombination in plant mitochondria exists, the biochemical basis for this activity has yet to be demonstrated. Given the observed rearrangements in plant mitochondrial genomes and the evidence for recombination activity in mammalian and yeast mitochondria, it is likely that plant mitochondria also contain a recombination activity. Here we provide the first report of a mitochondrion-targeted RecA homologue in plants. We show that this protein, which is homologous to chloroplast and E. coli RecA proteins, is specifically localized to mitochondria and partially complements the effects of DNA-damaging agents on an E. coli ΔrecA strain.
Materials and methods
Bacterial strains, growth conditions and plasmids
The XL1-Blue (Stratagene), DH5α (Life Technologies) and JM109 (Promega) strains of E. coli were used as hosts for routine cloning and isolation of ssDNA. The BL21 (DE3) strain [E. coli B F-, ompT , hsdS B (rB - mB -), gal dcm , (DE3)] was obtained from Novagen. A BLR(DE3) strain [E. coli B F-, ompT , hsdS B (rB - mB -), gal dcm , Δ(srl- recA)306::Tn10 (tetR) (DE3)} was used for overexpression of mtRecA and complementation analysis. The pCRT7 TOPO TA expression system was purchased from Invitrogen. The pGFPuv expression vector was obtained from Clontech. M13mp18 ssDNA and replicative form DNA were purchased from New England Biolabs. Bacterial cultures were grown in LB medium at 37°C unless indicated otherwise.
Sequence and phylogenetic analyses
The putative mtrecA gene was identified by a BLAST search of the Arabidopsis Gene Index in the TIGR (The Institute for Genomic Research, Rockville, Md.) database. Analysis of homologous sequences retrieved by the BLAST search was performed using the AlignX program from the Vector NTI suite (Informax), PHYLIP and CLUSTALW. Potential targeting presequences were predicted using the PSORT (Nakai and Horton 1999; Nakai 2000), TargetP (Emanuelsson et al. 2000), Predotar (http://genoplante-info.infobiogen.fr/predotar/index.html), MitoP (Scharfe et al. 1999; 2000) and ChloroP (Emanuelsson et al. 1999) protein signal prediction programs. The EST clone (Accession No. AV525538) was obtained from the Kazusa DNA Research Institute (Chiba, Japan). The sequence of the full-length cDNA corresponding to mtRecA was confirmed, annotated and submitted to the GenBank database (under Accession No. AY072877).
Bacterial expression of GFP with an N-terminal mitochondrial presequence fusion
The 81-bp region encoding the predicted N-terminal presequence (27 residues) was amplified by PCR using the following primers (restriction sites are underlined): 5´-AGCTTGCATGCCTGCAGGATGGGTCGACTCTCATGGGCCAGTCCG-3´ (forward; 5´ Pst I site) and 5´-TCGGGGTACCCCAACGCTTCTTCTTCCATTCAGCTG-3´ (reverse; 3´-KpnI site). This fragment was directionally cloned into pGFPuv to produce a sequence encoding a 27-amino acid N-terminal presequence-fused to GFP (pPS-GFP), and the DNA sequence of the construct was confirmed. XL1-Blue host cells were transformed with pPS-GFP and were cultured in LB medium with 50 μg/ml ampicillin for 16 h at 37°C. The cells were centrifuged at 6000× g, and the resulting pellet was resuspended in 30 ml of resuspension buffer (20 mM TRIS-HCl pH 7.5, 150 mM NaCl, 5 mM EDTA). Cells were disrupted by sonication in an ice-bath for 20 min (4-s pulses at 20-s intervals) to obtain the crude protein extract. Intact cells and insoluble material were removed by centrifugation at 4°C for 30 min at 15,000× g. The fluorescent supernatant containing the fusion protein was enriched by precipitation with ammonium chloride followed by organic extraction (Yakhnin et al. 1998).
In vitro import of the PS-GFP fusion protein into isolated mitochondria
Mitochondria were isolated from 100 g of young soybean (Glycine max cv. Williams '82) leaves by fractionation on discontinuous Percoll gradients as described below, and the quality of the mitochondria was determined by a cytochrome c reduction assay (Hrubec et al. 1985). Intact mitochondria (200 μg protein) were incubated with 100 μl of PS-GFP extract (1 mg/ml protein) for 30 min at 25°C in a final volume of 300 μl of import buffer (0.25 M mannitol, 50 mM KCl, 10 mM MOPS pH 7.4, 2 mM ATP, 2 mM NADH, 1 mM MnCl2, 5 mM K3PO4 and 1% BSA). The sample was then divided into 100-μl aliquots and placed on ice. Two aliquots were incubated with 50 μg/ml proteinase K for 20 min at 25°C and for 15 min on ice. The third aliquot was not treated with proteinase K. Following incubation the mitochondrial suspensions were washed twice with import buffer and re-isolated on a Percoll gradient. The residual Percoll was removed by washing the mitochondria twice in import buffer, and the pellet was diluted in 300 μl of import buffer.
