Abstract
This study evaluated, for the first time, the genetic diversity of Toxoplasma gondii isolates from free-range chickens from the state of Paraíba, Northeast Brazil. Tissue samples from 33 chickens from properties in five municipalities of Paraíba (Esperança, Olho d’Água, Malta, Monteiro, and Patos) were bioassayed in mice. The brains of mice infected with T. gondii cysts were used for DNA extraction and genotyping. Genotyping was performed using 11 PCR-RFLP markers and 15 microsatellite (MS) markers. Complete genotyping results were obtained for 29 isolates, with nine genotypes detected by RFLP and 15 genotypes identified by MS. Three genotypes (#273, #274, and #277) have only been recently identified from pigs in the region. Brazilian clonal types BrII and BrIII were identified from one isolate each. Clonal types I, II, and III were not detected by RFLP. Genotype #13 (Caribbean 1), detected in 48.3% (14/29) of isolates from four of the five municipalities investigated, was the most prevalent genotype in the state of Paraíba. However, the MS analysis showed that of these 14 isolates, only four were unique genotypes, and considering the distance between the municipalities from where they were collected, it is possible that only seven are independent isolates while the others are clones. The other genotypes were restricted to different microregions. The results indicate that the Caribbean 1 lineage of T. gondii is circulating widely in Northeast Brazil. The genotypic diversity of T. gondii in the state of Paraíba is high, and microsatellite analysis revealed this diversity with higher resolution than PCR-RFLP.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Toxoplasma gondii is a protozoan in the phylum Apicomplexa that is capable of infecting all species of birds and mammals (Tenter and Johnson 1997; Elmore et al. 2010). Felids are the definitive hosts where the sexual reproduction of the parasite occurs and are responsible for the production and excretion of oocysts in the feces. Accidental ingestion of oocysts from the environment is one of the most common forms of parasite transmission. Other routes of transmission include the ingestion of undercooked meat containing viable cysts and vertical transmission through transplacental passage of tachyzoites to the fetus. The different routes of transmission and the possibility of T. gondii permanently infecting its hosts make this parasite one of the most prevalent and widespread worldwide (Dubey et al. 2012; Sullivan and Jeffers 2012).
Most humans infected with T. gondii are asymptomatic. The prevalence of T. gondii infection depends on the ingestion of undercooked meat containing T. gondii cysts and/or the ingestion of T. gondii oocysts present on contaminated food, particularly fruits and vegetables (Dubey et al. 2012; Gangneux and Dardé 2012). Free-range chickens are the best indicator for soil contamination with T. gondii oocysts because they feed from the ground and in doing so become infected (Furuta et al. 2007; Dubey 2009). In addition, the isolation and genotyping of T. gondii from chickens can provide information on the strains that are circulating in a region.
The early studies of T. gondii using the SAG2 gene showed a limited genetic diversity and only three clonal lineages: I, II, and III (Howe and Sibley 1995). However, later studies using a larger number of molecular markers revealed the existence of many recombinant or atypical lineages, especially in South America (Ajzenberg et al. 2002; Dubey et al. 2007a; Ferreira et al. 2008). These markers generate valuable information about the diversity of the parasite, are simple, and have a high resolution in the identification of T. gondii isolates (Su et al. 2010). Polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) is the most used genotyping method, having contributed to the genotypic characterization of T. gondii isolates from animals and humans worldwide (Howe and Sibley 1995; Su et al. 2010). Even though microsatellite (MS) analysis has a higher resolving power with two levels of discrimination, the first one identifying the lineages and the second one distinguishing closely related samples belonging to the same lineage (Ajzenberg et al. 2010).
There are few data available regarding T. gondii genotypes found in Northeast Brazil, especially in the state of Paraíba. A high level of T. gondii antibodies and often viable parasites were found in free-range chickens from that Brazilian state (Feitosa et al. 2016). Thus, this study evaluated, for the first time, the genetic diversity of T. gondii isolates from free-range chickens raised in the state of Paraíba, Northeast Brazil.
Materials and methods
Samples
Thirty-three T. gondii isolates from free-range chickens raised in five municipalities in the state of Paraíba, Northeast Brazil, and described in detail by Feitosa et al. (2016), were used in the study. Briefly, each mouse had been subcutaneously inoculated with 1.0 mL of brain and heart homogenates from chickens in the bioassays. The brains of mice positive for T. gondii cysts through direct examination by means of a compound microscopy were used for genotyping. The brain samples were homogenized in 0.85% saline using a mortar and pestle, and then stored in 1.5-mL microtubes at −70 °C until DNA extraction.