As a control, chloroplasts were isolated from 50 g of soybean leaves on a continuous Percoll gradient as described by Perry et al. (1991). Intact chloroplasts were incubated with 100 μl of PS-GFP extract or control GFP extract lacking the presequence for 30 min at 25°C in a final volume of 300 μl of import buffer with occasional mixing. Samples were then treated with proteinase K for 20 min at 25°C and for 15 min on ice. Chloroplasts were reisolated on a continuous Percoll gradient as above, washed twice with import buffer, and resuspended in 100 μl of import buffer. Fluorescence and differential interference contrast (DIC) microscopy were used to visualize samples. Standard fluorescein/rhodamine filter sets were used to scan the samples and to differentiate chloroplasts from mitochondria. At this excitation wavelength, chloroplasts emit red autofluorescence.
Generation of polyclonal antibodies specific for mtRecA
We scanned the sequence of mtRecA for a region that is unique relative to RecA homologues from other organisms and could serve as an antigen to generate antibodies specific for this protein. While the regions corresponding to functional domains of the RecA homologues are highly conserved, the N-terminal region between the predicted mitochondrial targeting presequence and the protein-protein interaction domain of Arabidopsis mtRecA is unique. Moreover, this region shares no homology with ctRecA, or with any other predicted proteins from the complete Arabidopsis genome sequence. A 16-amino acid peptide (CELDEVPDDRKVAEKD) from this region was synthesized commercially. Two rabbits were immunized with this peptide and serum was recovered after 8 weeks (BioCarta, Carlsbad, Calif.) and used for Western analysis of proteins obtained from purified plant organelles.
Isolation of mitochondria and chloroplasts by centrifugation on Percoll gradients
Leaves from 3-week-old A. thaliana (cv. Columbia) seedlings were used for isolation of organelle proteins to determine the localization of the mtRecA protein. Plant tissue was homogenized in cold Xpl buffer (0.33 M sorbitol, 0.05 M HEPES pH 7.5, 2 mM EDTA, 1 mM MgCl2, 5 mM sodium ascorbate, and 0.25% w/v BSA; Weigel and Glazebrook 2002) in a blender containing a single double-edged razor blade. Three 5-s bursts were used to homogenize the material. The homogenate was filtered through three layers of Miracloth (Calbiochem), and the chloroplasts pelleted by centrifugation for 8 min at 3500 rpm. The resulting supernatant was transferred to clean centrifuge bottles for the recovery of mitochondria (see below). The chloroplast pellet was gently resuspended in a minimal volume of Xpl buffer, and layered onto a discontinuous 40%/80% Percoll gradient (Weigel and Glazebrook 2002). The gradient was centrifuged in a swinging bucket rotor at 7500 rpm for 8 min with the brake off. Intact chloroplasts were recovered from the 40/80% boundary. The chloroplast fraction was diluted in 20 ml of Xpl buffer and centrifuged in a swinging bucket rotor for 5 min at 3500 rpm to remove the Percoll. The final chloroplast pellet was resuspended in minimal volume of TE buffer and lysed by adding an equal volume of 2× SDS-PAGE loading buffer and heating at 95°C for 5 min. Proteins were fractionated by SDS-PAGE and transferred to a PVDF membrane for Western analysis as described below.
The supernatant from the first low-speed centrifugation step contains the mitochondrial fraction. This was centrifuged at 13,000 rpm for 10 min to pellet mitochondria. The pellet was resuspended in a minimal volume of Mito buffer (0.35 M mannitol, 0.03 M MOPS pH 7.3, 1 mM EDTA, 0.6% w/v polyvinylpyrrolidone, and 0.2% w/v BSA), diluted in the same buffer, and pelleted again (Weigel and Glazebrook 2002). This pellet was resuspended in Mito buffer and carefully layered onto a discontinuous 21%/27%/45%/60% Percoll gradient. The gradient was centrifuged in a swinging bucket rotor at 10,500 rpm for 30 min with the brake off. Fractions (1 ml) from the bottom third of the gradient, avoiding any pellet at the bottom, were pooled and centrifuged at 13,000 rpm for 10 min to recover mitochondria. The mitochondria were washed in Mito buffer to remove Percoll as described above, resuspended in TE buffer and lysed by the addition of an equal volume of 2× SDS-PAGE loading buffer and heating at 95°C for 5 min. Samples were fractionated by SDS-PAGE and transferred to a PVDF membrane for Western analysis as follows.
Western analysis of purified chloroplast and mitochondrial proteins
Duplicate gels were prepared, one for staining and one for transfer of proteins to membranes for Western analysis. Both unstained and prestained molecular weight markers were included for size determinations. Electrophoresis was carried out for 90 min at 125 V. One gel was stained in Coomassie Blue for 1 h, followed by destaining to visualize proteins. Proteins from the other gel were transferred to a PVDF membrane using a semi-dry electroblotter (BioRad).
Membranes were blocked in TBS (50 mM TRIS-HCl pH 7.4, 200 mM NaCl) containing 5% nonfat dry milk for 1 h at room temperature. The antiserum against mtRecA was diluted 1:2500 in 20 ml of blocking buffer, and the membrane incubated in this solution with gentle shaking overnight at 4°C. Polyclonal antibodies raised against chloroplast a/b binding protein (Rose Biotechnology, 1:2500 dilution) and monoclonal antibodies against mitochondrial porin (kindly provided by Dr. Tom Elthon, 1:100 dilution) were used to monitor the purity of the chloroplast and mitochondrial protein preparations. After incubation with antibody, membranes were washed three times (for 15 min each) with TBS, the second wash containing 0.1% Tween 20. Membranes were blocked again as above, followed by incubation for 1 h at room temperature with a 1:40,000 dilution of goat anti-rabbit antibody conjugated with horseradish peroxidase (HRP; Promega). The membrane was washed three times as above, followed by incubation for 5 min in SuperSignal West Pico Substrate working solution (Pierce). The membrane was wrapped in plastic wrap and exposed to X-ray film for 5 s–2 min.
Expression of the mtRecA protein in E. coli
The region encoding the mature mtRecA protein (1089 bp) was amplified by PCR and cloned into the pCRT7 TOPOTA vector (Invitrogen). The orientation of the gene was confirmed by sequencing. The resulting construct was called pT7mtrecA. E. coli BLR (DE3) was used as the host for over-expression of the recombinant protein. Over-expression was carried out by growth of the cells in Superbroth as indicated by the manufacturer (BD Biosciences). The culture was induced at 0.6 A600 with 1 mM IPTG and grown overnight at 37°C.
Immunodetection of over-expressed mtRecA
Equal amounts of protein, adjusted according to the optical density of the overnight cultures, were fractionated on a 4–15% TRIS-HCl polyacrylamide gel (BioRad). Proteins were transferred to PVDF membrane as above, and the membranes were blocked overnight with 5% (w/v) non-fat dry milk in TBS at 4°C and incubated for 2 h with polyclonal antibodies raised against chloroplast RecA in rabbits, followed by incubation with HRP-conjugated anti-rabbit antibody. Cross-reacting proteins were visualized by color development using an HRP-conjugate substrate (BioRad).
Measurement of bacterial resistance to DNA-damaging agents
Cells were grown overnight in LB medium with appropriate antibiotics. Fresh LB (10 ml) was inoculated with 50 μl of overnight culture and grown at 37°C for 4 h. Isopropyl β-D-thiogalactopyranoside (IPTG, 0.1 mM) was added and growth was allowed to continue for 30 min. The bacteria were then harvested and resuspended in 10 mM MgSO4 to a density of 3×108 colony-forming units (cfu) per ml. Cell suspensions were treated with different concentrations of the DNA-damaging agents methyl methanesulfonate (MMS) and mitomycin C according to Pang et al. (1992). Appropriate dilutions of treated cells were spread on LB plates in triplicate and incubated overnight at 37°C. The colony forming units were counted and the surviving fraction was calculated using the method described by Pang et al. (1992). The values presented are the means from three independent experiments.
Results
Identification of a putative Arabidopsis RecA homologue with a mitochondrial targeting presequence
A putative recA gene homologue that encodes a 394-residue (43.64 kDa) preprotein with a predicted 27-residue mitochondrial presequence was identified (identified as Atm in Fig. 1) by a BLAST search of the Arabidopsis nuclear genome database (DNA Accession No. AY072877, protein Accession No. AAF04410). The search yielded a single RecA homologue (hereafter referred to as mtrecA for the gene and mtRecA for the protein) that was predicted with high probability to be targeted to mitochondria by the PSORT (probability 0.705), Predotar (0.998), TargetP (0.861) and MITOP (0.9392) prediction programs. While the ChloroP program gave a probability of 0.491 for targeting of this gene product to chloroplasts, this program identified no targeting presequence. The other prediction programs yielded probabilities of 0.047 or less for import of mtRecA into chloroplasts, with no predicted targeting presequences. A total of four recA homologues were found in the Arabidopsis genome database, but the mtrecA gene was the only one with a clear prediction for a mitochondrial targeting sequence. The second homologue has a predicted chloroplast targeting sequence and has already been shown to be imported into chloroplasts (Cao et al. 1997; identified as Atc in Fig. 1), but the MitoP program gave a prediction of 0.700 for targeting to mitochondria with a putative 46 amino acid targeting presequence. The work presented by Cao et al. (1997) does not include experiments designed to determine whether this protein is imported into mitochondria, so it is possible that this protein could be targeted to both organelles (see Discussion). The other two rec A homologues have mixed, low or no organelle targeting predictions and lack clear targeting sequences (identified as At3 and At4 in Fig. 1), so it is not clear whether these proteins might be targeted to an organelle, or whether they remain in the cytoplasm. The At3 sequence, in addition to lacking a predicted targeting sequence, contains a number of additional amino acids at the C-terminal end, suggesting that this protein may be functionally distinct. The At4 sequence is much shorter than the others, and lacks a considerable number of amino acids at the N-terminal end and near the C-terminal end, particularly in regions involved in DNA and nucleotide binding.