DNA extraction
After thawing, a 300-μL aliquot of brain homogenate from each sample was separated; washed three times with TE buffer, pH 8.0 (10 mM Tris-HCl, 1 mM EDTA); and centrifuged at 12,000g for 5 min. DNA extraction was performed using WIZARD® Genomic Purification Kit (catalog A 1125; Promega, Madison, WI, USA).
Isolate genotyping
Genotyping of isolates was performed using 11 PCR-RFLP genetic markers: SAG1, SAG2 (5′3′ SAG2 and alt. SAG2), SAG3, BTUB, GRA6, c22-8, c29-2, L358, PK1, and Apico (Su et al. 2010) and marker CS3 (Pena et al. 2008). Clonal type I (RH), type II (PTG), and type III (CTG) samples and reference samples Cougar, MAS, and TgCatBr5 were used as positive controls. First, the target DNA sequences were amplified by multiplex PCR using the outer primers for all markers followed by nested PCR separately for each marker. The PCR products were diluted 1:1 in ultrapure water before being amplified in nested PCR. All protocols have been described elsewhere (Su et al. 2010; Pena et al. 2008).
To determine the RFLP pattern of each sample, 3 μL of nested PCR products was mixed with 17 μL of digestion reaction containing buffer and one unit of a given restriction enzyme. The samples were incubated at the proper temperature for each restriction enzyme according to the manufacturer’s recommendations. The restriction enzymes used are described in Su et al. (2006), except for CS3, which is described in Pena et al. (2008). The digested PCR products were resolved by electrophoresis in a 2.0–3.0% agarose gel containing 10 μL of SYBR® Safe for each 100 mL of solution with a molecular weight marker with multiple 100-base pair (bp) fragments. The bands were visualized under UV light on an image analyzer (Alpha Innotech Corporation, San Leandro, CA, USA).
The isolates were compared and classified according to the genotypes available in the ToxoDB database (http://toxodb.org/toxo/) and based on recent studies. The phylogenetic relationships of T. gondii isolates genotyped by PCR-RFLP were examined using SplitsTree4 software (Huson 1998; Huson and Bryant 2006).
The isolates were also genotyped using 15 microsatellite markers: TUB2, W35, TgMA, B18, B17, M33, IV.1 and X1.1, N60, N82, AA, N61, N83, M48, and M102 (Ajzenberg et al. 2010). The data were analyzed using GeneMapper® 4.1 software (Applied Biosystems, Foster City, CA, USA).
Results
Of the 33 isolates analyzed by PCR-RFLP, complete genotyping results were obtained for the same 29 samples with RFLP and MS markers (Supplementary material). Nine different RFLP genotypes and 15 MS genotypes were detected (Table 1). Two isolates from the state capital (Patos), TgCkBrPB9 and TgCkBrPB30, belonged to Brazilian clonal lineages BrII (#11) and BrIII (#8), respectively. Clonal archetypal lineages I, II, and III were not detected by RFLP.
Genotypes #48 (TgCkBrPB4,5,6) and #88 (TgCkBrPB7,8) had been previously identified in chickens, but from different Northeast states and as single isolates. Genotype #116 (TgCkBrPB1,2) had been previously detected in chickens and pigs in North and Northeast Brazil, respectively. Genotypes #273 (TgCkBrPB11,12,14), #274 (TgCkBrPB26), and #277 (TgCkBrPB7,8) have only recently been reported in Brazil in pig isolates from the state of Paraíba (Feitosa, unpublished data).
Genotype #13, detected in 48.3% (14/29) of isolates, was the most prevalent and well-distributed genotype in the state of Paraíba. These isolates came from four municipalities located 70–262 km apart, whereas the other genotypes were restricted to different microregions. Nevertheless, the MS analysis indicated the possibility of a high level of circulation of genotype #13 (Caribbean 1) clones due to the occurrence of identical genotypes in isolates from the same municipality and from nearby properties, including isolates TgCkBrPB15,16 (Esperança), TgCkBrPB10,13 (Monteiro), TgCkBrPB20,25 (Malta), and TgCkBrPB22,23,24,27 (Malta) (Supplementary material). Similarly, isolates TgCkBrPB1,2 (genotype #116) and TgCkBrPB7,8 (genotype #277) also appear to be clones from the same sample.
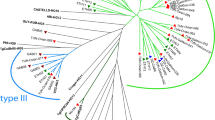
The genetic diversity of the 29 T. gondii isolates from chickens is summarized in Fig. 1. The analysis showed a great divergence of these isolates from type II strains and a clear clustering with type III and type BrIII strains.