Alignment of RecA protein sequences from Arabidopsis thaliana with RecA of bacterial origin. Atm, predicted A. thaliana RecA homologue targeted to mitochondria. Atc, A. thaliana RecA targeted to chloroplasts (Cao et al. 1997). At3, a third A. thaliana RecA homologue lacking a predicted targeting signal. At4, a fourth A. thaliana RecA homologue lacking predicted targeting signal. Syn, Synechocystis sp. PCC 6803 RecA; Msm, Mycobacterium smegmatis RecA; Eco, Escherichia coli RecA; Rpr, Rickettsia prowazekii RecA; Rpa, Rhodopseudomonas palustris RecA. Sequences at the N-terminal end of Atm and Atc highlighted in red correspond to the predicted targeting sequences for mitochondria and chloroplasts, respectively. The 16-amino acid sequence used for production of mtRecA-specific polyclonal antibodies is highlighted in green. Sequence identities are highlighted in yellow, and conservative changes are shaded gray. Conserved functional domains of RecA are underlined and labeled
Thorough analysis of upstream sequences beyond those shown in Fig. 1 identified no additional in-frame methionine codons that do not have intervening in-frame stop codons (data not shown), suggesting that the sequences shown are likely to be those of the actual protein products. A full-length cDNA clone of the mtrecA gene was obtained and the DNA sequence was confirmed. Northern analysis using the EST clone insert as probe at high stringency confirmed that this gene is transcribed in Arabidopsis plants (data not shown), as expected given the origin of the clone. The mtrecA gene shows a very different organization to the ctrecA gene with respect to the number, size and location of exons and introns (Fig. 2A). Comparisons of the predicted Arabidopsis mtRecA sequence with RecA sequences from several different organisms indicated that it is most closely related to the Arabidopsis RecA targeted to chloroplasts and to members of the alpha-Proteobacteria (Fig. 2B).
Organization of Arabidopsis genes for mitochondrial (M) and chloroplast (C) targeted rec A genes ( A), and phylogenetic analysis of RecA homologues ( B). A The angled arrows on the left align the genes at the predicted transcription start sites. The filled boxes represent exons, and the lines between boxes represent introns and non-coding flanking sequences. B A sequence alignment was constructed with RecA sequences from the organisms listed. Phylogenetic analysis was performed and bootstrap values (1000 replicates each) are indicated, with values based on parsimony above each line, and values based on distance analysis shown below each line. MtRecA showed highest homology to ctRecA and RecA from Synechocystis, a cyanobacterium, followed by RecA proteins from proteobacteria (alpha subdivision)
The mtRecA presequence targets GFP into isolated mitochondria
The selectively amplified mitochondrial presequence region of Arabidopsis mtrecA (81 bases) encoding the predicted 27-amino acid targeting peptide sequence was cloned into a GFP fusion vector to produce a presequence-GFP fusion protein, PS-GFP. In vitro import studies with purified soybean mitochondria showed specific import of PS-GFP into mitochondria (Fig. 3E), and not into chloroplasts. A control experiment with the same protein and purified chloroplasts confirmed that the fusion is not imported into isolated chloroplasts (Fig. 3C). The red autofluorescence results from the chlorophyll in the chloroplasts and did not co-localize with GFP fluorescence. No fluorescence was detected in mitochondria incubated with GFP lacking a presequence (Fig. 3A), demonstrating that a targeting sequence is necessary for the localization of GFP to mitochondria. Samples that had not been exposed to proteinase K treatment or reisolation on Percoll gradients showed a considerable background fluorescence that did not colocalize with organelles (data not shown), indicating that digestion with proteinase K and reisolation on Percoll gradients was sufficient to remove GFP not localized within mitochondria. DIC images of the same fields were used to correlate localized fluorescence with mitochondria (Fig. 3B, D and F).
Import of mtRecA into isolated mitochondria. Intact soybean mitochondria and chloroplasts isolated by Percoll gradient centrifugation were incubated with a recombinant PS-GFP fusion protein, followed by two washes with incubation buffer and treatment with proteinase K. Mitochondria (panels A and E) and chloroplasts (panel C) were observed by fluorescence microscopy (Nikon E600) to detect GFP. Each field was analyzed by differential interference contrast (DIC) microscopy to confirm the location of individual mitochondria and chloroplasts (panels B, D and F). The bars represent 10 μm
Western analysis confirms the mitochondrial localization of mtRecA
The polyclonal antibody prepared against a synthetic 16-amino acid peptide representing a unique region near the N-terminal end of the mature mtRecA protein recognized a single specific polypeptide of the expected size in total protein extracts from purified Arabidopsis mitochondria, and did not cross-react with the purified chloroplast sample (Fig. 4). Preimmune serum did not react with any of the proteins on the membrane (not shown). As a control for the purity of the organelle fractions and the specificity of the antibody, an antibody against chloroplast a/b binding protein was tested and gave a signal only with the chloroplast protein sample and not with mitochondria (Fig. 4). As a positive control for mitochondria, antibody against mitochondrial outer membrane porin was tested and gave a signal only for the mitochondrial sample and not the chloroplast sample (Fig. 4). The antiserum raised against the synthetic fragment of mtRecA showed no cross-reaction with purified E. coli RecA protein, a wild-type E. coli strain expressing native RecA, or the ΔrecA strain of E. coli used for overexpression of mtRecA, but did react strongly with a protein of the expected size in a total protein extract of the E. coli strain overexpressing mtRecA (data not shown).
Localization of mtRecA in mitochondria by Western analysis. Total chloroplast (lanes marked C) and mitochondrial (lanes marked M) proteins were fractionated by SDS-PAGE, transferred to PVDF membrane, and analyzed using monoclonal antibodies raised against mitochondrial porin (porin, 29.2 kDa), or polyclonal antibodies against the chlorophyll a/b binding protein (chl a/b, 25.2 kDa) or against the 16-amino acid sequence from the N-terminal end of the mtRecA protein (mtRecA, 39.7 kDa). Lane L, HRP-conjugated molecular weight markers (Invitrogen), with sizes indicated on the left
Overexpression and analysis of the mitochondrially targeted Arabidopsis RecA protein
Histidine-tagged mtRecA protein from E. coli was batch purified on a Ni2+-NTA agarose column and analyzed by SDS-PAGE and Western blotting. The majority of overexpressed mature mtRecA protein was found in the soluble protein fraction (Fig. 5). Antibodies against the chloroplast RecA protein (kindly provided by A. Jagendorf, Cornell Univ.) were found to cross-react with the over-expressed mtRecA protein (Fig. 5) as expected. The 6×His-tag added ~3.5 kDa to the molecular weight of the protein.
Western analysis of over-expressed mtRecA protein. Total soluble and insoluble proteins from cells over-expressing pT7mtrecA were analyzed using polyclonal antibodies against ctRecA. Lane 1, total proteins from the host strain (BLR) control; lane 2, purified E. coli RecA (38 kDa); lane 3, total proteins from an uninduced culture; lane 4, total soluble proteins from an induced culture; lane 5, total insoluble proteins from an induced culture. The arrow indicates the expected size of the 6×His-tagged mtRecA protein. The locations of size markers are indicated along the left side of the panel
Partial complementation of an E. coli ΔrecA strain by mtRecA
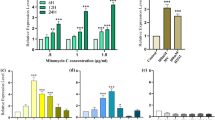
Arabidopsis mtrecA, when expressed in E. coli under the control of an inducible T7 promoter in a high-copy number plasmid, conferred resistance to the DNA-alkylating agent methylmethanesulfonate (MMS), and the DNA-crosslinking agent mitomycin C (Fig. 6A and B, respectively). Comparison of surviving fractions of the Δ recA strain (BLR) with the same strain bearing the mtrecA plasmid showed that induction of mtrecA expression with IPTG provided up to a 188-fold increase in resistance to MMS and an almost 230-fold enhancement of resistance to DNA damage by mitomycin C. Derepression by IPTG was carried out at one-tenth the concentration used for over-expression to avoid the formation of inclusion bodies. Plots of IPTG levels against the survival rates of the treated cultures showed that 0.1 mM IPTG produced survival rates comparable to those of the uninduced control rec A+ strain. The levels of mtRecA expressed in the presence of 0.1 mM IPTG were analyzed by SDS-PAGE and observed to be comparable to the levels in the wild-type strain (not shown).
Partial complementation of E. coli ΔrecA by the mtRecA protein. The E. coli strains C600 ( recA +; filled circles) and BLR (ΔrecA, open circles) were the control strains used in the experiment. BLR ΔrecA was transformed with either pT7mtrecA (open and filled squares) or with the plasmid pET5a (filled triangles). The pT7mtrecA-containing BLR culture was either grown in the absence (open squares), or presence (filled squares) of 0.1 mM IPTG. The surviving fraction was calculated by dividing cfu in treated resuspensions by cfu in untreated suspensions (Pang et al. 1992). A Cultures treated with MMS (methyl methanesulfonate). B Cultures treated with mitomycin C
Discussion
An Arabidopsis homologue of the E. coli rec A gene encoding a mitochondrial target peptide has been identified. Protein sequence alignment indicated 45% similarity and 36% identity between mtRecA and chloroplast RecA (ctRecA) and 47–52% similarity and 32–36% identity with selected alpha-proteobacterial RecA proteins. The majority of RecA signature motifs were found to be conserved in mtRecA, as in the case of ctRecA, while some of these regions are truncated or missing in the third and fourth Arabidopsis RecA homologues, which also appear to lack organelle targeting presequences (Fig. 1). This suggests conservation of functional domains and the possibility of conserved biochemical activity of the mtRecA protein in Arabidopsis.