Phylogenetic network of the T. gondii isolates from chickens from Paraíba state, Northeast Brazil, using PCR-RFLP data. The samples in bold were obtained in the present study. The following were included: archetypal reference genotypes (GTI = type I, PTG = type II, and CTG = type III), lineage type BrI, and isolates from pigs that had previously been described in the same region
Discussion
Genotyping results revealed a high genetic diversity of T. gondii in chickens from the state of Paraíba, Northeast Brazil, supporting previous studies that reported similar findings. This diversity is best represented by the MS analysis, which due to its higher resolving power identified 15 genotypes in the 29 isolates analyzed against the nine RFLP genotypes. Pena et al. (2013) identified 11 genotypes in 44 isolates from free-range chickens from the state of Espírito Santo, and Sousa et al. (2016) found four genotypes in five isolates from the state of Maranhão. In Brazil, these findings are common, unlike other countries where the genetic diversity of T. gondii is low. For instance, Dubey and Su (2009) identified only 18 genotypes of 253 isolates in the USA, whereas genotyping of 149 isolates identified 58 genotypes in Brazil (Pena et al. 2008).
Despite the great genetic diversity of T. gondii previously identified in Brazil, Pena et al. (2008) suggested that there are four clonal types circulating among different hosts and regions in the country: BrI, II, III, and IV. Type BrIV lineage has only been found in southern Brazil. In the current study, only two isolates were types BrII and BrIII. These Brazilian clonal types have been identified in cats, chickens, dogs, sheep, and newborn humans in South and Southeast Brazil (Dubey et al. 2007b, 2008; Silva et al. 2011; Carneiro et al. 2013), but this is the first time that clonal type BrII has been identified in the Northeast and clonal type BrIII has only been recently identified from pigs in the region (Feitosa, unpublished data), highlighting its high level of circulation in Brazil.
The clonal spread of T. gondii can be explained by its modes of transmission, which include transmission by ingestion of meat of intermediate hosts without the parasite passing through the definitive host and undergoing meiosis and genetic recombination. This recombination can occur in the intestine of the definitive host (cat) if it gets infected with different strains of the parasite simultaneously or in a very short time, enabling the effective crossing of gametes and the production of progenies that are genetically different from the original parental strains (Sibley and Ajioka 2008; Boothroyd and Grigg 2002). According to Feitosa et al. (2014), most domestic cats in the study region have free access to the outdoors and engage in hunting activity, providing the perfect environment for the occurrence of recombinant and novel genotypes in the study region.
Samples with the same genotype are not necessarily epidemiologically related and may comprise independent samples. The MS analysis can more accurately identify this relationship, because PCR-RFLP detects less variation in each locus when compared to microsatellite genotyping. Ajzenberg et al. (2002) found 3–16 alleles per locus in a population of 83 T. gondii isolates and only 2–4 alleles for each PCR-RFLP marker, suggesting that the microsatellite technique has a superior discriminatory power than PCR-RFLP.
The genotype with the highest frequency was #13 (Caribbean 1), which accounted for approximately half of the isolates, indicating that it is circulating freely among chickens in Paraíba. Nevertheless, the occurrence of a large number of possible clones as indicated by the MS analysis and the origin of isolates underscores the role of chickens as indicators for environmental contamination with oocysts and the widespread distribution of oocysts in the soil after excretion by cats. In other Northeast states, including Ceará, Rio Grande do Norte, Pernambuco, Alagoas, Sergipe, and Bahia, this genotype has been identified in chickens (Dubey et al. 2008). Genotype #13 has also been identified in other animals, including monkeys and pigs, also in Northeast Brazil (Pena et al. 2011; Clementino Andrade et al. 2013). The only state in the Northeast region where genotyping of T. gondii isolates has been conducted without any records of this genotype is Maranhão (Dubey et al. 2008; Sousa et al. 2016). Even though Maranhão is a Northeast state, its climate characteristics differ from those of the other states, because unlike the others that have a predominantly semi-arid climate, it is in a transition zone between the semi-arid and humid equatorial climate of the Amazon forest, which could result in different T. gondii epidemiology, routes of spread, and prevalent genotypes in the state.
Conclusion
T. gondii has high genotypic diversity in the state of Paraíba, Brazil; genotype #13 can be considered a frequent clonal type in Northeast Brazil, and due to the epidemiological features of transmission, the chances are great of finding novel non-archetypal genotypes in the study region.