The 27-residue N-terminal presequence is predicted to provide specific targeting into mitochondria. Our results using isolated mitochondria and chloroplasts for in vitro import assays with a protein comprising this presequence fused to GFP confirmed that the mitochondrial presequence specifically targets the protein to the mitochondria and not to chloroplasts (Fig. 3). There are reports of co-targeting of some nucleus-encoded proteins to both chloroplasts and mitochondria (Chow et al. 1997; Silva-Filho et al. 1997; Akashi et al. 1998; Small et al. 1998; Kobayashi et al. 2001; Watanabe et al. 2001; Hedtke et al. 2002); however, in some of these cases this dual targeting is due to the use of alternate methionine start codons, which yields presequences with different targeting capabilities (Small et al. 1998; Kobayashi et al. 2001; Watanabe et al. 2001). Analysis of the sequence upstream of the mtrecA gene identified no in-frame methionine codons without intervening stop codons, suggesting that the first methionine codon shown in Fig. 1 is the start codon for our protein. To address the possibility of dual targeting or artifacts due to the properties of the fusion constructs, an antibody against a unique 16-amino acid segment of mtRecA was used for Western analysis of proteins from mitochondria and chloroplasts purified on Percoll gradients. The results support the finding that this protein is specifically localized in mitochondria and not in chloroplasts (Fig. 4).
A RecA homologue targeted to chloroplasts has been identified previously, and a role in homologous recombination/repair activity has been suggested for it (Cerutti et al. 1992). This protein was shown to be imported into and processed by isolated chloroplasts (Cao et al. 1997), but no experiments showing that this protein is not imported into mitochondria were included in this study, so it is possible that this protein could be dual targeted. The other two Arabidopsis rec A homologues (Fig. 1) may also be targeted to mitochondria and/or chloroplasts, as the prediction programs are unable to identify any consensus sequence for targeting. Import experiments will need to be done to clarify these questions. The import data from our experiments indicate that mtRecA is imported into mitochondria and not into chloroplasts, but additional experiments will be required to determine if this is the only RecA homologue imported into Arabidopsis mitochondria.
Western analysis using polyclonal antibodies against chloroplast RecA indicated that Arabidopsis mtRecA shares antigenic determinants with chloroplast RecA (Fig. 5). This observation is consistent with the phylogenetic analysis of RecA protein sequences from several different groups of organisms, which indicate that mtRecA is more closely related to ctRecA. The antibodies raised against the unique 16-amino acid region of mtRecA did not cross-react with the chloroplast protein fraction (Fig. 4) or with purified E. coli RecA (not shown), indicating that this part of mtRecA is indeed unique. Furthermore, analysis of the coding sequences of the genes for the chloroplast-targeted and mitochondria-targeted RecAs indicate significant differences in the number, locations and sizes of exons and introns (Fig. 2A), suggesting that these genes are not the result of a recent gene duplication with subsequent development of targeting sequences specific for each organelle.
When a bacterial host strain deleted for its endogenous recA gene was transformed with a DNA construct containing the mature mtrecA coding sequence downstream of a T7 promoter, the cells were able to withstand DNA damage significantly better than the strains which did not harbor mtrecA. Recombinant strains that were induced with 0.1 mM IPTG had a higher survival rate than the uninduced strains. The level of survival in uninduced strains carrying the mtrecA construct can be attributed to basal-level expression of the mtrecA gene from the lac promoter, which appears to have allowed complementation of the deletion at a level about two orders of magnitude less than that seen with the induced sample. Based on the above observations we conclude that mtRecA is a functional homologue of E. coli RecA that is targeted to the mitochondria in Arabidopsis.
In considering a possible role for mtrecA in plant mitochondria, further analysis of the Arabidopsis genome database was carried out to identify homologues of the RecBCD helicase/nuclease proteins that are involved in early events of recombination in E. coli (Kowalczykowski 2000). However, no obvious homologues were found. The recBCD genes are absent in many bacteria, including those considered to be most closely related to the ancestors of mitochondria (Andersson et al. 1998).
In bacteria a second helicase, identified as RecQ, functions in some recombination pathways that do not involve RecBCD. The RecQ protein is present in many species and is actively involved in recombinational DNA repair (Wu and Hickson 2001). In E. coli, RecQ works in conjunction with RecA and SSB, and in this combination RecQ has been shown to initiate recombination events in vitro (Harmon and Kowalczykowski 1998). In eukaryotes, RecQ appears to be involved in the maintenance of nuclear genome stability, and possibly functions as an S-phase replication checkpoint (Frei and Gasser 2000). RecQ homologues exist in yeast and in humans, where at least five genes have been identified, and three of these human genes have been associated with inherited diseases. Nine RecQ helicase homologues have been identified in Arabidopsis (Hartung et al. 2000; our own more recent analysis). Computer analysis with the protein signal prediction programs did not identify any of these genes as having a high probability for mitochondrial targeting, and from what is known about RecQ homologues in yeast and humans, at least some of these may be involved in nuclear genome stability. However, targeting of each will need to be tested to determine the functional location of each of these gene products.
We have also identified two distinct putative ssb genes in the Arabidopsis genome database with high probabilities for mitochondrial targeting. SSB is known to stimulate RecA-dependent recombination activity (Harmon and Kowalczykowski 1998). We have preliminary data that shows that one of these is indeed targeted specifically to mitochondria (Edmondson and Nielsen, unpublished data). The presence of a mitochondrially targeted RecA as presented here, together with the putative mitochondrially targeted SSB and the possibility that RecQ and other associated proteins may also be imported into mitochondria, suggests that recombinational repair may be active in Arabidopsis mitochondria. Studies with the Arabidopsis ssb and rec Q genes are currently in progress.