References
Ajzenberg D, Bañuls AL, Tibayrenc M, Dardé ML (2002) Microsatellite analysis of Toxoplasma gondii shows considerable polymorphism structured into two main clonal groups. Int J Parasitol 32:27–38. doi:10.1016/S0020-7519(01)00301-0
Ajzenberg D, Collinet F, Mercier A, Vignoles P, Dardé ML (2010) Genotyping of Toxoplasma gondii isolates with 15 microsatellite markers in a single multiplex PCR assay. J Clin Microbiol 48:4641–4645. doi:10.1128/JCM.01152-10
Boothroyd JC, Grigg ME (2002) Population biology of Toxoplasma gondii and its relevance to human infection: do different strains cause different disease? Curr Opin Microbiol 5(4):438–442. doi:10.1016/S1369-5274(02)00349-1
Carneiro AC, Andrade GM, Costa JG, Pinheiro BV, Vasconcelos-Santos DV, Ferreira AM, Su C, Januário JN, Vitor RW (2013) Genetic characterization of Toxoplasma gondii revealed highly diverse genotypes for isolates from newborns with congenital toxoplasmosis in southeastern Brazil. J Clin Microbiol 51:901–907. doi:10.1128/JCM.02502-12
Clementino Andrade MM, Pinheiro BV, Cunha MM, Carneiro ACAV, Andrade Neto VF, Vitor RWA (2013) New genotypes of Toxoplasma gondii obtained from farm animals in Northeast Brazil. Res Vet Sci 94:587–589. doi:10.1016/j.rvsc.2013.01.006
Dubey PB (2009) Toxoplasmosis in sheep—the last 20 years. Vet Parasitol 163:1–14. doi:10.1016/j.vetpar.2009.02.026
Dubey JP, Su C (2009) Population biology of Toxoplasma gondii: what’s out and where did they come from. Mem Inst Oswaldo Cruz 104:190–195. doi:10.1590/S0074-02762009000200011
Dubey JP, Sundar N, Gennari SM, Minervino AH, Farias NA, Ruas JL, Dos Santos TR, Cavalcante GT, Kwok OC, Su C (2007a) Biologic and genetic comparison of Toxoplasma gondii isolates in free-range chickens from the northern Para state and the southern state Rio Grande do Sul, Brazil revealed highly diverse and distinct parasite populations. Vet Parasitol 143:182–188. doi:10.1016/j.vetpar.2006.08.024
Dubey JP, Gennari SM, Sundar N, Vianna MC, Bandini LM, Yai LE, Kwok CH, Su C (2007b) Diverse and atypical genotypes identified in Toxoplasma gondii from dogs in São Paulo, Brazil. J Parasitol 93:60–64. doi:10.1645/GE-972R.1
Dubey JP, Velmurugan GV, Chockalingan A, Pena HFJ, Leifer CA, Gennari SM, Bahia-Oliveira LMG, Su C (2008) Genetic diversity of Toxoplasma gondii isolates from chickens from Brazil. Vet Parasitol 157:299–305. doi:10.1016/j.vetpar.2008.07.036
Dubey JP, Lago EG, Gennari SM, Su C, Jones JL (2012) Toxoplasmosis in humans and animals in Brazil: high prevalence, high burden of disease, and epidemiology. Parasitology 139:1375–1424. doi:10.1017/S0031182012000765
Elmore SA, Jones JL, Conrad PA, Patton S, Lindsay DS, Dubey JP (2010) Toxoplasma gondii: epidemiology, feline clinical aspects, and prevention. Trends Parasitol 26(4):190–196. doi:10.1016/j.pt.2010.01.009
Feitosa TF, Vilela VLR, Dantas ES, Souto DVO, Pena HFJ, Athayde ACR, Azevedo SS (2014) Toxoplasma gondii and Neospora caninum in domestic cats from the Brazilian semi-arid: seroprevalence and risk factors. Arq Bras Med Vet Zootec 66:1060–1066. doi:10.1590/1678-6696
Feitosa TF, Vilela VLR, Almeida-Neto JL, Santos A, De Morais DF, Athayde ACR, Azevedo SS, Pena HFJ (2016) First study on seroepidemiology and isolation of Toxoplasma gondii in free-range chickens in the semi-arid region of Paraíba state, Brazil. Parasitol Res 115:3983–3990. doi:10.1007/s00436-016-5164-5
Ferreira IM, Vidal JE, Costa-Silva TA, Meira CS, Hiramoto RM, Penalva de Oliveira AC, Pereira-Chioccola VL (2008) Toxoplasma gondii: genotyping of strains from Brazilian AIDS patients with cerebral toxoplasmosis by multilocus PCR-RFLP markers. Exp Parasitol 118:221–227. doi:10.1016/j.exppara.2007.08.006
Furuta PI, Mineo TW, Carrasco AO, Godoy GS, Pinto AA, Machado RZ (2007) Neospora caninum infection in birds: experimental infections in chicken and embryonated eggs. Parasitology 134:1931–1939. doi:10.1017/S0031182007003344