References
Akashi K, Grandjean O, Small I (1998) Potential dual targeting of an Arabidopsis archaebacterial-like histidyl-tRNA synthetase to mitochondria and chloroplasts. FEBS Lett 431:39–44
Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UC, Podowski RM, Naslund AK, Eriksson AS, Winkler HH, Kurland CG (1998) The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133–140
Angulo JF, Schwencke J, Moreau PL, Moustacchi E, Devoret R (1985) A yeast protein analogous to Escherichia coli RecA protein whose cellular level is enhanced after UV irradiation. Mol Gen Genet 201:20–24
Cao J, Combs C, Jagendorf AT (1997) The chloroplast-located homolog of bacterial DNA recombinase. Plant Cell Physiol 38:1319–1325
Cerutti H, Osman M, Grandoni P, Jagendorf AT (1992) A homolog of Escherichia coli RecA protein in plastids of higher plants. Proc Natl Acad Sci USA 89:8068–8072
Chow KS, Singh DP, Roper JM, Smith AG (1997) A single precursor protein for ferrochelatase-I from Arabidopsis is imported in vitro into both chloroplasts and mitochondria. J Biol Chem 272:27565–27561
Conley CA, Hanson MR (1995) How do alterations in plant mitochondrial genomes disrupt pollen development? J Bioenerg Biomembr 27:447–457
Cox MM (2000) Recombinational DNA repair in bacteria and the RecA protein. Prog Nucleic Acid Res Mol Biol 63:311–366
Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8:978–984
Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300:1005–1016
Foury F, Lahaye A (1987) Cloning and sequencing of the PIF gene involved in repair and recombination of yeast mitochondrial DNA. EMBO J 6:1441–1449
Frei C, Gasser SM (2000) RecQ helicases: the DNA replication checkpoint connection. J Cell Sci 113:2641–2646
Geiss KT, Abbas GM, Makaroff CA (1994) Intron loss from the NADH dehydrogenase subunit 4 gene of lettuce mitochondrial DNA: evidence for homologous recombination of a cDNA intermediate. Mol Gen Genet 243:97–105
Gutierres S, Combettes B, De Paepe R, Mirande M, Lelandais C, Vedel F, Chetrit P (1999) In the Nicotiana sylvestris CMSII mutant, a recombination-mediated change 5´ to the first exon of the mitochondrial nad1 gene is associated with lack of the NADH:ubiquinone oxidoreductase (complex I) NAD1 subunit. Eur J Biochem 261:361–370
Harmon FG, Kowalczykowski SC (1998) RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev 12:1134–1144
Hartung F, Plchova H, Puchta H (2000) Molecular characterization of RecQ homologues in Arabidopsis thaliana. Nucleic Acids Res 28:4275–4282
Hedtke B, Legen J, Weihe A, Hermann RG, Börner T (2002) Six active phage-type RNA polymerase genes in Nicotiana tabacum. Plant J 30:625–637
Hong EL, Shinohara A, Bishop DK (2001) Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single- strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J Biol Chem 276:41906–41912
Hrubec T, Robinson J, Donaldson R (1985) Isolation of mitochondria from soybean leaves on discontinuous Percoll gradients. Plant Physiol 77:1010–1012
Kajander OA, Karhunen PJ, Holt IJ, Jacobs HT (2001) Prominent mitochondrial DNA recombination intermediates in human heart muscle. EMBO Rep 2:1007–1012
Klein M, Eckert-Ossenkopp U, Schmiedeberg I, Brandt P, Unseld M, Brennicke A, Schuster W (1994) Physical mapping of the mitochondrial genome of Arabidopsis thaliana by cosmid and YAC clones. Plant J 6:447–455
Kobayashi Y, Dokiya Y, Sugita M (2001) Dual targeting of phage-type RNA polymerase to both mitochondria and plastids is due to alternative translation initiation in single transcripts. Biochem Biophys Res Comm 289:1106–1113
Kowalczykowski SC (2000) Initiation of genetic recombination and recombination-dependent replication. Trends Biochem Sci 25:156–165
Larsson NG, Clayton DA (1995) Molecular genetic aspects of human mitochondrial disorders. Ann Rev Genet 29:151–178
Ling F, Makishima F, Morishima N, Shibata T (1995) A nuclear mutation defective in mitochondrial recombination in yeast. EMBO J 14:4090–4101
Lockshon D, Zweifel SG, Freeman-Cook LL, Lorimer HE, Brewer BJ, Fangman WL (1995) A role for recombination junctions in the segregation of mitochondrial DNA in yeast. Cell 81:947–955
MacAlpine DM, Perlman PS, Butow RA (1998) The high mobility group protein Abf2p influences the level of yeast mitochondrial DNA recombination intermediates in vivo. Proc Natl Acad Sci USA 95:6739–6743
Manna F, Massardo DR, Del Giudice L, Buonocore A, Nappo AG, Alifano P, Schafer B, Wolf K (1991) The mitochondrial genome of Schizosaccharomyces pombe. Stimulation of intra-chromosomal recombination in Escherichia coli by the gene product of the first cox1 intron. Curr Genet 19:295–299
Marienfeld J, Unseld M, Brandt P, Brennicke A (1996) Genomic recombination of the mitochondrial atp6 gene in Arabidopsis thaliana at the protein processing site creates two different presequences. DNA Res 3:287–290
Marienfeld JR, Unseld M, Brandt P, Brennicke A (1997) Mosaic open reading frames in the Arabidopsis thaliana mitochondrial genome. Biol Chem 378:859–862
Muise RC, Hauswirth WW (1995) Selective DNA amplification regulates transcript levels in plant mitochondria. Curr Genet 28:113–121
Nakai K (2000) Protein sorting signals and prediction of subcellular localization. Adv Protein Chem 54:277–344
Nakai K, Horton P (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci 24:34–36
Ogawa T, Shinohara A, Nabetani A, Ikeya T, Yu X, Egelman EH, Ogawa H (1993) RecA-like recombination proteins in eukaryotes: functions and structures of RAD51 genes. Cold Spring Harb Symp Quant Biol 58:567–576
Pang Q, Hays JB, Rajagopal I (1992) A plant cDNA that partially complements Escherichia coli rec A mutations predicts a polypeptide not strongly homologous to RecA proteins. Proc Natl Acad Sci USA 89:8073–8077
Perry SE, Li HM, Keegstra K (1991) In vitro reconstitution of protein transport into chloroplasts. Methods Cell Biol 34:327–343
Sato S, Hotta Y, Tabata S (1995) Structural analysis of a recA -like gene in the genome of Arabidopsis thaliana. DNA Res 2:89–93
Scharfe C, Zaccaria P, Hoertnagel K, Jaksch J, Klopstock T, Lill R, Prokisch H, Gerbitz KD, Mewes HW, Meitinger T (1999) MITOP, database for mitochondria-related proteins, genes and diseases. Nucleic Acids Res 27:153–155
Scharfe C, Zaccaria P, Hoertnagel K, Jaksch J, Klopstock T, Dembowski M, Lill R, Prokisch H, Gerbitz KD, Neupert W, Mewes HW, Meitinger T (2000) MITOP, the mitochondrial proteome database: 2000 update. Nucleic Acids Res 28:155–158
Sena EP, Revet B, Moustacchi E (1986) In vivo homologous recombination intermediates of yeast mitochondrial DNA analyzed by electron microscopy. Mol Gen Genet 202:421–428
Silva-Filho MD, Wieers MC, Flugge UI, Chaumont F, Boutry M (1997) Different in vitro and in vivo targeting properties of the transit peptide of a chloroplast envelope inner membrane protein. J Biol Chem 272:15264–15269
Small I, Wintz H, Akashi K, Mireau H (1998) Two birds with one stone: genes that encode products targeted to two or more compartments. Plant Mol Biol 38:265–277
Small WC, McAlister-Henn L (1997) Metabolic effects of altering redundant targeting signals for yeast mitochondrial malate dehydrogenase. Arch Biochem Biophys 344:53–60
Thyagarajan B, Padua RA, Campbell C (1996) Mammalian mitochondria possess homologous DNA recombination activity. J Biol Chem 271:27536–27543
Unseld M, Marienfeld JR, Brandt P, Brennicke A (1997) The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet 15:57–61
Watanabe N, Che FS, Iwano M, Takayama S, Yoshida S, Isogai A (2001) Dual targeting of spinach protoporphyrinogen oxidase II to mitochondria and chloroplasts by alternative use of two in-frame initiation codons. J Biol Chem 276:20474–20481
Weigel D, Glazebrook J (2002) Arabidopsis: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
Wu L, Hickson ID (2001) RecQ helicases and topoisomerases: components of a conserved complex for the regulation of genetic recombination. Cell Mol Life Sci 58:894–901
Yakhnin AV, Vinokurov LM, Surin AK, Alakhov YB (1998) Green fluorescent protein purification by organic extraction. Protein Expr Purif 14:382–386
Acknowledgements
We thank Drs. Narendra Singh, William McCleary and Allan Caplan for helpful discussions, Dr. Andre T. Jagendorf for the polyclonal antibodies against chloroplast RecA and Dr. Tom Elthon for monoclonal antibodies against mitochondrial porin. We also thank Dr. David McClellan for assistance with the phylogenetic analyses, and Paul Gunn and Sean Echols for help with various aspects of the work. We acknowledge support from the NIH, the COSAM Dean's Research Initiative (Auburn University), the Alabama Agricultural Experiment Station, and the Mentoring Environment Grant Program of the BYU Office of Research and Creative Activities
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. G. Herrmann
Rights and permissions
About this article
Cite this article
Khazi, F.R., Edmondson, A.C. & Nielsen, B.L. An Arabidopsis homologue of bacterial RecA that complements an E. coli recA deletion is targeted to plant mitochondria. Mol Gen Genomics 269, 454–463 (2003). https://doi.org/10.1007/s00438-003-0859-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-003-0859-6