Gangneux FR, Dardé ML (2012) Epidemiology of and diagnostic strategies for toxoplasmosis. Clin Microbiol Rev 25:264–296. doi:10.1128/CMR.05013-11
Howe DK, Sibley LD (1995) Toxoplasma gondii comprises three clonal lineages: correlation of parasite genotype with human disease. J Infect Dis 172:1561–1566
Huson DH (1998) Splits tree: a program for analyzing and visualizing evolutionary data. Bioinformatics 14:68–73
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. doi:10.1093/molbev/msj030
Pena HFJ, Gennari SM, Dubey JP, Su C (2008) Population structure and mouse-virulence of Toxoplasma gondii in Brazil. Int J Parasitol 38:561–569. doi:10.1016/j.ijpara.2007.09.004
Pena HF, Marvulo MF, Horta MC, Silva MA, Silva JC, Siqueira DB, Lima PA, Vitaliano SN, Gennari SM (2011) Isolation and genetic characterisation of Toxoplasma gondii from a red-handed howler monkey (Alouatta belzebul), a jaguarundi (Puma yagouaroundi), and a black-eared opossum (Didelphis aurita) from Brazil. Vet Parasitol 175(3–4):377–381. doi:10.1016/j.vetpar.2010.10.015
Pena HFJ, Vitaliano SN, Beltrame MA, Pereira FE, Gennari SM, Soares RM (2013) PCR-RFLP genotyping of Toxoplasma gondii from chickens from Espirito Santo state, southeast region, Brazil: new genotypes and a new SAG3 marker allele. Vet Parasitol 192:111–117. doi:10.1016/j.vetpar.2012.10.004
Sibley LD, Ajioka JW (2008) Population structure of Toxoplasma gondii: clonal expansion driven by infrequent recombination and selective sweeps. Annu Rev Microbiol 62:329–351. doi:10.1146/annurev.micro.62.081307.162925
Silva RC, Langoni H, Su C, Da Silva AV (2011) Genotypic characterization of Toxoplasma gondii in sheep from Brazilian slaughterhouses: new atypical genotypes and the clonal type II strain identified. Vet Parasitol 175(1–2):173–177. doi:10.1016/j.vetpar.2010.09.021
Sousa IC, Pena HF, Santos LS, Gennari SM, Costa FN (2016) First isolation and genotyping of Toxoplasma gondii from free-range chickens on São Luis Island, Maranhão state, Brazil, with a new genotype described. Vet Parasitol 223:159–164. doi:10.1016/j.vetpar.2016.04.041
Su C, Zhang X, Dubey JP (2006) Genotyping of Toxoplasma gondii by multilocus PCR-RFLP markers: a high resolution and simple method for identification of parasites. Int J Parasitol 36:841–848. doi:10.1016/j.ijpara.2006.03.003
Su C, Shwab EK, Zhou P, Zhu XQ, Dubey JP (2010) Moving towards an integrated approach to molecular detection and identification of Toxoplasma gondii. Parasitology 137:1–11. doi:10.1017/S0031182009991065
Sullivan WJ Jr, Jeffers V (2012) Mechanisms of Toxoplasma gondii persistence and latency. FEMS Microbiol Rev 36(3):717–733. doi:10.1111/j.1574-6976.2011.00305.x
Tenter AM, Johnson AM (1997) Phylogeny of the tissue-cyst forming coccidia. Adv Parasitol 39:69–139. doi:10.1016/S0065-308X(08)60045-7
Acknowledgments
This work was supported by the Coordination for the Improvement of Higher Education Personnel-CAPES, Brazil (grant number 1841/2016), and the National Council for Scientific and Technological Development (grant number 474737/2012-8, H.F.J.P.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics
This experiment was conducted in accordance with the laws in force in Brazil and was approved by the ethics committee of the Federal University of Campina Grande (UFCG), under protocol 01-2012/05-03-2012.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(PDF 553 kb)
Rights and permissions
About this article
Cite this article
Feitosa, T.F., Vilela, V.L.R., de Almeida-neto, J.L. et al. First report of typical Brazilian Toxoplasma gondii genotypes from isolates of free-range chickens (Gallus gallus domesticus) circulating in the state of Paraíba, Northeast Brazil. Parasitol Res 116, 2265–2270 (2017). https://doi.org/10.1007/s00436-017-5531-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-017-5531-x